Isolation of Three Metallothionein Genes and Their Roles in Mediating Cadmium Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. cDNA Library Construction and Cd Tolerance Gene Screening
2.3. Bioinformatics Analysis
2.4. Yeast Experiments
2.5. Construction of Vector for Subcellular Localization Analysis and Fluorescence Observation
2.6. Determination of Cd Content
2.7. Gene Expression Analysis
2.8. Sequence Information
3. Results
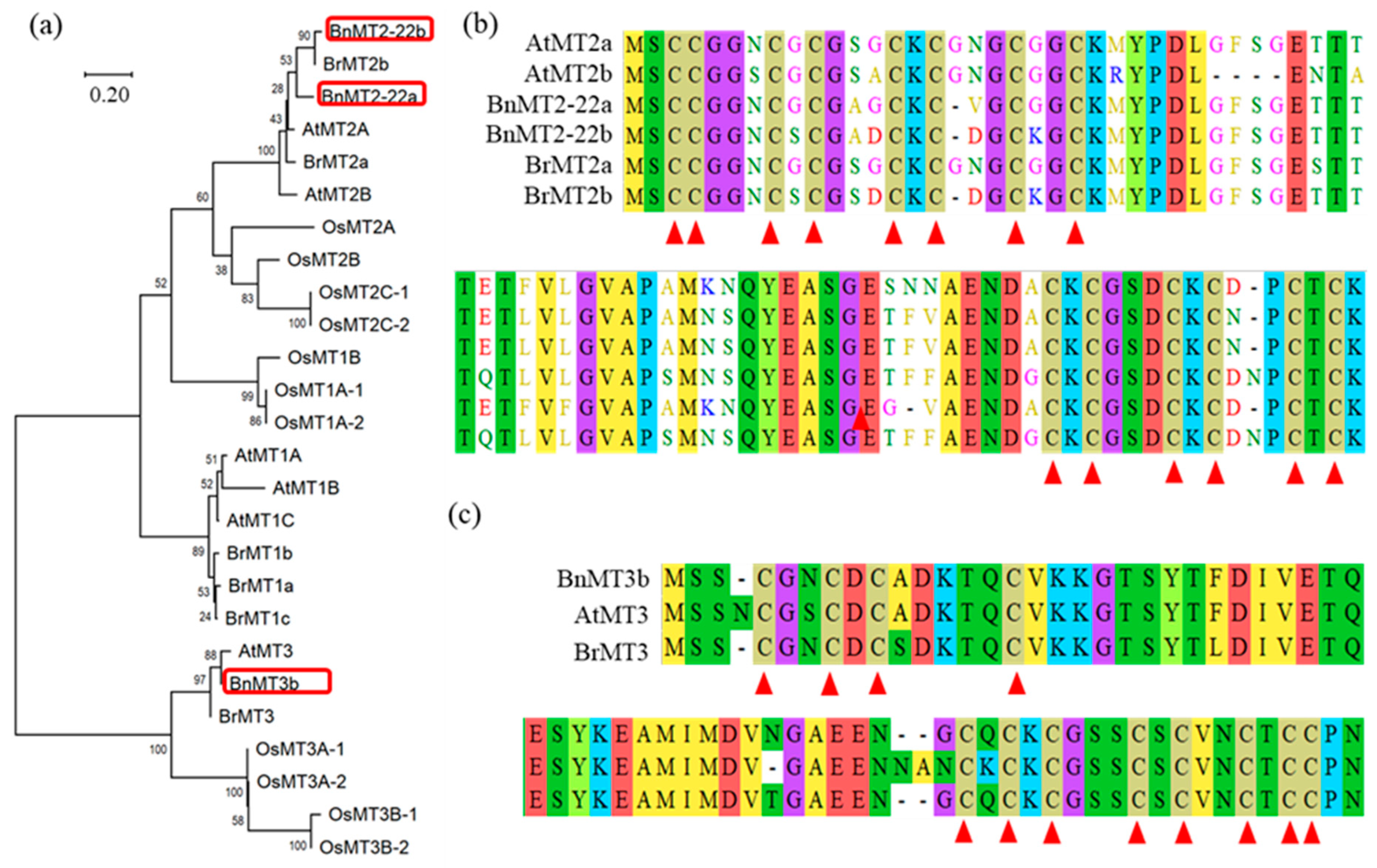
3.1. Cloning and Bioinformatics Analysis of BnMT2-22a, BnMT2-22b and BnMT3b
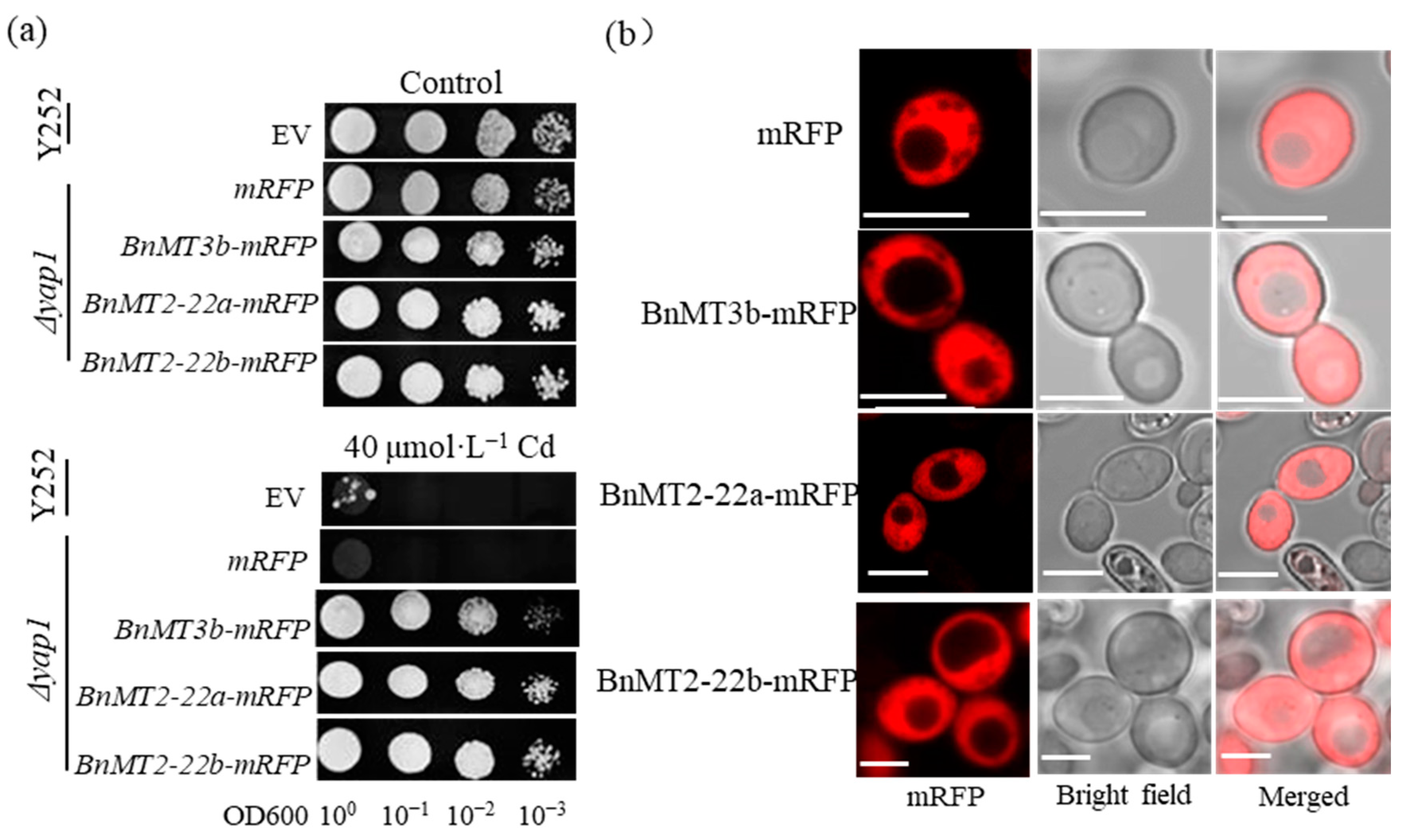
3.2. BnMT2-22a, BnMT2-22b and BnMT3b Mediate the Yeast Tolerance to Cd and Cu
3.3. BnMT2-22a, BnMT2-22b and BnMT3b Localized in the Cytosol
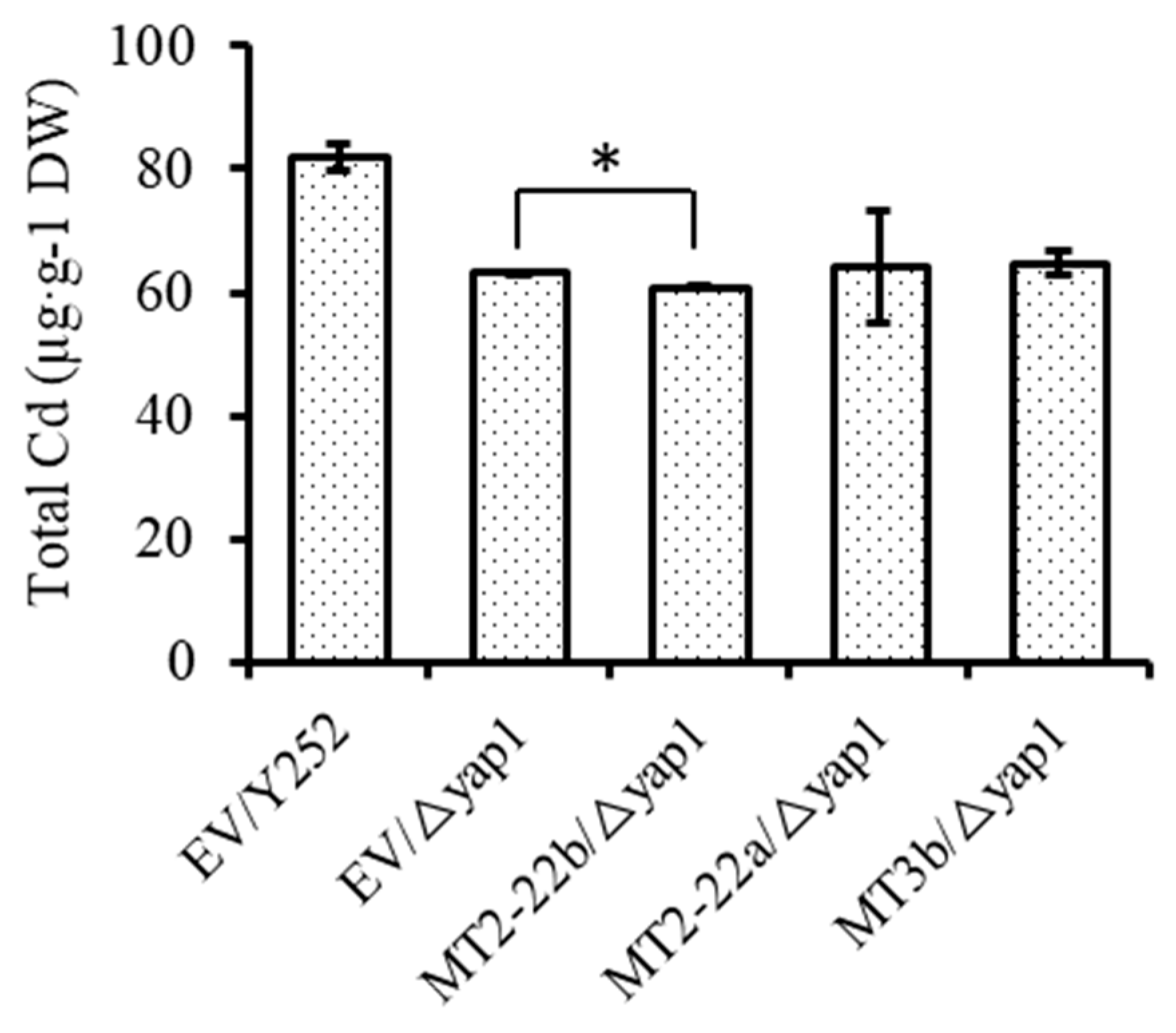
3.4. BnMT2-22a, BnMT2-22b, BnMT3b Hardly Affect the Accumulation of Cd in Cells
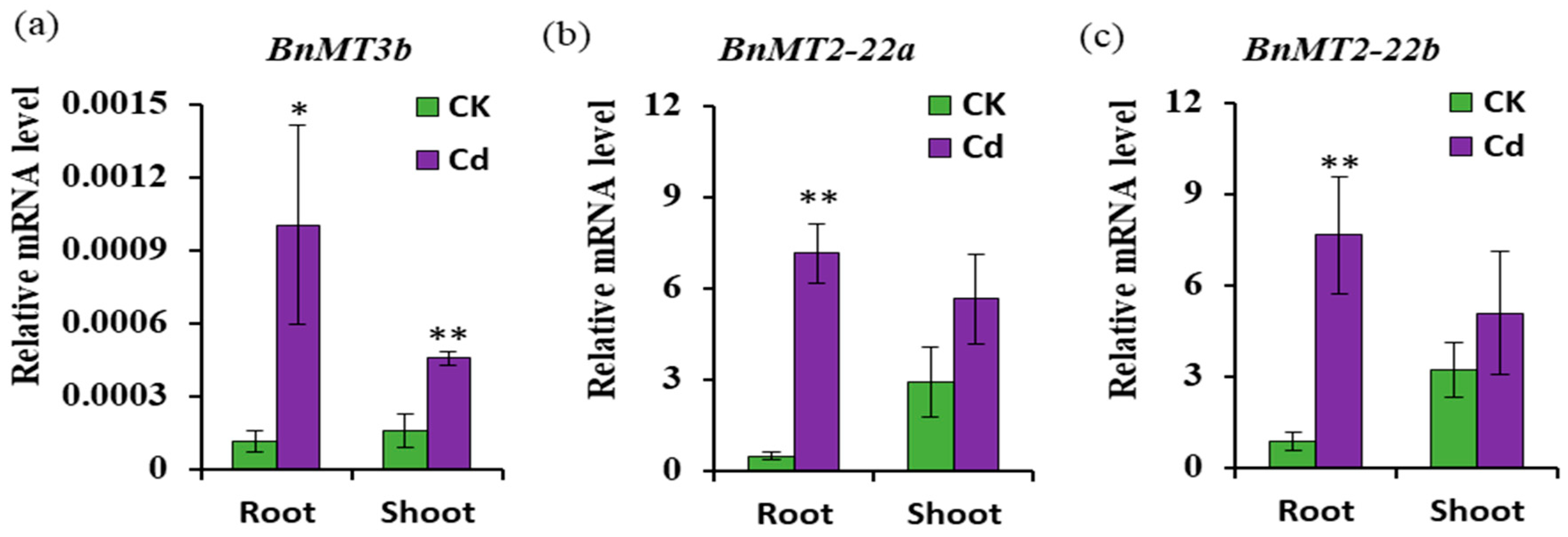
3.5. Expression of BnMT3b, BnMT2-22a and BnMT2-22b Response to Cd Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, B.; Tao, Q.; Zhou, Y.; Gill, R.; Ali, S.; Rafiq, M.T.; Xu, L.; Zhou, W. 5-Aminolevolinic acid mitigates the cadmium-induced changes in Brassica napus as revealed by the biochemical and ultra-structural evaluation of roots. Ecotoxicol. Environ. Saf. 2013, 92, 271–280. [Google Scholar] [CrossRef]
- Ali, B.; Gill, R.A.; Yang, S.; Gill, M.B.; Ali, S.; Rafiq, M.T.; Zhou, W. Hydrogen sulfide alleviates cadmium-induced morpho-physiological and ultrastructural changes in Brassica napus. Ecotoxicol. Environ. Saf. 2014, 110, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Yang, L.; Xu, C.; Shi, Z.; Shao, J.; Xian, M.; Chen, J. Cadmium disrupts the balance between hydrogen peroxide and superoxide radical by regulating endogenous hydrogen sulfide in the root tip of Brassica rapa. Front. Plant Sci. 2017, 8, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Zhao, X.; Sum, X.; Tan, Q.; Tang, Y.; Nie, Z.; Qu, C.; Chen, Z.; Hu, C. Antioxidant enzyme systems and the ascorbate-glutathione cycle as contributing factors to cadmium accumulation and tolerance in two oilseed rape cultivars Brassica napus L. under moderate cadmium stress. Chemosphere 2015, 138, 526–536. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, T.; Tang, T.; Song, H.; Guan, C.; Huang, J.; Hua, Y. A multiomics approach reveals the pivotal role of subcellular reallocation in determining rapeseed resistance to cadmium toxicity. J. Exp. Bot. 2019, 70, 5437–5455. [Google Scholar] [CrossRef]
- Peng, J.S.; Guan, Y.H.; Lin, X.J.; Xu, X.J.; Xiao, L.; Wang, H.H.; Meng, S. Comparative understanding of metal hyperaccumulation in plants: A mini-review. Environ. Geochem. Health 2021, 43, 1599–1607. [Google Scholar] [CrossRef]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leszczyszyn, O.I.; Imam, H.T.; Blindauer, C.A. Diversity and distribution of plant metallothioneins: A review of structure properties and functions. Metallomics 2013, 5, 1146–1169. [Google Scholar] [CrossRef]
- Freisinger, E. Structural features specific to plant metallothioneins. J. Bio. Inorg. Chem. 2011, 16, 1035–1045. [Google Scholar] [CrossRef] [Green Version]
- Zimeri, A.M.; Dhankher, O.P.; Mccaig, B.; Meagher, R.B. The plant MT1 metallothioneins are stabilized by binding cadmiums and are required for cadmium tolerance and accumulation. Plant Mol. Bio. 2005, 58, 839–855. [Google Scholar] [CrossRef]
- Benatti, M.R.; Yookongkaew, N.; Meetam, M.; Guo, W.J.; Punyasuk, N.; Abuqamar, S.; Goldsbrough, P. Metallothionein deficiency impacts copper accumulation and redistribution in leaves and seeds of Arabidopsis. New Phytol. 2014, 202, 940–951. [Google Scholar] [CrossRef]
- Guo, W.J.; Meetam, M.; Goldsbrough, P.B. Examining the specific contributions of individual Arabidopsis metallothioneins to copper distribution and metal tolerance. Plant Physiol. 2008, 146, 1697–1706. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Shim, D.; Song, W.; Hwang, I.; Lee, Y. Arabidopsis metallothioneins 2a and 3 enhance resistance to cadmium when expressed in Vicia faba guard cells. Plant Mol. Bio. 2004, 54, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Liu, Y.; Chen, H.; Li, G.; Zhang, X.; Zhao, J. Type 4 metallothionein genes are involved in regulating Zn ion accumulation in late embryo and in controlling early seedling growth in Arabidopsis. Plant Cell Environ. 2012, 35, 770–789. [Google Scholar] [CrossRef]
- Blanvillain, R.; Kim, J.H.; Wu, S.; Lima, A.; Ow, D.W. OXIDATIVE STRESS 3 is a chromatin-associated factor involved in tolerance to heavy metals and oxidative stress. Plant J. 2009, 57, 654–665. [Google Scholar] [CrossRef]
- Clemens, S.; Kim, E.J.; Neumann, D.; Schroeder, J.I. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999, 18, 3325–3333. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.Y.; Kim, D.Y.; Shim, D.; Song, W.Y.; Lee, J.; Schroeder, J.I.; Kim, S.; Moran, N.; Lee, Y. Expression of the novel wheat gene TM20 confers enhanced cadmium tolerance to bakers’ yeast. J. Bio. Chem. 2008, 283, 15893–15902. [Google Scholar] [CrossRef] [Green Version]
- Papoyan, A.; Kochian, L.V. Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance. Characterization of a novel heavy metal transporting ATPase. Plant Physiol. 2004, 136, 3814–3823. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.S.; Ding, G.; Meng, S.; Yi, H.Y.; Gong, J.M. Enhanced metal tolerance correlates with heterotypic variation in SpMTL, a metallothionein-like protein from the hyperaccumulator Sedum plumbizincicola. Plant Cell Environ. 2017, 40, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Martinoia, E.; Lee, J.; Kim, D.; Kim, D.Y.; Vogt, E.; Shim, D.; Choi, K.S.; Hwang, I.; Lee, Y. A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 2004, 135, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Shim, D.; Hwang, J.U.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, A.E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef] [Green Version]
- Vatamaniuk, O.K.; Mari, S.; Lu, Y.P.; Rea, P.A. AtPCS1, a phytochelatin synthase from Arabidopsis: Isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. USA 1999, 96, 7110–7115. [Google Scholar] [CrossRef] [Green Version]
- Wemmie, J.A.; Wu, A.L.; Harshman, K.D.; Parker, C.S.; Moye-Rowley, W.S. Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J. Biol. Chem. 1994, 269, 14690–14697. [Google Scholar] [CrossRef]
- MacDiarmid, C.W.; Gaither, L.A.; Eide, D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2845–2855. [Google Scholar] [CrossRef] [Green Version]
- Welch, J.; Fogel, S.; Buchman, C.; Karin, M. The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J. 1989, 8, 255–260. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 38–41. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, M.; Wang, S.; Ma, G.; Huang, X.; Qiao, C.; Wang, R.; Xu, X.; Liang, Y.; Lu, K.; et al. Genome-Wide characterization and analysis of metallothionein family genes that function in metal stress tolerance in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2181. [Google Scholar] [CrossRef] [Green Version]
- Elble, R. A simple and efficient procedure for transformation of yeasts. Biotechniques 1992, 13, 18–20. [Google Scholar]
- Peng, J.S.; Wang, Y.J.; Ding, G.; Ma, H.L.; Zhang, Y.J.; Gong, J.M. A pivotal role of cell wall in cadmium accumulation in the crassulaceae hyperaccumulator Sedum plumbizincicola. Mol. Plant. 2017, 10, 771–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.H.; Xu, Z.J.; Peng, J.S.; Zhao, J.; Zhang, G.B.; Xie, J.; Yi, Z.X.; Zhang, J.H.; Gong, J.M.; Ye, N.H.; et al. OsProT1 and OsProT3 function to mediate proline- and γ-aminobutyric acid-specific transport in yeast and are differentially expressed in rice (Oryza sativa L.). Rice 2019, 12, 79. [Google Scholar] [CrossRef]
- Peng, J.S.; Yi, H.Y.; Gong, J.M. Isolation and characterization of cadmium tolerant gene SpMT2 in the hyperaccumulator Sedum plumbizincicola. Sheng Wu Gong Cheng Xue Bao 2020, 36, 541–548. [Google Scholar] [PubMed]
- An, Z.; Li, C.; Zu, Y.; Du, Y.; Wachter, A.; Gromes, R.; Rausch, T. Expression of BjMT2, a metallothionein 2 from Brassica juncea, increases copper and cadmium tolerance in Escherichia coli and Arabidopsis thaliana, but inhibits root elongation in Arabidopsis thaliana seedlings. J. Exp. Bot. 2006, 57, 3575–3582. [Google Scholar]
- Liu, J.; Zhang, J.; Kim, S.H.; Lee, H.S.; Marinoia, E.; Song, W.Y. Characterization of Brassica rapa metallothionein and phytochelatin synthase genes potentially involved in heavy metal detoxification. PLoS ONE 2021, 16, e0252899. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.-H.; Zhang, X.-J.; Tang, T.-W.; Hu, H.-L.; Bai, N.-N.; Zhang, D.-W.; Meng, S.; Peng, J.-S. Isolation of Three Metallothionein Genes and Their Roles in Mediating Cadmium Resistance. Agronomy 2022, 12, 2971. https://doi.org/10.3390/agronomy12122971
Zhang P-H, Zhang X-J, Tang T-W, Hu H-L, Bai N-N, Zhang D-W, Meng S, Peng J-S. Isolation of Three Metallothionein Genes and Their Roles in Mediating Cadmium Resistance. Agronomy. 2022; 12(12):2971. https://doi.org/10.3390/agronomy12122971
Chicago/Turabian StyleZhang, Pei-Hong, Xue-Jie Zhang, Ting-Wei Tang, Heng-Liang Hu, Ning-Ning Bai, Da-Wei Zhang, Shuan Meng, and Jia-Shi Peng. 2022. "Isolation of Three Metallothionein Genes and Their Roles in Mediating Cadmium Resistance" Agronomy 12, no. 12: 2971. https://doi.org/10.3390/agronomy12122971
APA StyleZhang, P.-H., Zhang, X.-J., Tang, T.-W., Hu, H.-L., Bai, N.-N., Zhang, D.-W., Meng, S., & Peng, J.-S. (2022). Isolation of Three Metallothionein Genes and Their Roles in Mediating Cadmium Resistance. Agronomy, 12(12), 2971. https://doi.org/10.3390/agronomy12122971





