The Progress towards Novel Herbicide Modes of Action and Targeted Herbicide Development
Abstract
1. Introduction
2. Herbicide Resistance Risk
3. Limiting Factors in Studying Novel Herbicide MoAs
4. Novel Potential Herbicide Targets
4.1. The Discovery of New Targets Based on Known Herbicides
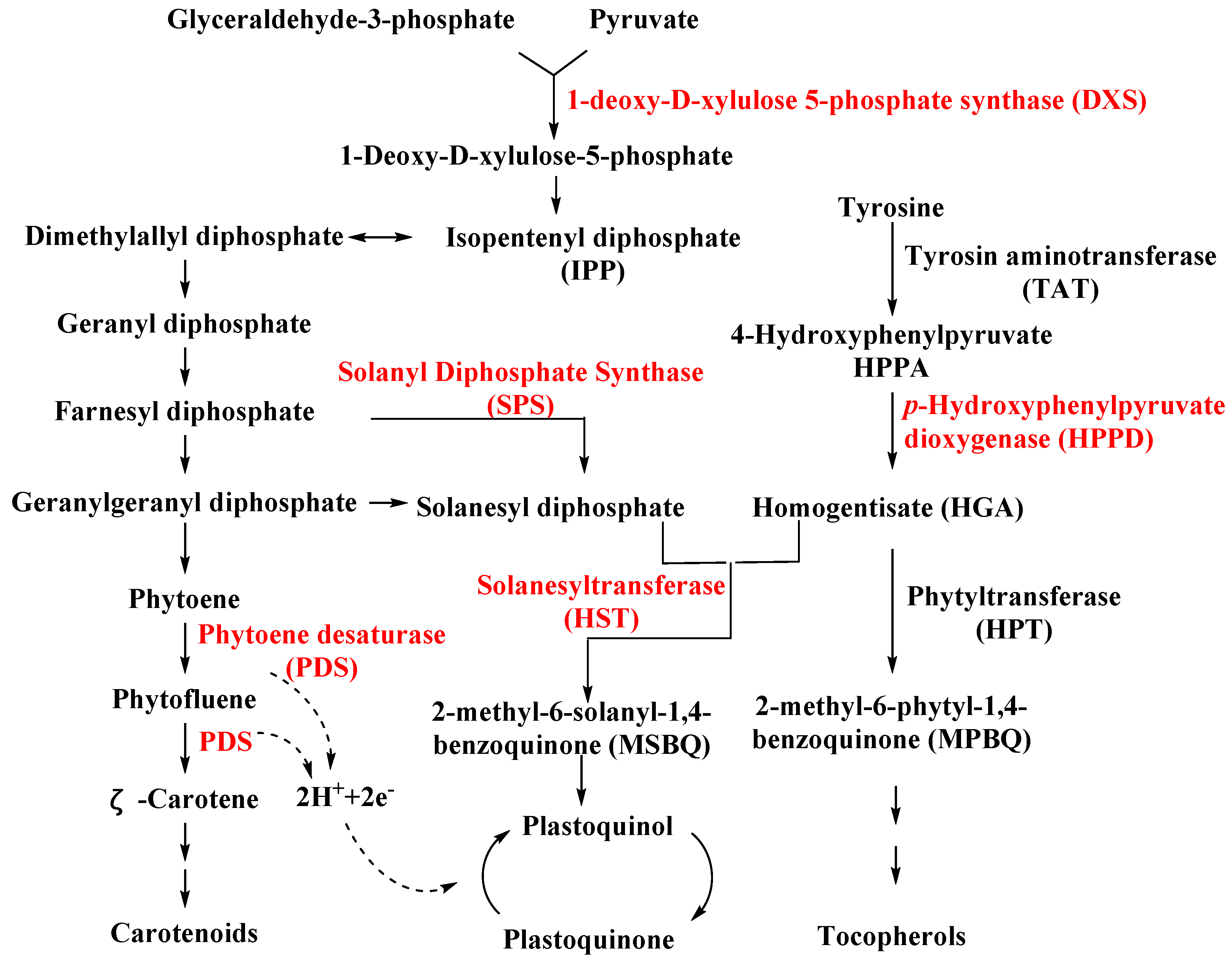
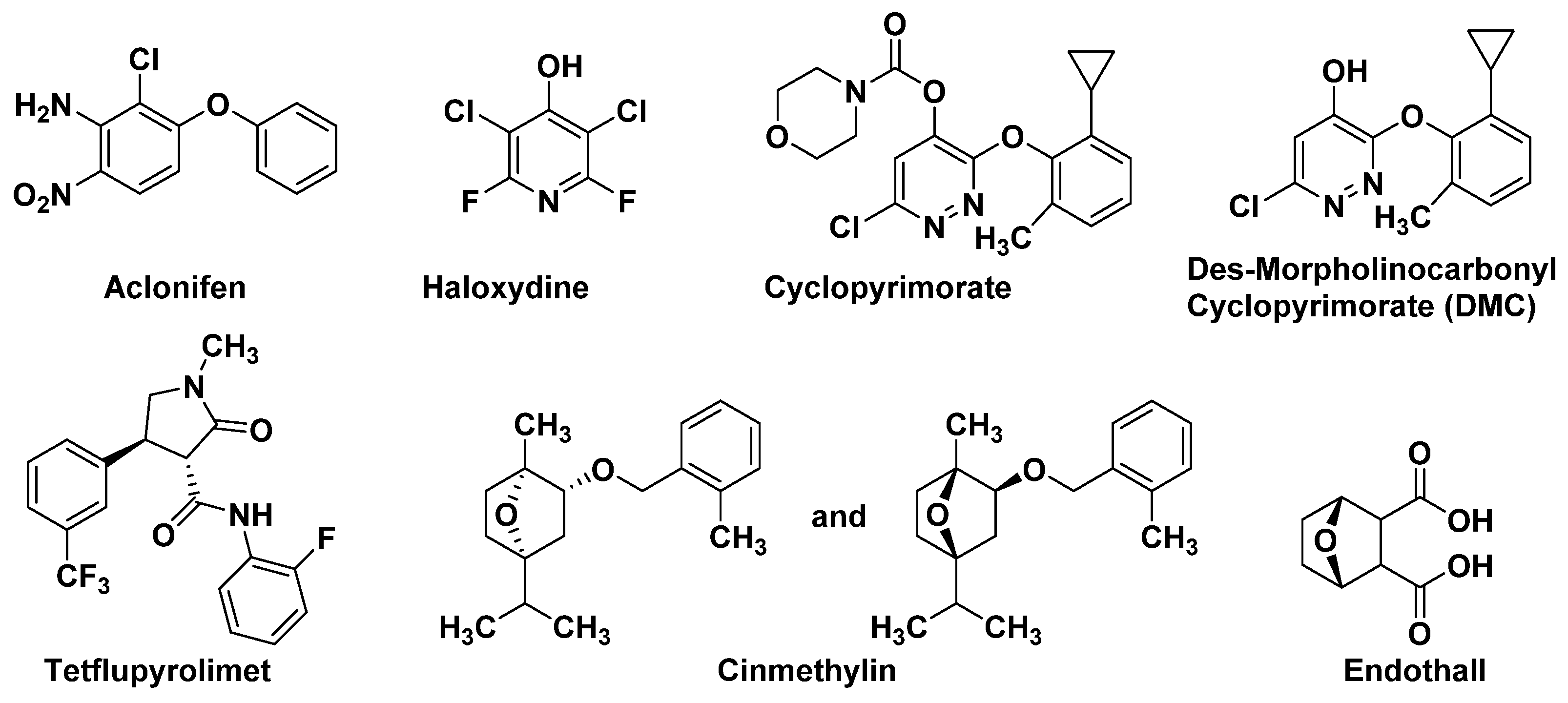
4.1.1. Solanyl Diphosphate Synthase (SPS)
4.1.2. Homogentisate Solanesyltransferase (HST)
4.1.3. Dihydroorotate Dehydrogenase (DHODH)
4.1.4. Fatty Acid Thioesterase (FAT)
4.1.5. Serine/Threonine Protein Phosphatases (PPs)
4.2. Exploring New Herbicide MoAs Based on Natural Products
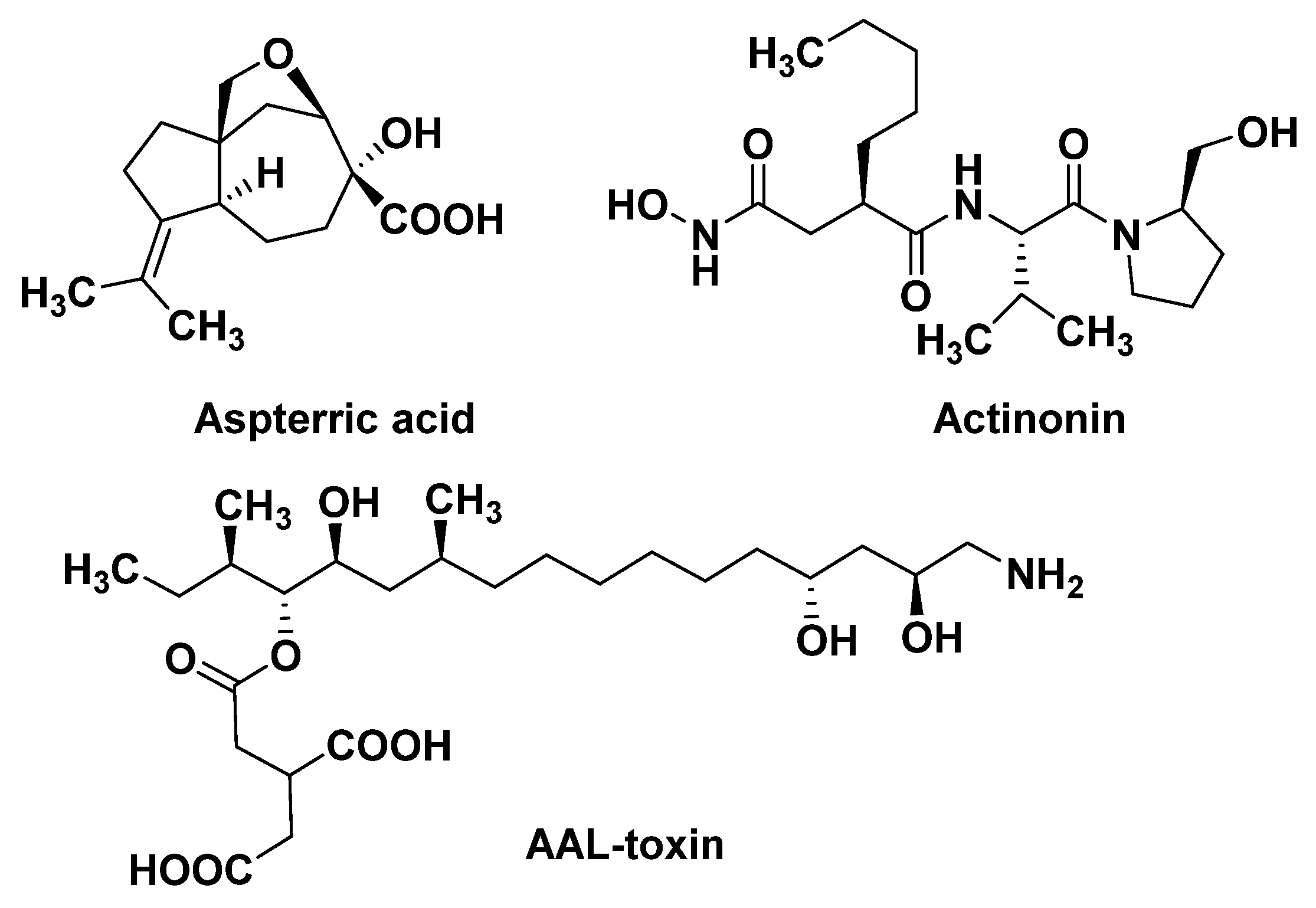
4.2.1. Dihydroxy-Acid Dehydratase (DHAD)
4.2.2. Plastid Peptide Deformylase (PDEF)
4.2.3. Ceramide Synthase
4.3. Exploration of Potential Herbicide MoAs Based on Their Biochemical Mechanisms
4.3.1. DNA Gyrase
4.3.2. Dihydrofolate Reductase (DHFR)
5. Targeted Herbicide Discovery and Development
6. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mascarelli, A. Growing up with pesticides. Science 2013, 341, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Sparks, T.C.; Lorsbach, B.A. Perspectives on the agrochemical industry and agrochemical discovery. Pest Manag. Sci. 2017, 73, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Enserink, M.; Hines, P.J.; Vignieri, S.N.; Wigginton, N.S.; Yeston, J.S. Smarter pest control. The pesticide paradox. Introduction. Science 2013, 341, 728–729. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Dong, J.; Lin, H.Y.; Wang, M.Y.; Li, X.K.; Zheng, B.F.; Chen, Q.; Hao, G.F.; Yang, W.C.; Yang, G.F. Pyrazole-isoindoline-1,3-dione hybrid: A promising scaffold for 4-hydroxyphenylpyruvate dioxygenase inhibitors. J. Agric. Food Chem. 2019, 67, 10844–10852. [Google Scholar] [CrossRef]
- Lamberth, C.; Jeanmart, S.; Luksch, T.; Plant, A. Current challenges and trends in the discovery of agrochemicals. Science 2013, 341, 742–746. [Google Scholar] [CrossRef]
- Duke, S.O.; Owens, D.K.; Dayan, F.E. The growing need for biochemical bioherbicides. ACS Symp. Ser. 2014, 1172, 31–43. [Google Scholar] [CrossRef]
- Service, R.F. Agriculture. What happens when weed killers stop killing? Science 2013, 341, 1329. [Google Scholar] [CrossRef]
- Peterson, G.E. The discovery and development of 2,4-D. Agric. Hist. Soc. 1967, 41, 243–254. [Google Scholar]
- Sterling, T.M.; Hall, J.C. Mechanism of action of auxins and the kinetics of cellular growth. In Herbicide Activity: Biochemistry and Molecular Biology; Roe, R.M., Burton, J.D., Kuhr, R.J., Eds.; IOS Press: Amsterdam, The Netherlands, 1997; pp. 111–141. [Google Scholar]
- Wang, Q.; Ge, L.; Zhao, N.; Zhang, L.; You, L.; Wang, D.; Liu, W.; Wang, J. A Trp-574-Leu mutation in the acetolactate synthase (ALS) gene of Lithospermum arvense L. confers broad-spectrum resistance to ALS inhibitors. Pestic. Biochem. Physiol. 2019, 158, 12–17. [Google Scholar] [CrossRef]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop Prot. 2012, 35, 15–28. [Google Scholar] [CrossRef]
- Sparks, T.C. Insecticide discovery: An evaluation and analysis. Pestic. Biochem. Physiol. 2013, 107, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Perry, E.D.; Ciliberto, F.; Hennessy, D.A.; Moschini, G. Genetically engineered crops and pesticide use in U.S. maize and soybeans. Sci. Adv. 2016, 2, e1600850. [Google Scholar] [CrossRef] [PubMed]
- Copping, L.G.; Menn, J.J. Biopesticides: A review of their action, applications and efficacy. Pest Manag. Sci. 2000, 56, 651–676. [Google Scholar] [CrossRef]
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef]
- Brookes, G.; Barfoot, P. Environmental impacts of genetically modified (GM) crop use 1996–2015: Impacts on pesticide use and carbon emissions. GM Crops Food 2017, 8, 117–147. [Google Scholar] [CrossRef]
- Duke, S.O. The history and current status of glyphosate. Pest Manag. Sci. 2018, 74, 1027–1034. [Google Scholar] [CrossRef]
- Owen, M.D.; Zelaya, I.A. Herbicide-resistant crops and weed resistance to herbicides. Pest Manag. Sci. 2005, 61, 301–311. [Google Scholar] [CrossRef]
- Reddy, K.N. Weed control and species shift in bromoxynil- and glyphosate-resistant cotton (Gossypium hirsutum) rotation systems. Weed Technol. 2004, 18, 131–139. [Google Scholar] [CrossRef]
- Cao, M.X.; Huang, J.Q.; Wei, Z.M.; Yao, Q.H.; Wan, C.Z.; Lu, J.A. Engineering higher yield and herbicide resistance in rice by agrobacterium-mediated multiple gene transformation. Crop Sci. 2004, 44, 2206–2213. [Google Scholar] [CrossRef]
- Appleby, A.P. A history of weed control in the United States and Canada—A sequel. Weed Sci. 2005, 53, 762–768. [Google Scholar] [CrossRef]
- Duke, S.O. Why have no new herbicide modes of action appeared in recent years? Pest Manag. Sci. 2012, 68, 505–512. [Google Scholar] [CrossRef]
- Tranel, P.J.; Riggins, C.W.; Bell, M.S.; Hager, A.G. Herbicide resistances in Amaranthus tuberculatus: A call for new options. J. Agric. Food Chem. 2011, 59, 5808–5812. [Google Scholar] [CrossRef] [PubMed]
- Tresch, S.; Schmotz, J.; Grossmann, K. Probing mode of action in plant cell cycle by the herbicide endothall, a protein phosphatase inhibitor. Pestic. Biochem. Physiol. 2011, 99, 86–95. [Google Scholar] [CrossRef]
- Kahlau, S.; Schroder, F.; Freigang, J.; Laber, B.; Lange, G.; Passon, D.; Kleessen, S.; Lohse, M.; Schulz, A.; von Koskull-Doring, P.; et al. Aclonifen targets solanesyl diphosphate synthase, representing a novel mode of action for herbicides. Pest Manag. Sci. 2020, 76, 3377–3388. [Google Scholar] [CrossRef]
- Liu, M.; Lu, S. Plastoquinone and ubiquinone in plants: Biosynthesis, physiological function and metabolic engineering. Front. Plant Sci. 2016, 7, 1898. [Google Scholar] [CrossRef]
- Ohara, K.; Sasaki, K.; Yazaki, K. Two solanesyl diphosphate synthases with different subcellular localizations and their respective physiological roles in Oryza sativa. J. Exp. Bot. 2010, 61, 2683–2692. [Google Scholar] [CrossRef]
- Dayan, F.E.; Haesaert, G.; van Leeuwen, T.; Holden-Dye, L.; Crossthwaite, A.; Nauen, R. Pesticides modes of action and resistance: A perspective from the 2019 IUPAC congress. Outlooks Pest Manag. 2019, 30, 157–163. [Google Scholar] [CrossRef]
- Jun, L.; Saiki, R.; Tatsumi, K.; Nakagawa, T.; Kawamukai, M. Identification and subcellular localization of two solanesyl diphosphate synthases from Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1882–1888. [Google Scholar] [CrossRef]
- Sadre, R.; Frentzen, M.; Saeed, M.; Hawkes, T. Catalytic reactions of the homogentisate prenyl transferase involved in plastoquinone-9 biosynthesis. J. Biol. Chem. 2010, 285, 18191–18198. [Google Scholar] [CrossRef]
- Collakova, E.; DellaPenna, D. Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol. 2003, 131, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Collakova, E.; DellaPenna, D. Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and arabidopsis. Plant Physiol. 2001, 127, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Shino, M.; Hamada, T.; Shigematsu, Y.; Hirase, K.; Banba, S. Action mechanism of bleaching herbicide cyclopyrimorate, a novel homogentisate solanesyltransferase inhibitor. J. Pestic. Sci. 2018, 43, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, A.; Knecht, W.; Piskur, J.; Löffler, M. Plant dihydroorotate dehydrogenase differs significantly in substrate specificity and inhibition from the animal enzymes. FEBS Lett. 2002, 529, 346–350. [Google Scholar] [CrossRef]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef]
- Campe, R.; Hollenbach, E.; Kammerer, L.; Hendriks, J.; Hoffken, H.W.; Kraus, H.; Lerchl, J.; Mietzner, T.; Tresch, S.; Witschel, M.; et al. A new herbicidal site of action: Cinmethylin binds to acyl-ACP thioesterase and inhibits plant fatty acid biosynthesis. Pestic. Biochem. Physiol. 2018, 148, 116–125. [Google Scholar] [CrossRef]
- Umetsu, N.; Shirai, Y. Development of novel pesticides in the 21st century. J. Pestic. Sci. 2020, 45, 54–74. [Google Scholar] [CrossRef]
- Dayan, F.E.; Romagni, J.G.; Duke, S.O. Herbicides, cinmethylin. In Encyclopedia of Agrochemicals; Plimmer, J.R., Gammon, D.W., Ragsdale, N.N., Eds.; John Wiley & Sons: New York, NY, USA, 2003; pp. 754–757. [Google Scholar]
- Uhrig, R.G.; Labandera, A.M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef]
- Bajsa, J.; Pan, Z.; Dayan, F.E.; Owens, D.K.; Duke, S.O. Validation of serine/threonine protein phosphatase as the herbicide target site of endothall. Pestic. Biochem. Physiol. 2012, 102, 38–44. [Google Scholar] [CrossRef]
- Duke, S.O.; Stidham, M.A.; Dayan, F.E. A novel genomic approach to herbicide and herbicide mode of action discovery. Pest Manag. Sci. 2019, 75, 314–317. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S.E.; Tang, Y. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Z.; Patel, D.V.; Hackbarth, C.J.; Wang, W.; Dreyer, G.; Young, D.C.; Margolis, P.S.; Wu, C.; Ni, Z.J.; Trias, J.; et al. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 2000, 39, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Bisson, C.; Britton, K.L.; Sedelnikova, S.E.; Rodgers, H.F.; Eadsforth, T.C.; Viner, R.C.; Hawkes, T.R.; Baker, P.J.; Rice, D.W. Crystal structures reveal that the reaction mechanism of imidazoleglycerol-phosphate dehydratase is controlled by switching Mn(II) coordination. Structure 2015, 23, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Duke, S.O. Natural compounds as next-generation herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef]
- Adams, J.M. On the release of the formyl group from nascent protein. J. Mol. Biol. 1968, 33, 571–589. [Google Scholar] [CrossRef]
- Serero, A.; Giglione, C.; Sardini, A.; Martinez-Sanz, J.; Meinnel, T. An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway. J. Biol. Chem. 2003, 278, 52953–52963. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Merrill, A.H. Ceramide synthase. Methods Enzymol. 2000, 311, 15–21. [Google Scholar] [PubMed]
- Gechev, T.S.; Gadjev, I.Z.; Hille, J. An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell. Mol. Life Sci. 2004, 61, 1185–1197. [Google Scholar]
- Duke, S.O.; Dayan, F.E. Clues to new herbicide mechanisms of action from natural sources. ACS Symp. Ser. 2013, 1141, 203–215. [Google Scholar] [CrossRef]
- Hsiao, P.; Sanjaya; Su, R.C.; Teixeira da Silva, J.A.; Chan, M.T. Plant native tryptophan synthase beta 1 gene is a non-antibiotic selection marker for plant transformation. Planta 2007, 225, 897–906. [Google Scholar] [CrossRef]
- Dayan, F.E.; Ferreira, D.; Wang, Y.H.; Khan, I.A.; McInroy, J.A.; Pan, Z. A pathogenic fungi diphenyl ether phytotoxin targets plant enoyl (acyl carrier protein) reductase. Plant Physiol. 2008, 147, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Templeton, M.D.; Reinhardt, L.A.; Collyer, C.A.; Mitchell, R.E.; Cleland, W.W. Kinetic analysis of the L-ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola that is resistant to the transition state analogue (R)-N delta-(N′-sulfodiaminophosphinyl)-L-ornithine. Biochemistry 2005, 44, 4408–4415. [Google Scholar] [CrossRef] [PubMed]
- Groth, G. Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc. Natl. Acad. Sci. USA 2002, 99, 3464–3468. [Google Scholar] [CrossRef] [PubMed]
- Meiss, E.; Konno, H.; Groth, G.; Hisabori, T. Molecular processes of inhibition and stimulation of ATP synthase caused by the phytotoxin tentoxin. J. Biol. Chem. 2008, 283, 24594–24599. [Google Scholar] [CrossRef]
- Hou, C.X.; Dirk, L.M.; Pattanaik, S.; Das, N.C.; Maiti, I.B.; Houtz, R.L.; Williams, M.A. Plant peptide deformylase: A novel selectable marker and herbicide target based on essential cotranslational chloroplast protein processing. Plant Biotechnol. J. 2007, 5, 275–281. [Google Scholar] [CrossRef]
- Driouich, A.; Jauneau, A.; Staehelin, L.A. 7-Dehydrobrefeldin A, a naturally occurring brefeldin a derivative, inhibits secretion and causes a cis-to-trans breakdown of golgi stacks in plant cells. Plant Physiol. 1997, 113, 487–492. [Google Scholar] [CrossRef][Green Version]
- Hejli, A.M.; Koster, K.L. The allelochemical sorgoleone inhibits root H+-ATPase and water uptake. J. Chem. Ecol. 2004, 30, 2181–2191. [Google Scholar] [CrossRef]
- Danielsen, E.M. Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J. Biol. Chem. 1990, 29, 493–498. [Google Scholar]
- Wallace, M.D.; Waraich, N.F.; Debowski, A.W.; Corral, M.G.; Maxwell, A.; Mylne, J.S.; Stubbs, K.A. Developing ciprofloxacin analogues against plant DNA gyrase: A novel herbicide mode of action. Chem. Commun. 2018, 54, 1869–1872. [Google Scholar] [CrossRef]
- Evans-Roberts, K.M.; Mitchenall, L.A.; Wall, M.K.; Leroux, J.; Mylne, J.S.; Maxwell, A. DNA gyrase is the target for the quinolone drug ciprofloxacin in Arabidopsis thaliana. J. Biol. Chem. 2016, 291, 3136–3144. [Google Scholar] [CrossRef]
- Dayan, F.E. Current Status and Future Prospects in Herbicide Discovery. Plants 2019, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.M.; Danenberg, P.V.; Johnston, P.G.; Lenz, H.J.; Ladner, R.D. Standing the test of time: Targeting thymidylate biosynthesis in cancer therapy. Nat. Rev. Clin. Oncol. 2014, 11, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Corral, M.G.; Haywood, J.; Stehl, L.H.; Stubbs, K.A.; Murcha, M.W.; Mylne, J.S. Targeting plant DIHYDROFOLATE REDUCTASE with antifolates and mechanisms for genetic resistance. Plant J. 2018, 95, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Peter, E.; Rothbart, M.; Oelze, M.L.; Shalygo, N.; Dietz, K.J.; Grimm, B. Mg protoporphyrin monomethylester cyclase deficiency and effects on tetrapyrrole metabolism in different light conditions. Plant Cell Physiol. 2010, 51, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Maimann, S.; Wagner, C.; Kreft, O.; Zeh, M.; Willmitzer, L.; Hofgen, R.; Hesse, H. Transgenic potato plants reveal the indispensable role of cystathionine beta-lyase in plant growth and development. Plant J. 2000, 23, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Silverman, D.N.; Lindskog, S. The catalytic mechanism of carbonic anhydrase: Implications of a rate-limiting protolysis of water. Acc. Chem. Res. 2002, 21, 30–36. [Google Scholar] [CrossRef]
- Kochetov, G.A.; Solovjeva, O.N. Structure and functioning mechanism of transketolase. Biochim. Biophys. Acta 2014, 1844, 1608–1618. [Google Scholar] [CrossRef]
- Zheng, Y.; Tian, L.; Liu, H.; Pan, Q.; Zhan, J.; Huang, W. Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regul. 2009, 58, 251–260. [Google Scholar] [CrossRef]
- Xiao, Z.; Morris-Natschke, S.L.; Lee, K.H. Strategies for the optimization of natural leads to anticancer drugs or drug candidates. Med. Res. Rev. 2016, 36, 32–91. [Google Scholar] [CrossRef]
- Cantrell, C.L.; Dayan, F.E.; Duke, S.O. Natural products as sources for new pesticides. J. Nat. Prod. 2012, 75, 1231–1242. [Google Scholar] [CrossRef]
- Beaudegnies, R.; Edmunds, A.J.F.; Fraser, T.E.M.; Hall, R.G.; Hawkes, T.R.; Mitchell, G.; Schaetzer, J.; Wendeborn, S.; Wibley, J. Herbicidal 4-hydroxyphenylpyruvate dioxygenase inhibitors—A review of the triketone chemistry story from a Syngenta perspective. Bioorg. Med. Chem. 2009, 17, 4134–4152. [Google Scholar] [CrossRef] [PubMed]
- Ohno, R.; Nagaoka, M.; Hirai, K.; Uchida, A.; Kochi, S.-i.; Yamada, O.; Tokumura, J. Synthesis and insecticidal activity of novel 1-alkyl-3-sulfonyloxypyrazole-4-carboxamide derivatives. J. Pestic. Sci. 2010, 35, 15–22. [Google Scholar] [CrossRef]
- Guan, A.; Liu, C.; Yang, X.; Dekeyser, M. Application of the intermediate derivatization approach in agrochemical discovery. Chem. Rev. 2014, 114, 7079–7107. [Google Scholar] [CrossRef] [PubMed]
- Taft, C.A.; Da Silva, V.B.; Da Silva, C.H. Current topics in computer-aided drug design. J. Pharm. Sci. 2008, 97, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, I.D. Structure-based strategies for drug design and discovery. Science 1992, 257, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Olliaro, P.L.; Gottlieb, M.; Wirth, D.F. Plasmodium falciparum proteinases: Targeted drug development. Parasitol. Today 1996, 12, 413–414. [Google Scholar] [CrossRef]
- Albert, A. Chemical aspects of selective toxicity. Nature 1958, 182, 421–422. [Google Scholar] [CrossRef]
- Rautio, J.; Kumpulainen, H.; Heimbach, T.; Oliyai, R.; Oh, D.; Jarvinen, T.; Savolainen, J. Prodrugs: Design and clinical applications. Nat. Rev. Drug Discov. 2008, 7, 255–270. [Google Scholar] [CrossRef]
- Mishra, A.P.; Chandra, S.; Tiwari, R.; Srivastava, A.; Tiwari, G. Therapeutic Potential of Prodrugs towards Targeted Drug Delivery. Open Med. Chem. J. 2018, 12, 111–123. [Google Scholar] [CrossRef]
- Zawilska, J.B.; Wojcieszak, J.; Olejniczak, A.B. Prodrugs: A challenge for the drug development. Pharmacol. Rep. 2013, 65, 1–14. [Google Scholar] [CrossRef]
- Huttunen, K.M.; Raunio, H.; Rautio, J. Prodrugs—From serendipity to rational design. Pharmacol. Rev. 2011, 63, 750–771. [Google Scholar] [CrossRef]
- Graf, N.; Lippard, S.J. Redox activation of metal-based prodrugs as a strategy for drug delivery. Adv. Drug Deliv. Rev. 2012, 64, 993–1004. [Google Scholar] [CrossRef]
- Singh, P.P.; Joshi, S.; Russell, P.J.; Verma, N.D.; Wang, X.; Khatri, A. Molecular chemotherapy and chemotherapy: A new front against late-stage hormone-refractory prostate cancer. Clin. Cancer Res. 2011, 17, 4006–4018. [Google Scholar] [CrossRef] [PubMed]
- Kokil, G.R.; Rewatkar, P.V. Bioprecursor prodrugs: Molecular modification of the active principle. Mini-Rev. Med. Chem. 2010, 10, 1316–1330. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, N.R. The rationality for using prodrug approach in drug discovery programs for new xenobiotics: Opportunities and challenges. Eur. J. Drug Metab. Pharmacokinet. 2011, 36, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Kratz, F.; Müller, I.A.; Ryppa, C.; Warnecke, A. Prodrug strategies in anticancer chemotherapy. ChemMedChem 2008, 3, 20–53. [Google Scholar] [CrossRef]
- Hsieh, P.W.; Hung, C.F.; Fang, J.Y. Current prodrug design for drug discovery. Curr. Pharm. Des. 2009, 15, 2236–2250. [Google Scholar] [CrossRef]
- Jana, S.; Mandlekar, S.; Marathe, P. Prodrug design to improve pharmacokinetic and drug delivery properties: Challenges to the discovery scientists. Curr. Med. Chem. 2010, 17, 3874–3908. [Google Scholar] [CrossRef]




| Herbicide Group | Example Group | Dicots | Monocots | Total |
|---|---|---|---|---|
| Inhibition of Acetolactate Synthase | Chlorsulfuron | 105 | 66 | 171 |
| PSII inhibitors-Serine 264 Binders | Chlorotoluron | 53 | 34 | 87 |
| Inhibition of Enolpyruvyl Shikimate Phosphate Synthase | Glyphosate | 27 | 29 | 56 |
| Inhibition of Acetyl CoA Carboxylase | Sethoxydim | 0 | 50 | 50 |
| Auxin Mimics | 2,4-D | 34 | 8 | 42 |
| PS I Electron Diversion | Paraquat | 22 | 10 | 32 |
| Inhibition of Protoporphyrinogen Oxidase | Oxyfluorfen | 10 | 4 | 14 |
| Very Long-Chain Fatty Acid Synthesis inhibitors | Butachlor | 2 | 11 | 13 |
| Inhibition of Microtubule Assembly 2 | Trifluralin | 2 | 10 | 12 |
| Inhibition of Lycopene Cyclase | Amitrole | 1 | 5 | 6 |
| Inhibition of Glutamine Synthetase | Glufosinate-ammonium | 1 | 5 | 6 |
| Phytoene Desaturase inhibitors | Diflufenican | 4 | 1 | 5 |
| PSII inhibitors-Histidine 215 Binders | Bromoxynil | 3 | 1 | 4 |
| Inhibition of Cellulose Synthesis | Dichlobenil | 0 | 4 | 4 |
| Inhibition of Hydroxyphenyl Pyruvate Dioxygenase | Isoxaflutole | 3 | 0 | 3 |
| Inhibition of Microtubule Assembly | Clomazone | 0 | 3 | 3 |
| Antimicrotubule mitotic disrupter | Flamprop-methyl | 0 | 3 | 3 |
| Inhibition of Microtubule Organization | Propham | 0 | 1 | 1 |
| Nucleic acid inhibitors | MSMA | 1 | 0 | 1 |
| Unknown | Endothall | 0 | 1 | 1 |
| Cell elongation inhibitors | Difenzoquat | 0 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, B.; Hu, Y.; Wang, W.; Yan, W.; Ye, Y. The Progress towards Novel Herbicide Modes of Action and Targeted Herbicide Development. Agronomy 2022, 12, 2792. https://doi.org/10.3390/agronomy12112792
He B, Hu Y, Wang W, Yan W, Ye Y. The Progress towards Novel Herbicide Modes of Action and Targeted Herbicide Development. Agronomy. 2022; 12(11):2792. https://doi.org/10.3390/agronomy12112792
Chicago/Turabian StyleHe, Bo, Yanhao Hu, Wen Wang, Wei Yan, and Yonghao Ye. 2022. "The Progress towards Novel Herbicide Modes of Action and Targeted Herbicide Development" Agronomy 12, no. 11: 2792. https://doi.org/10.3390/agronomy12112792
APA StyleHe, B., Hu, Y., Wang, W., Yan, W., & Ye, Y. (2022). The Progress towards Novel Herbicide Modes of Action and Targeted Herbicide Development. Agronomy, 12(11), 2792. https://doi.org/10.3390/agronomy12112792