Identification of SNPs and Candidate Genes Associated with Salt Stress in Two Korean Sorghum Cultivars and Understanding Selection Pressures in the Breeding Process
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
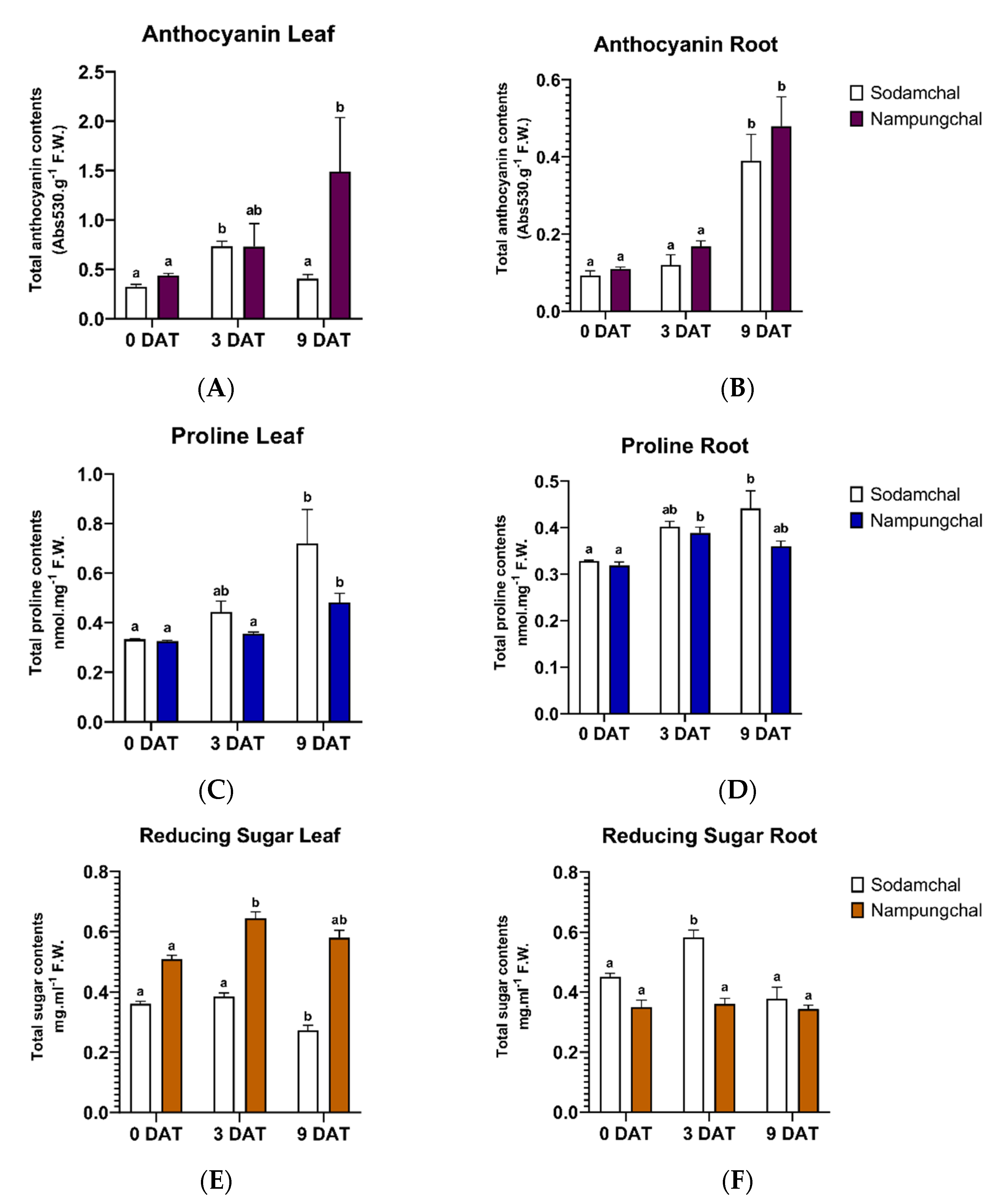
2.2. Quantification of the Anthocyanin Contents
2.3. Quantification of the Proline Contents
2.4. Quantification of the Reducing Sugar Contents
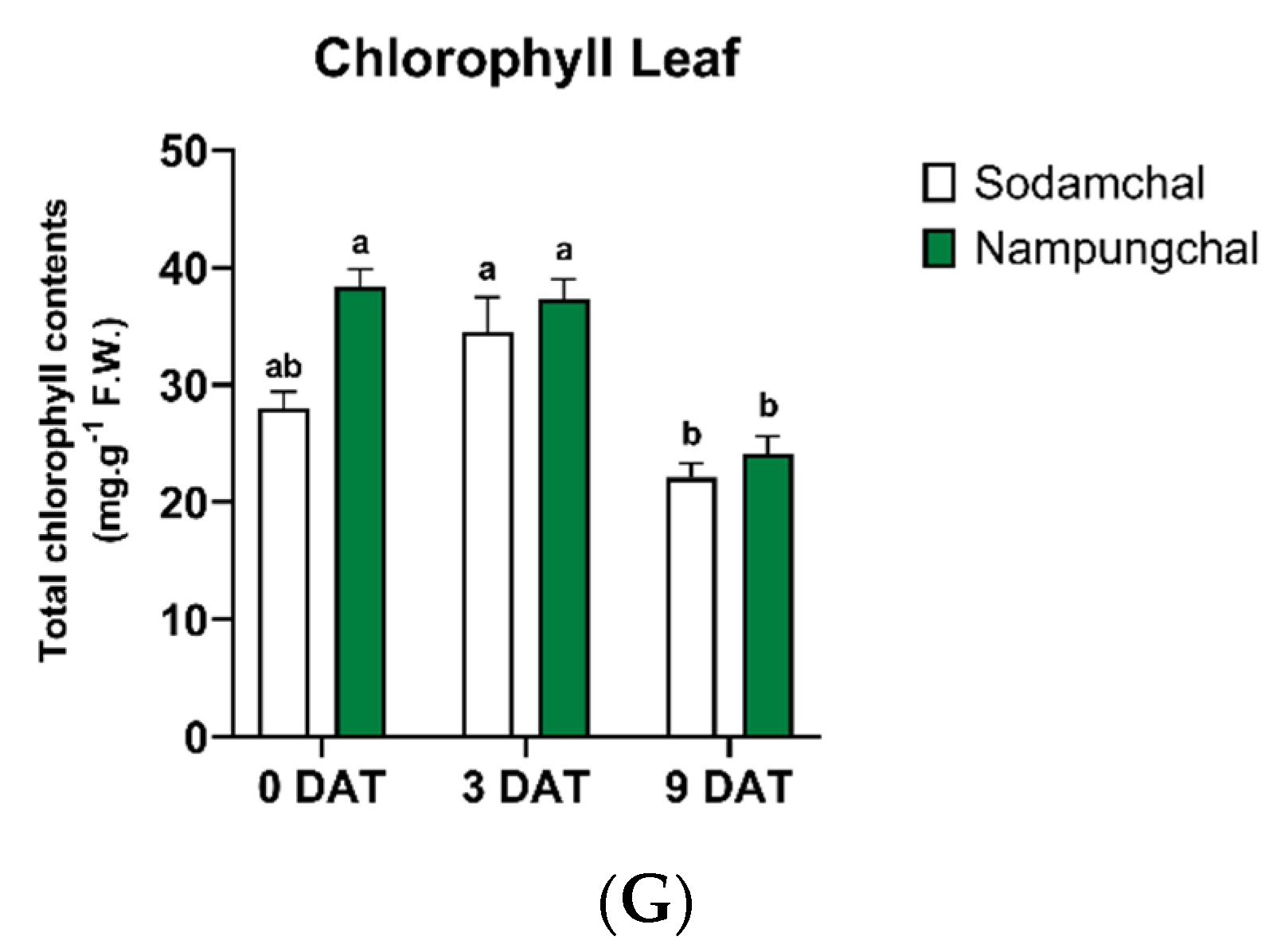
2.5. Quantification of the Chlorophyll Contents
2.6. RNA Isolation
2.7. Construction of the QuantSeq Library for DEG Analyses
2.8. Construction of the RNASeq Library and Sequencing for SNP Analyses
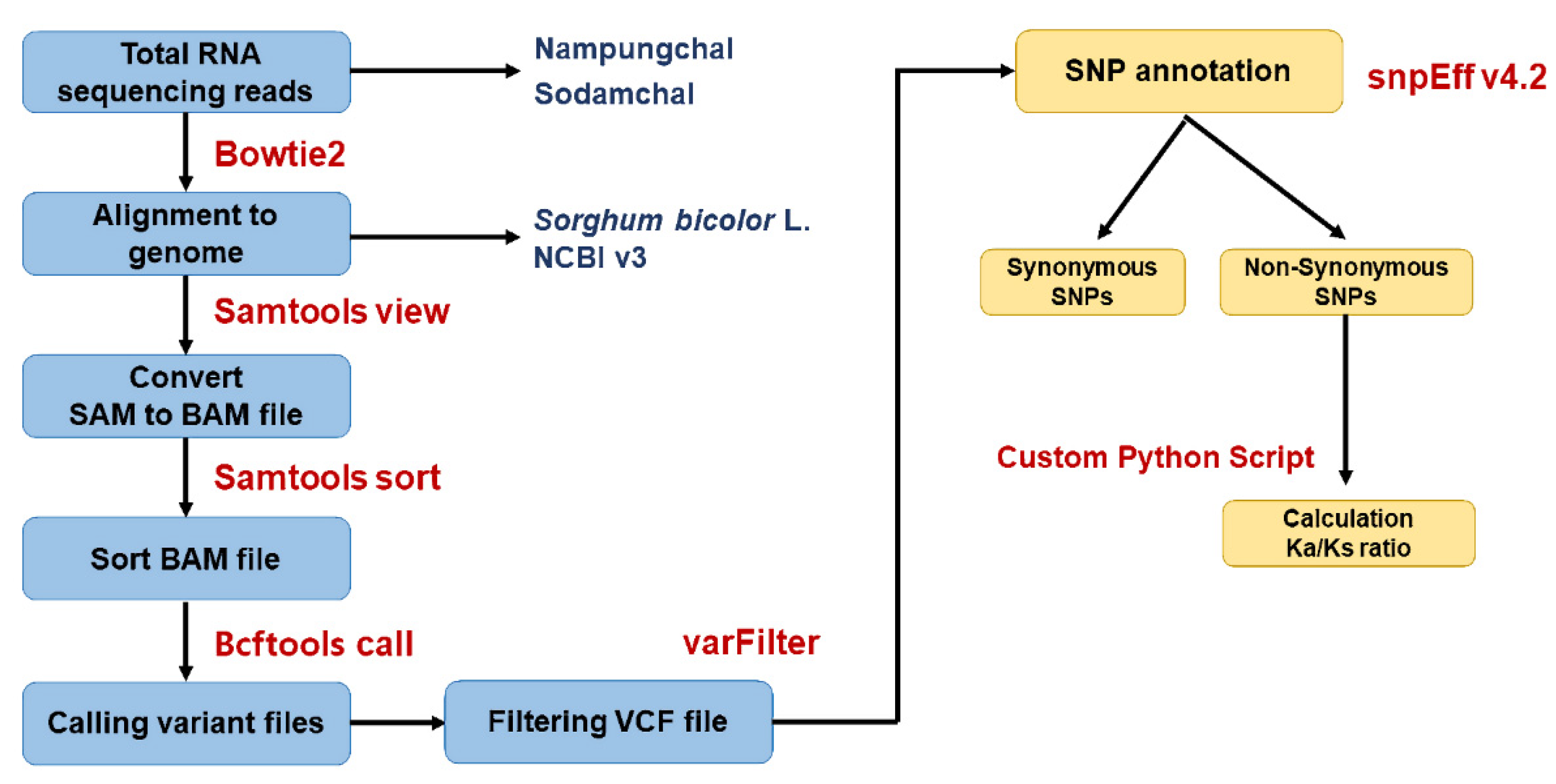
2.9. Sequencing Data Analysis
2.10. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes Database (KEGG) Pathway Enrichment Analysis of DEGs
2.11. Analysis of Quantitative Real-Time PCR (qRT-PCR)
2.12. SNP Classification and Selection Pressure
2.13. Statistical Data Analysis
3. Results
3.1. Growth Performance and Physiological Response to Salt Stress
3.1.1. Quantification of the Anthocyanin Contents
3.1.2. Quantification of the Proline Contents
3.1.3. Quantification of the Reducing Sugar Contents
3.1.4. Quantification of the Chlorophyll Contents
3.2. Analysis of Gene Expression in Two Sorghum Cultivars Using QuantSeq Data
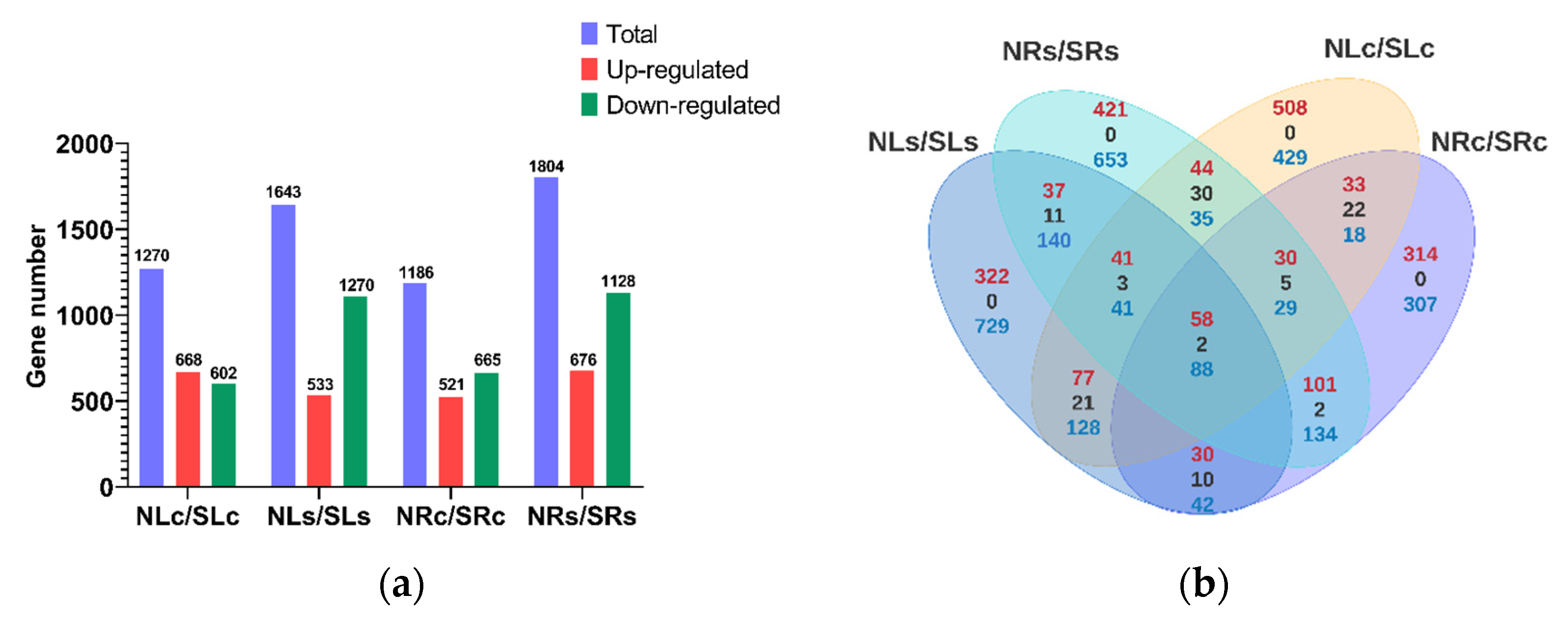
3.2.1. Differences in Gene Expression Profiles of the Two Sorghum Cultivars under Salt Stress
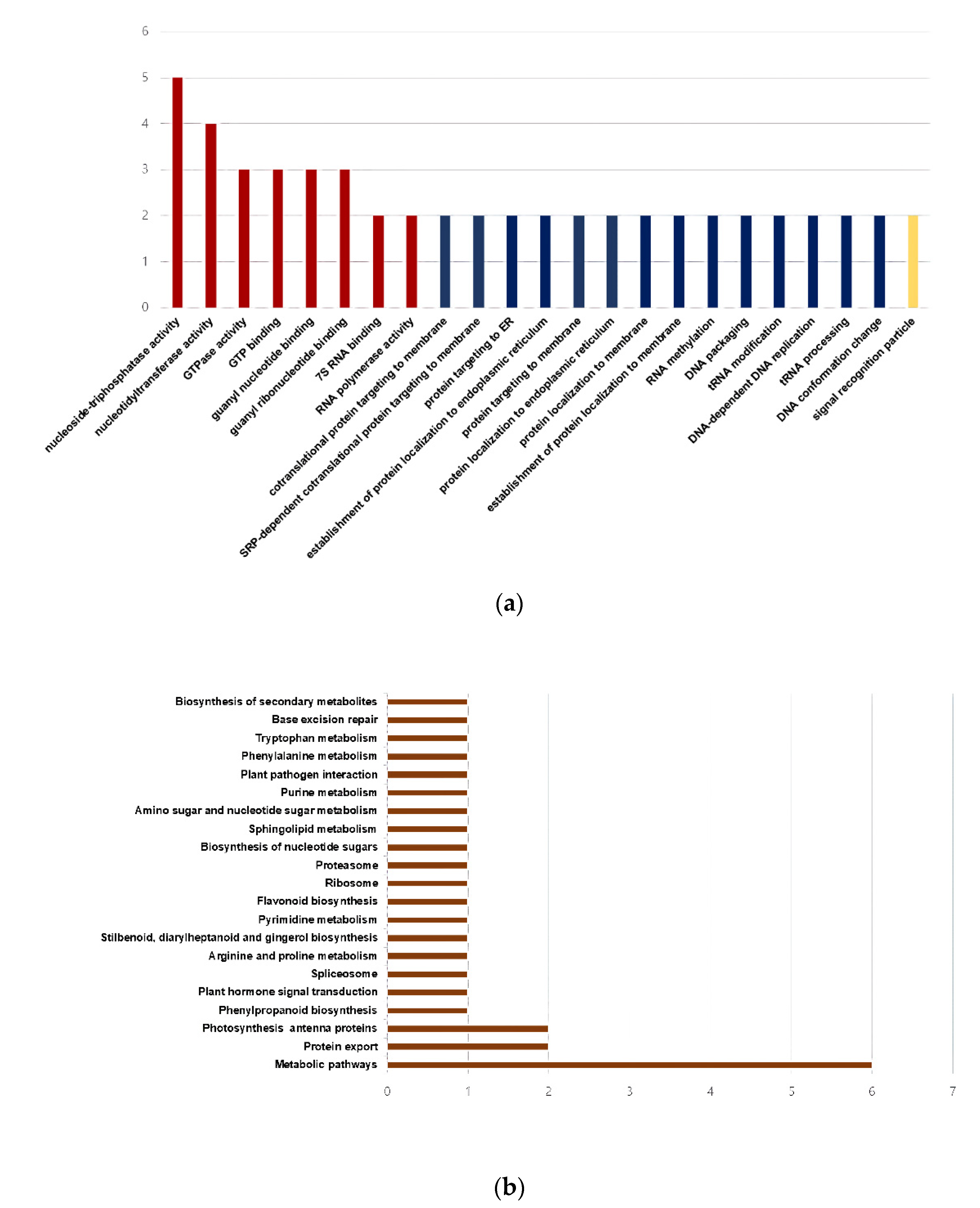
3.2.2. GO Classification and KEGG Pathway Analysis of the DEGs between the Two Sorghum Cultivars
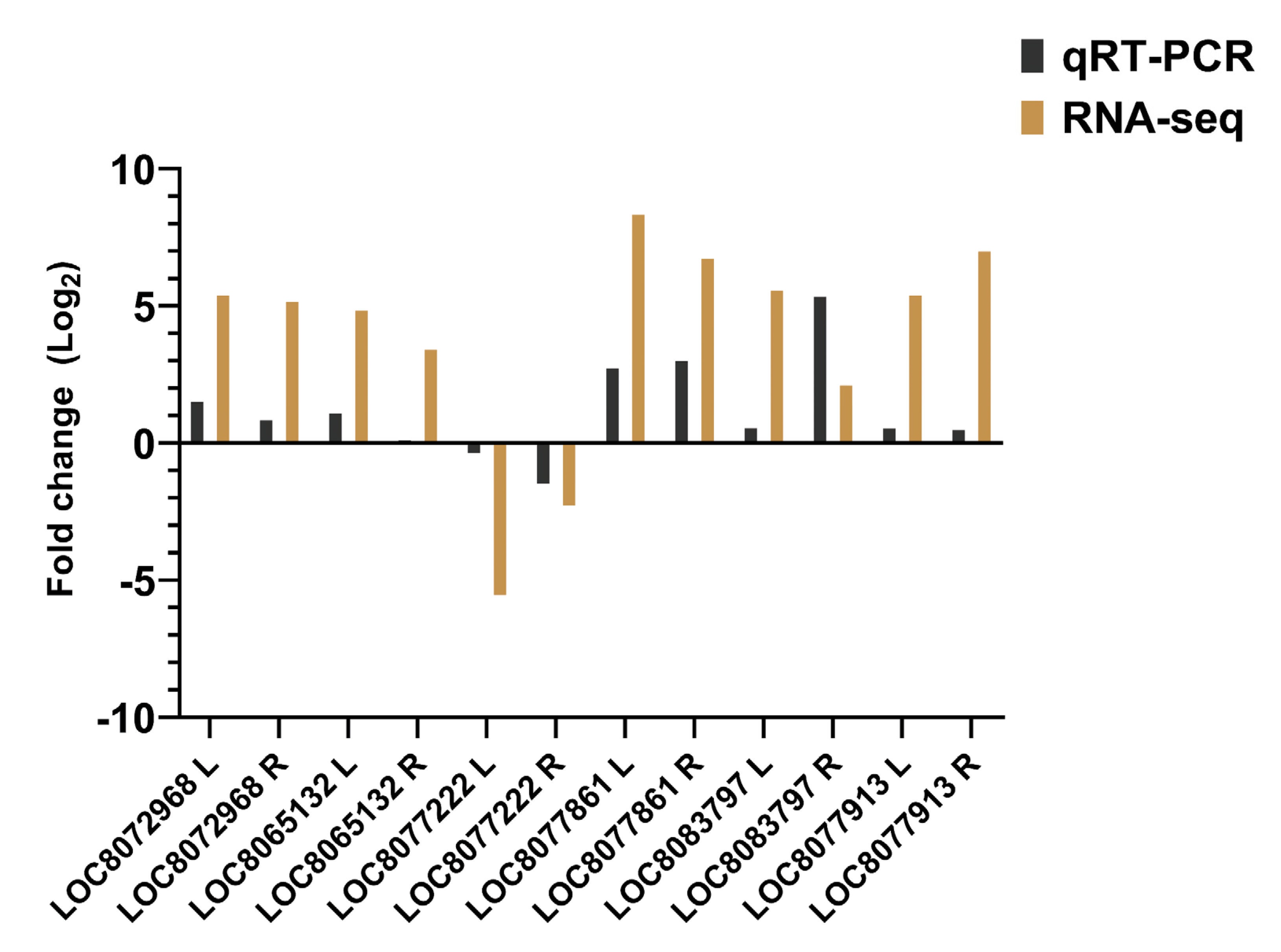
3.2.3. Validation of Differentially Expressed Genes by qRT-PCR
3.3. Variants Analysis in Two Sorghum Cultivars
3.3.1. Identification and Classification of SNPs from the Two Sorghum Cultivars
3.3.2. Identification of DEGs with Non-Synonymous SNPs
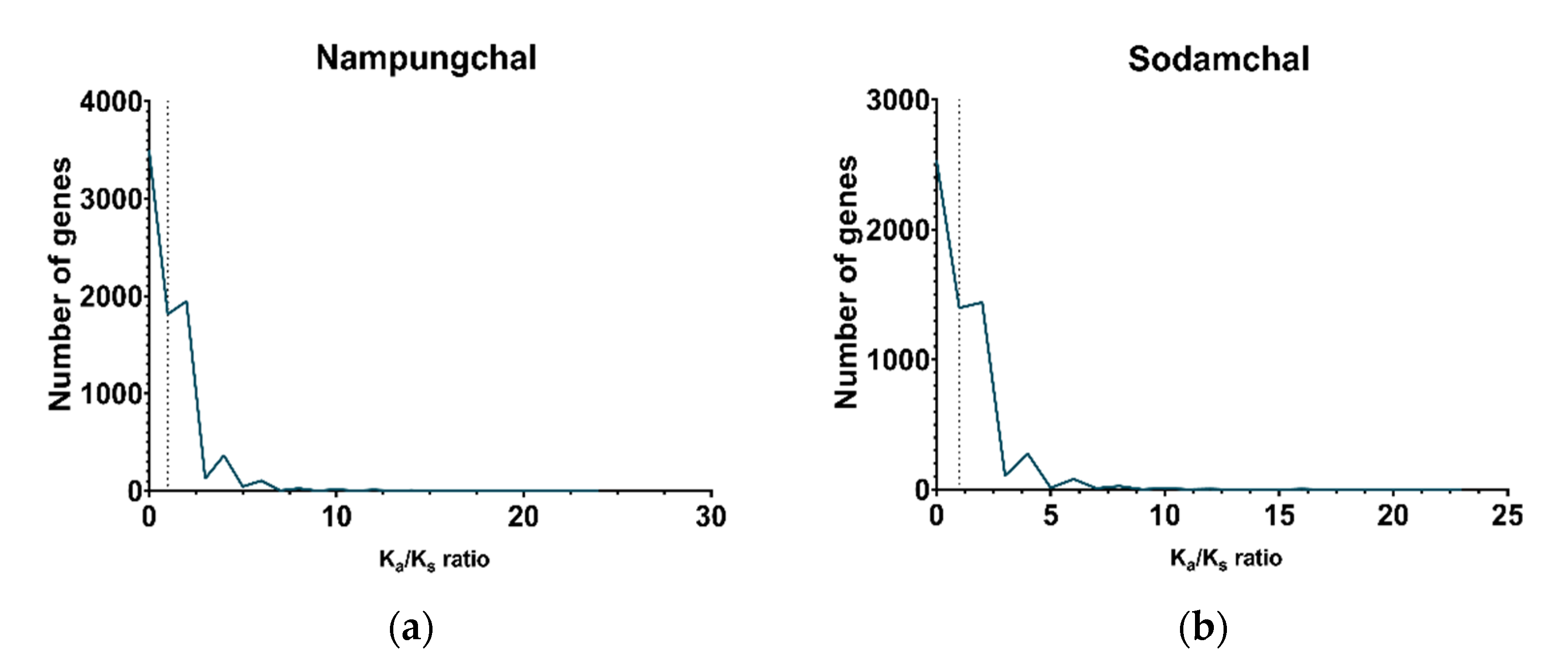
3.3.3. Selection Pressure of the Genes Calculated as the Ratio of Ka/Ks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruinsma, J. The Resource Outlook to 2050 by How Much Do Land, Water and Crop Yields Need to increase by 2050? In How to feed the World in 2050, Proceedings of the Technical Meeting of Experts, Rome, Italy, 24–26 June 2009; FAO Expert Meeting on How to Feed the World in 2050; FAO: Rome, Italy, 2009; pp. 1–33. [Google Scholar]
- Cominelli, E.; Conti, L.; Tonelli, C.; Galbiati, M. Challenges and perspectives to improve crop drought and salinity tolerance. New Biotechnol. 2013, 30, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- de Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; de Abreu, C.E.B.; Gomes-Filho, E. Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ. Exp. Bot. 2006, 56, 87–94. [Google Scholar] [CrossRef]
- Henderson, A.N.; Crim, P.M.; Cumming, J.R.; Hawkins, J.S. Phenotypic and physiological responses to salt exposure in Sorghum reveal diversity among domesticated landraces. Am. J. Bot. 2020, 107, 983–992. [Google Scholar]
- Isayenkov, S.V. Physiological and molecular aspects of salt stress in plants. Cytol. Genet. 2012, 46, 302–318. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Rahnama, A.; James, R.A.; Poustini, K.; Munns, R. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 2010, 37, 255–263. [Google Scholar] [CrossRef]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, K.; Zheng, Y.; Wang, Y.; Wang, J.; Liao, H. Disruption of AtWNK8 Enhances Tolerance of Arabidopsis to Salt and Osmotic Stresses via Modulating Proline Content and Activities of Catalase and Peroxidase. Int. J. Mol. Sci. 2013, 14, 7032–7047. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B.R. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Naeem, N.; Jacqueline, M.; Brian, W.; Yuan, Z.-C. Comparative transcriptomic analysis reveals different responses of Arabidopsis thaliana roots and shoots to infection by Agrobacterium tumefaciens in a hydroponic co-cultivation system. Physiol. Mol. Plant Pathol. 2019, 106, 109–119. [Google Scholar]
- Zhang, W.; Yang, L.; Li, M.; Ma, B.; Yan, C.; Chen, J. Omics-Based Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes during Early Infestation by Small Brown Planthopper. Int. J. Mol. Sci. 2015, 16, 28746–28764. [Google Scholar] [CrossRef] [PubMed]
- Kalaitzis, P.; Bazakos, C.; Manioudaki, M. Comparative Transcriptome Analysis of Two Olive Cultivars in Response to NaCl Stress. In Proceedings of the II International Symposium on Horticulture in Europe, Angers, France, 1–5 July 2012; Volume 1099, pp. 801–806. [Google Scholar]
- Beritognolo, I.; Harfouche, A.; Brilli, F.; Prosperini, G.; Gaudet, M.; Brosche, M.; Salani, F.; Kuzminsky, E.; Auvinen, P.; Paulin, L.; et al. Comparative study of transcriptional and physiological responses to salinity stress in two contrasting Populus alba L. genotypes. Tree Physiol. 2011, 31, 1335–1355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, D.; Majee, A.; Singh, S.; Roohi; Asif, M.H.; Sane, A.P.; Sane, V.A. Identification of tomato root growth regulatory genes and transcription factors through comparative transcriptomic profiling of different tissues. Physiol. Mol. Biol. Plants 2021, 27, 1173–1189. [Google Scholar] [CrossRef] [PubMed]
- Almodares, A.; Hadi, M.R. Production of bioethanol from sweet sorghum: A review. Afr. J. Agric. Res. 2009, 4, 772–780. [Google Scholar]
- Gnansounou, E.; Dauriat, A.; Wyman, C.E. Refining sweet sorghum to ethanol and sugar: Economic trade-offs in the context of North China. Bioresour. Technol. 2005, 96, 985–1002. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Kang, C.; Sok, L.; Kwon, S. Screening for Fittest Miscellaneous Cereals for Reclaimed Land and Functionality Improvement of Sorghum bicolor Cultivated in Reclaimed Land. Korean J. Crop Sci. 2019, 64, 109–126. [Google Scholar]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A Bhlh-Type Transcription Factor, Aba-Inducible Bhlh-Type Transcription Factor/JA-Associated Myc2-Like1, Acts as a Repressor to Negatively Regulate Jasmonate Signaling in Arabidopsis. Plant Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, 80. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, R.P.; Srinivas, R.D.; Sivasakthi, K.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Evaluation of Sorghum [Sorghum bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time PCR Data Normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Bierne, N.; Eyre-Walker, A. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: Implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics 2003, 165, 1587–1597. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Dhar, Y.V.; Asif, M.H.; Bag, S.K. In Silico identification of SNP diversity in cultivated and wild tomato species: Insight from molecular simulations. Sci. Rep. 2016, 6, 38715. [Google Scholar] [CrossRef]
- Dai, L.Y.; Zhang, L.J.; Jiang, S.J.; De Yin, K. Saline and alkaline stress genotypic tolerance in sweet sorghum is linked to sodium distribution. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2014, 64, 471–481. [Google Scholar] [CrossRef]
- Marco, A.; Thomas, F.D.; Snorre, B.H.; Hughes, N.M.; Leather, S.R.; Lee, D.W.; Lev-Yadun, S.; Manetas, Y.; Ougham, H.J.; Schaberg, P.G.; et al. Unravelling the evolution of autumn colours: An interdisciplinary approach. Trends Ecol. Evol. 2009, 24, 166–173. [Google Scholar]
- Shimada, S.; Inoue, Y.T.; Sakuta, M. Anthocyanidin synthase in non-anthocyanin-producing caryophyllales species. Plant J. 2005, 44, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Chutipaijit, S.; Cha-um, S.; Sompornpailin, K. High contents of proline and anthocyanin increase protective response to salinity in Oryza sativa L. spp. indica. Aust. J. Crop Sci. 2011, 5, 1191–1198. [Google Scholar]
- Oh, J.E.; Kim, Y.H.; Kwon, Y.R.; Lee, H. Enhanced Level of Anthocyanin Leads to Increased Salt Tolerance in Arabidopsis PAP1-D Plants upon Sucrose Treatment. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 79–88. [Google Scholar] [CrossRef]
- Mbarki, S.; Sytar, O.; Zivcak, M.; Abdelly, C.; Cerda, A.; Brestic, M. Anthocyanins of Coloured Wheat Genotypes in Specific Response to SalStress. Molecules 2018, 23, 1518. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.M.F. Protection of plasma membrane of onion epidermal cells by glycinebetaine and proline against NaCl stress. Plant Physiol. Biochem. 1998, 36, 767–772. [Google Scholar] [CrossRef]
- Abdelaziz, M.N.; Xuan, T.D.; Mekawy, A.M.M.; Wang, H.; Khanh, T.D. Relationship of Salinity Tolerance to Na+ Exclusion, Proline Accumulation, and Antioxidant Enzyme Activity in Rice Seedlings. Agriculture 2018, 8, 166. [Google Scholar] [CrossRef]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of salt stress on ion concentration, proline content, antioxidant enzyme activities and gene expression in tomato cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef]
- Martinez, C.A.; Maestri, M.; Lani, E.G. In vitro salt tolerance and proline accumulation in Andean potato (Solanum spp.) differing in frost resistance. Plant Sci. 1996, 116, 177–184. [Google Scholar] [CrossRef]
- Lutts, S.; Majerus, V.; Kinet, J.M. NaCl effects on proline metabolism in rice (Oryza sativa) seedlings. Physiol. Plant. 1999, 105, 450–458. [Google Scholar] [CrossRef]
- de Lacerda, C.F.; Cambraia, J.; Oliva, M.A.; Ruiz, H.A.; Prisco, J.T. Solute accumulation and distribution during shoot and leaf development in two sorghum genotypes under salt stress. Environ. Exp. Bot. 2003, 49, 107–120. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Zhifang, G.; Loescher, W.H. Expression of a celery mannose 6-phosphate reductase in Arabidopsis thaliana enhances salt tolerance and induces biosynthesis of both mannitol and a glucosyl-mannitol dimer. Plant Cell Environ. 2003, 26, 275–283. [Google Scholar] [CrossRef]
- Murakeözy, E.P.; Nagy, Z.; Duhazé, C.; Bouchereau, A.; Tuba, Z. Seasonal changes in the levels of compatible osmolytes in three halophytic species of inland saline vegetation in Hungary. J. Plant Physiol. 2003, 160, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Khavari-Nejad, R.A.; Mostofi, Y. Effects of NaCl on photosynthetic pigments, saccharides, and chloroplast ultrastructure in leaves of tomato cultivars. Photosynthetica 1998, 35, 151–154. [Google Scholar] [CrossRef]
- Ashraf, M.; Tufail, M. Variation in Salinity Tolerance in Sunflower (Helianthus annuus L.). J. Agron. Crop Sci.-Z. Fur Acker Und Pflanzenbau 1995, 174, 351–362. [Google Scholar] [CrossRef]
- Hema, V.; Pattathil, S.; Romit, C.; George, T. Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.)—differential response in salt-tolerant and sensitive varieties. Plant Sci. 2003, 165, 1411–1418. [Google Scholar]
- Verma, S.; Mishra, S.N. Putrescine alleviation of growth in salt stressed Brassica juncea by inducing antioxidative defense system. J. Plant Physiol. 2005, 162, 669–677. [Google Scholar] [CrossRef]
- Sairam, R.K.; Tyagi, A. Physiology and molecular biology of salinity stress tolerance in plants. Curr. Sci. 2004, 86, 407–421. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Xuan, T.D.; Minh, T.N.; Anh, L.H.; Khanh, T.D. Allelopathic momilactones A and B are implied in rice drought and salinity tolerance, not weed resistance. Agron. Sustain. Dev. 2016, 36, 52. [Google Scholar] [CrossRef]
- Sahid, S.; Roy, C.; Paul, S.; Datta, R. Rice lectin protein r40c1 imparts drought tolerance by modulating S-adenosylmethionine synthase 2, stress-associated protein 8 and chromatin-associated proteins. J. Exp. Bot. 2020, 71, 7331–7346. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.T.; Figueroa, C.R. Jasmonates and Plant Salt Stress: Molecular Players, Physiological Effects, and Improving Tolerance by Using Genome-Associated Tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef] [PubMed]
- Qun, W.; Shao, H.; Xu, Z.; Liu, J.; Zhang, D.; Huang, Y. Salinity Tolerance Mechanism of Osmotin and Osmotin-like Proteins: A Promising Candidate for Enhancing Plant Salt Tolerance. Curr. Genom. 2017, 18, 553–556. [Google Scholar]
- Clark, G.; Brown, K.A.; Tripathy, M.K.; Roux, S.J. Recent Advances Clarifying the Structure and Function of Plant Apyrases (Nucleoside Triphosphate Diphosphohydrolases). Int. J. Mol. Sci. 2021, 22, 3283. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Z.; Xia, X.Z.; Zhao, M.G.; Tian, Q.Y.; Zhang, W.H. Expression of a Medicago falcata small GTPase gene, MfARL1 enhanced tolerance to salt stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2013, 63, 227–235. [Google Scholar] [CrossRef]
- Garrido-Cardenas, J.A.; Mesa-Valle, C.; Manzano-Agugliaro, F. Trends in plant research using molecular markers. Planta 2018, 247, 543–557. [Google Scholar] [CrossRef]
- Han, G.; Qiao, Z.; Li, Y.; Yang, Z.; Wang, C.; Zhang, Y.; Liu, L.; Wang, B. RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 877011. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Zhang, F.; Zhang, G.; Jiang, X.; Yu, H.; Hou, B. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Mei, C.; Liang, S.; Yu, Y.T.; Lu, K.; Wu, Z.; Wang, X.F.; Zhang, D.P. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 2015, 88, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Kryazhimskiy, S.; Plotkin, J.B. The Population Genetics of dN/dS. PLoS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.-E.; Hou, P.; Xiao, F.; Liu, Y. Overexpressing a novel RING-H2 finger protein gene, OsRHP1, enhances drought and salt tolerance in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 357–365. [Google Scholar] [CrossRef]
- Liu, N.; Shang, W.; Li, C.; Jia, L.; Wang, X.; Xing, G.; Zheng, W. Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 2018, 8, 170231. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Li, C.; Wang, T.; Zhang, P.; Chen, K. PnLRR-RLK27, a novel leucine-rich repeats receptor-like protein kinase from the Antarctic moss Pohlia nutans, positively regulates salinity and oxidation-stress tolerance. PLoS ONE 2017, 12, e0172869. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, X.; Hu, B.; Wan, Y.; Xie, J. YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 2013, 6, 7. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W.Y. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Wang, W.; Xu, M.; Liu, X.; Tu, J. The Rice Eukaryotic Translation Initiation Factor 3 Subunit e (OselF3e) Influences Organ Size and Pollen Maturation. Front. Plant Sci. 2016, 7, 1399. [Google Scholar]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed]

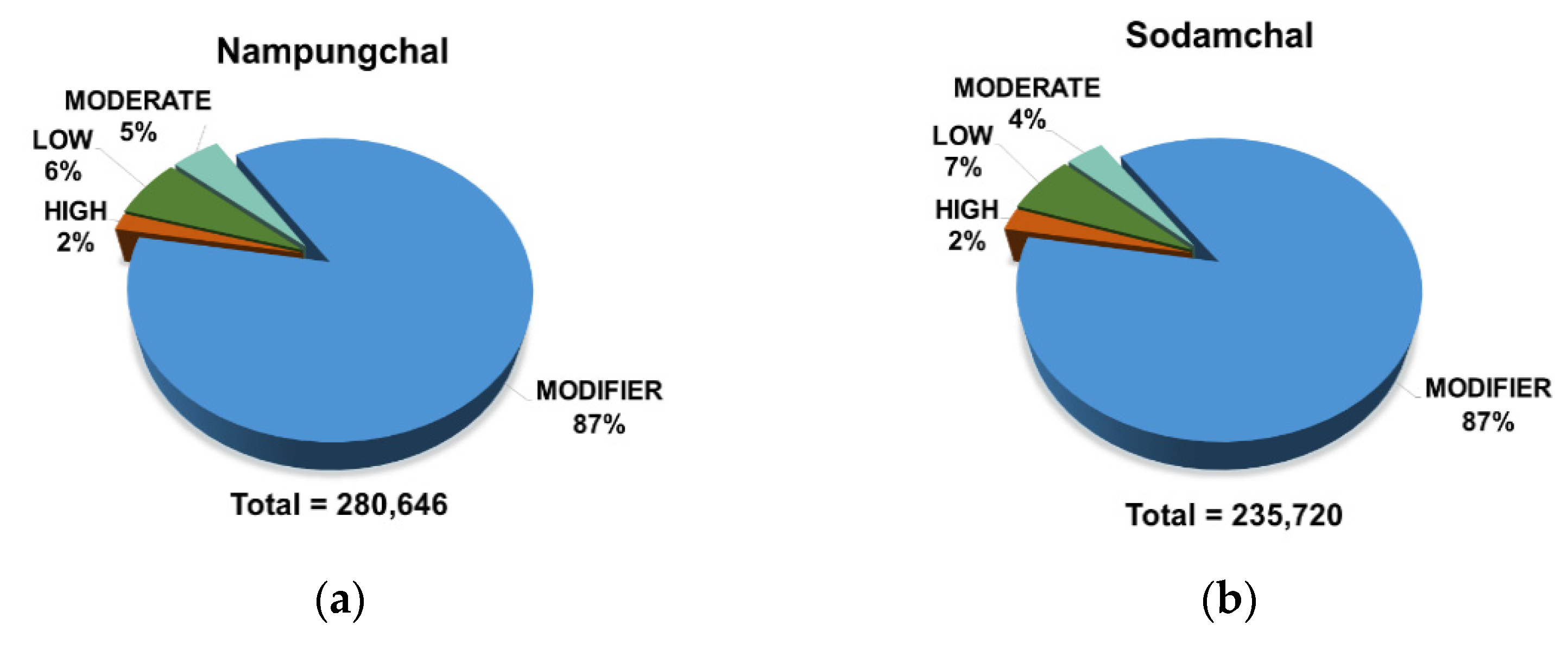
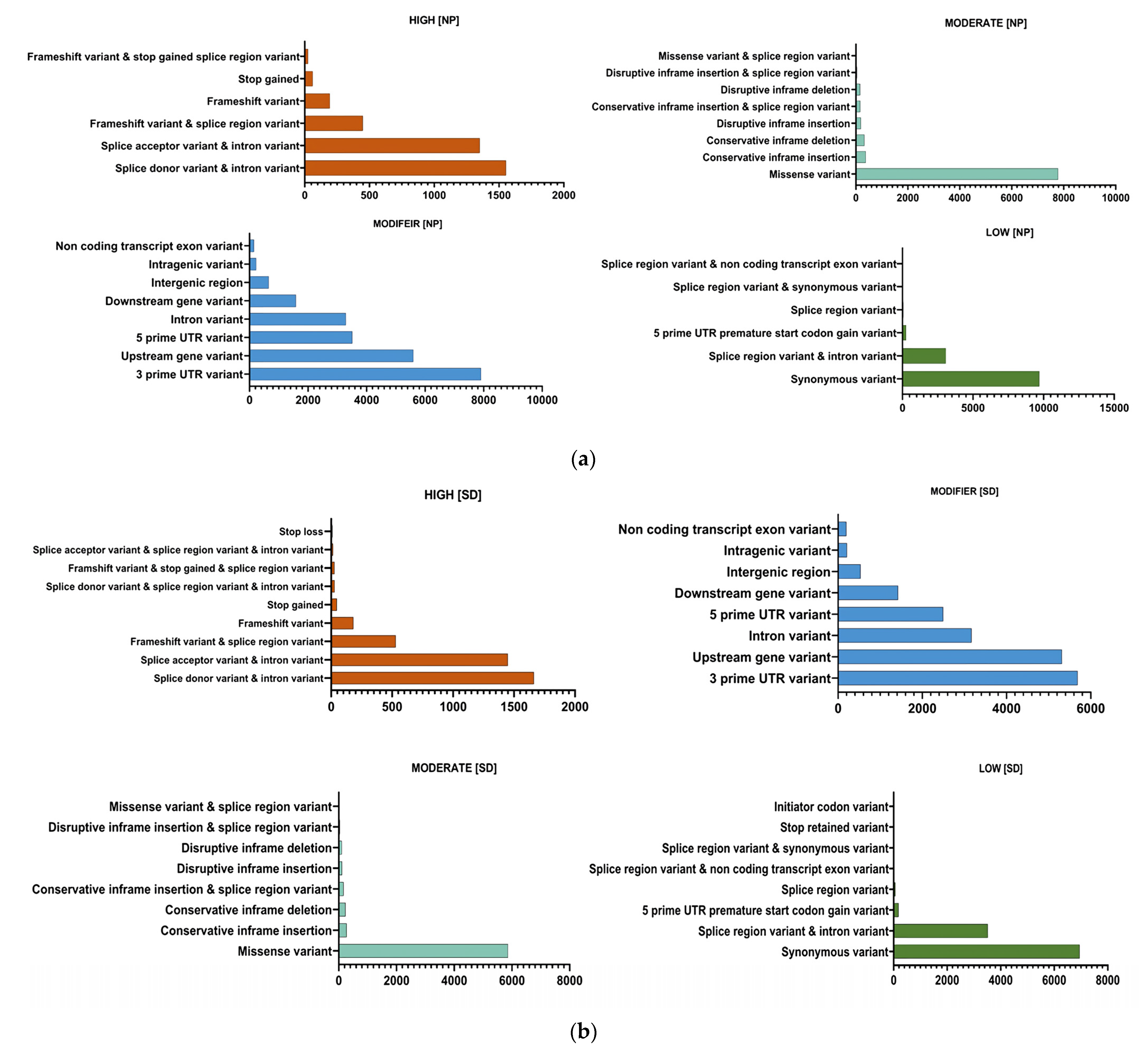
| Type of Genotypes | Type of Region | Type of Effect | Frequency of Variants |
|---|---|---|---|
| Nampungchal | Coding | Synonymous | 13,968 |
| Non-synonymous | 11,527 | ||
| UTR | 16,732 | ||
| Splice region | 4624 | ||
| Intergenic | 2771 | ||
| Intron | 25,534 | ||
| Sodamchal | Coding | Synonymous | 9600 |
| Non-synonymous | 8431 | ||
| UTR | 12,215 | ||
| Splice region | 10,239 | ||
| Intergenic | 2276 | ||
| Intron | 17,008 |
| Gene Symbol | Ka/Ks | High | Low | Moderate | Modifier | Description |
|---|---|---|---|---|---|---|
| LOC8058550 | 10 | 3 | 1 | 5 | 5 | Signal-recognition particle 54 kDa protein, chloroplastic |
| LOC8079195 | 4 | 0 | 1 | 2 | 4 | E3 ubiquitin-protein ligase EL5 |
| LOC8067380 | 4 | 0 | 0 | 2 | 1 | Uncharacterized LOC8067380 |
| LOC8060343 | 4 | 0 | 1 | 6 | 2 | Uncharacterized LOC8060343 |
| LOC110434949 | 4 | 0 | 2 | 2 | 0 | Uncharacterized LOC110434949 |
| LOC110429564 | 4 | 0 | 0 | 2 | 0 | Uncharacterized LOC110429564 |
| LOC8057624 | 2.8 | 1 | 2 | 7 | 19 | Protein HASTY 1 |
| LOC8059007 | 2 | 0 | 0 | 1 | 0 | Eukaryotic translation initiation factor 3 subunit G |
| LOC8066766 | 2 | 0 | 0 | 1 | 3 | Proactivator polypeptide-like 1 |
| LOC8055639 | 2 | 0 | 0 | 2 | 6 | Ethylene-responsive transcription factor ERF104 |
| LOC8066157 | 2 | 0 | 2 | 5 | 0 | Uncharacterized LOC8066157 |
| LOC110432337 | 2 | 0 | 0 | 1 | 1 | Uncharacterized LOC110432337 |
| LOC110432325 | 2 | 0 | 0 | 1 | 1 | Uncharacterized LOC110432325 |
| LOC8057276 | 1.2 | 0 | 2 | 3 | 1 | F-box/FBD/LRR-repeat protein At4g00160 |
| LOC8074840 | 1.171717 | 1 | 50 | 58 | 7 | Disease resistance protein RGA2 |
| LOC8068782 | 1 | 0 | 0 | 1 | 2 | Jasmonate O-methyltransferase |
| LOC8065187 | 1 | 1 | 1 | 1 | 0 | Chaperone protein dnaJ 13 |
| LOC8062760 | 0.8 | 0 | 2 | 2 | 6 | Uncharacterized protein C227.17c |
| LOC8085912 | 0.666667 | 0 | 1 | 1 | 6 | TIP41-like protein |
| LOC8076019 | 0.666667 | 0 | 1 | 1 | 0 | RING-H2 finger protein ATL8 |
| LOC8072250 | 0.666667 | 0 | 1 | 1 | 3 | Histone H2B.1 |
| LOC8066486 | 0.545455 | 0 | 6 | 3 | 12 | Uncharacterized LOC8066486 |
| LOC8072272 | 0.470588 | 1 | 9 | 5 | 9 | Momilactone A synthase |
| LOC8071656 | 0.444444 | 0 | 4 | 2 | 2 | SPX domain-containing protein 1 |
| LOC8054095 | 0.4 | 0 | 2 | 1 | 0 | Nicotianamine aminotransferase A |
| LOC8082038 | 0.285714 | 0 | 3 | 1 | 0 | Probable LRR receptor-like serine/threonine-protein kinase At4g36180 |
| LOC8078184 | 0 | 0 | 1 | 0 | 21 | Momilactone A synthase |
| LOC8074201 | 0 | 0 | 1 | 0 | 0 | Mediator of RNA polymerase II transcription subunit 8 |
| LOC8077219 | 0 | 0 | 1 | 0 | 8 | Chlorophyll a-b binding protein 1, chloroplastic |
| LOC110429806 | 0 | 0 | 1 | 0 | 4 | CBS domain-containing protein CBSX1, chloroplastic-like |
| LOC8066950 | 0 | 0 | 1 | 0 | 2 | Uncharacterized LOC8066950 |
| LOC8063059 | 0 | 0 | 2 | 0 | 15 | Uncharacterized LOC8063059 |
| LOC110433303 | 0 | 0 | 1 | 0 | 0 | Uncharacterized LOC110433303 |
| LOC110429560 | 0 | 1 | 1 | 0 | 0 | Uncharacterized LOC110429560 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, D.; Lee, S.; Choi, S.; Kang, Y.; Kim, C. Identification of SNPs and Candidate Genes Associated with Salt Stress in Two Korean Sorghum Cultivars and Understanding Selection Pressures in the Breeding Process. Agronomy 2022, 12, 2511. https://doi.org/10.3390/agronomy12102511
Jeon D, Lee S, Choi S, Kang Y, Kim C. Identification of SNPs and Candidate Genes Associated with Salt Stress in Two Korean Sorghum Cultivars and Understanding Selection Pressures in the Breeding Process. Agronomy. 2022; 12(10):2511. https://doi.org/10.3390/agronomy12102511
Chicago/Turabian StyleJeon, Donghyun, Solji Lee, Sehyun Choi, Yuna Kang, and Changsoo Kim. 2022. "Identification of SNPs and Candidate Genes Associated with Salt Stress in Two Korean Sorghum Cultivars and Understanding Selection Pressures in the Breeding Process" Agronomy 12, no. 10: 2511. https://doi.org/10.3390/agronomy12102511
APA StyleJeon, D., Lee, S., Choi, S., Kang, Y., & Kim, C. (2022). Identification of SNPs and Candidate Genes Associated with Salt Stress in Two Korean Sorghum Cultivars and Understanding Selection Pressures in the Breeding Process. Agronomy, 12(10), 2511. https://doi.org/10.3390/agronomy12102511









