Abstract
Although winter cover crops (WCCs) have demonstrated positive effects on soil properties, relatively little is known about the responses of the soil and plant microbiomes to the introduction of WCCs and their associated management. Our objective was to evaluate the effects of WCC suppression methods on the rhizosphere microbiome of oats under field conditions. Rhizospheric soil was extracted to quantify the abundances of amoA gene of ammonia-oxidizing bacteria and archaea, and nitrite reductase genes (nirK and nirS), and to determine potential nitrification activity. The bacterial 16S rRNA V4 region and fungal ITS regions were sequenced with the Illumina MiSeq system. Overall, our results indicated that the composition of the bacterial and fungal communities of the rhizosphere were sensitive to the WCC suppression methods. Some bacterial genera, including fungal antagonists and chitin degraders, and two fungi associated with plant potential pathogens, were favored by both suppression methods, yet both methods negatively affected other genera associated with plant growth promotion characteristics. Our work contributes to a more complete understanding of the interactions between WCC management practices, soil properties, and microbial communities in the rhizosphere, which is essential for choosing management strategies that maintain soil health and promote environmental sustainability.
Keywords:
oats; glyphosate; roller-crimper; rhizospheric soil; fungi; bacteria; nitrifiers; denitrifiers 1. Introduction
Crop rotations deploying no-till (NT) systems emerged in response to soil degradation problems caused by agricultural intensification [1]. Winter cover crops (WCCs) are introduced to complement the effects of NT during the fallow season due to the multiple ecosystem services they can provide, such as accumulation of soil organic matter, water quality regulation, suppression of weed growth, pest control, enhancement of soil microbial abundance, activity, and diversity [2,3,4,5].
WCC growth is terminated by either mechanical (e.g., rolling) or chemical methods before the planting of the main crop. Rolling with a roller-crimper implies rolling over the cover crop and crushing the plant stems at a specific growth stage near maturity, preventing regrowth, and depositing the residue uniformly on the soil surface [6]. Rolling replaces the use of herbicides and, consequently, decreases the environmental impact compared to traditional chemical suppression [7]. The most widely used suppression method, chemical control with glyphosate (N-[phosphonomethyl]glycine) is deployed annually on about 1.4 billion hectares worldwide [8]. In recent years, numerous environmental and public health problems have been linked to the widespread use of glyphosate [9].
These suppression methods can affect the plant metabolism, resulting in variations in root exudation, modifying interactions between plants and microorganisms. Due to its systemic action, glyphosate enters the plants through the cuticles of leaves and rapidly translocate to the rest of the plant tissues [10]. Residues of glyphosate that accumulate in root tissues can ultimately end up in the rhizosphere [11,12]. In recent greenhouse experiment, Allegrini et al. [13] observed a greater abundance of Betaproteobacteria and the genus Mesorhizobium in the rhizosphere of oats (Avena sativa L.) treated with glyphosate, while mechanical cutting favored the Verrucomicrobia phylum and the Gaiella genus. In turn, Schlatter et al. [14] reported that glyphosate has little impact on fungal diversity in wheat (Triticum aestivum L.) grown under greenhouse conditions, however, glyphosate may change interactions among fungi competing for senescent roots. More recently, Lupwayi et al. [15] reported that 7 years of glyphosate applications had little effect on bacterial communities in the rhizosphere of durum wheat (Triticum turgidum var. durum Desf.) cultivated in field plots. In their study, the herbicide treatment reduced the relative abundance of Alphaproteobacteria and the Opitutus genus (Verrucomicrobia) [15]. On the other hand, it has been reported that mechanical suppression increases the release of organic compounds in the rhizodeposits, probably as a mechanism of tolerance to defoliation stress [16]. Guo et al. [17] found that annual clipping of forbs (Ambrosia trifida, Solanum carolinense, and Euphorbia dentate) and grasses (Tridens flavus, Sporobolus compositus, and Sorghum halepense) increased the abundance of genes associated with denitrification, with a consequent decrease of soil nitrate (NO3−) content. Furthermore, the relative abundance of some important microbial taxa, including Actinobacteria, Bacteroidetes, Zygomycota, and Ascomycota, were significantly reduced [17].
Various studies have shown that WCCs influence N losses through nitrous oxide (N2O) emissions or nitrate leaching from the soil [18,19], but there have only been a few reported effects of WCC suppression methods on the microorganisms involved in the biogeochemical cycle of soil N [13,20,21]. In a greenhouse study, Allegrini et al. [13] observed that the abundance of ammonia-oxidizing bacteria (AOB) and archaea (AOA) decreased in the rhizosphere of oats suppressed with glyphosate compared to mechanical suppression (plant tissue cut resembling mowing). Recently, Allegrini et al. [20] also reported lower abundance of archaeal amoA in the the rhizosphere of glyphosate-desiccated oats compared to mowed plants. After one growing season of a WCC mixture in a field setting, Romdhane et al., [21] evaluated the effect of chemical vs. mechanical suppression on microbial diversity and abundances, with special focus on N-cycling guilds in bulk soil. The authors found that CC management affected the abundance of denitrifiers in soil, while no effect was observed on the total bacterial abundance and soil nutrients when cover crops were terminated using rolling and glyphosate termination.
Therefore, based on previous literature, we hypothesized the method of suppression of WCC affects the plant root environment, resulting in changes in the rhizospheric microbiome of senescent plants in the field. Our goal was to evaluate the rolling (mechanical with a roller crimper) and chemical (with glyphosate) WCC suppression methods on the rhizosphere microbial community of oats under field conditions using Illumina sequencing of bacterial 16S rRNA gene and fungal ITS amplicons, quantifying amoA-, nirK-, and nirS-carrying microbial groups, and determining potential nitrification activity in the rhizosphere and ancillary properties of the bulk soil. Results from this integrated approach will provide critical information to identify WCC management strategies that are more sustainable.
2. Materials and Methods
2.1. Study Site and Experimental Design
The research trial was established in 2018 at the Napostá Experimental Field (Universidad Nacional del Sur and MDA-PBA agreement, Figure 1), located in Bahía Blanca, Argentina (38°25′39″ S, 62°17′41″ W). The mean annual rainfall was 654 mm for the average 1959–2014 period, with two-thirds of the rainfall concentrated in the autumn and spring. There is a dry season in late winter and a semi-dry season in mid-summer (January and February). In this study, the precipitation values for 2018 and 2019 were 580 and 506 mm, respectively. The dominant soil is a Petrocalcic Paleustoll (Ap-A2-AC-2Ck-3Ckm) according to the USDA Soil Taxonomy classification [22]. Before introducing WCC, these plots had an average topsoil pH of 6.7 (1:2.5 soil:water), 4.24% soil organic matter determined by dry combustion with an automatic analyzer (LECO, St. Joseph, MI, USA), 8 mg kg−1 available P [23], 14.4 mg kg−1 of NH4+, and 10.6 mg kg−1 of NO3− as available N forms.
Figure 1.
Location of the experimental site in Bahía Blanca, Buenos Aires Province, Argentina.
The study was conducted for two consecutive years, in 2018–2019. An area of 15 × 15 m was delimited in soil with no WCC history. Sixteen plots (2.25 × 1.56 m) were established and the suppression methods (R: rolling; DQ, chemical suppression; SS, no suppression) were arranged in a randomized complete block design with four replicates and two plots per block of each treatment. The plots were manually sowed in rows spaced 15.6 cm apart with oats (Avena sativa L. var. Cristal INTA), with a planting density of 250 plants m−2 on 3 June 2018 and on 13 May 2019. Urea nitrogen (N) fertilizer (46% N) was surface applied in all plots at planting (40 kg N ha−1).
Oat growth was suppressed at the Z3.1 stage [24], which corresponded to 144 and 164 days after planting in 2018 and 2019, respectively. Suppression occurred either mechanically by rolling (R) the WCC with a roller-crimper, or chemically (DQ), with a commercial formulation of glyphosate (N-[phosphonomethyl]glycine as active ingredient, a.i.) using a knapsack sprayer with an extendible double nozzle with flat fan nozzle. The ESKOBA FULL II (Red Surcos S. A., 662 g a.i. L−1, monopotassium salt) and CREDIT FULL (Nufarm S. A., 700 g a.i. L−1, mixture of salts) commercial formulations of glyphosate were used in 2018 and 2019, respectively. The working solution was prepared immediately before application, by dissolving 15 mL of the herbicide in 1 L of distilled water following the recommendations on the label for applications with knapsack equipment. A polyethylene shield was used to avoid spray drift to adjacent plots. The roller-crimper was constructed following the design of Ashford and Reeves [25], using a drum roller 0.5 m wide and weighing 120 kg with straight blunt metal blades (80 mm height), which crushed and crimped cover crop stems without cutting them (Figure 2).

Figure 2.
Roller-crimper.
2.2. Soil Physicochemical Analysis
Bulk soil samples were collected from each experimental unit at a depth of 0–10 cm using a manual auger, both before suppression (SS) as well as two weeks after treatments were applied (R and DQ). Soil samples were then air dried, sieved (<2 mm), and sent to the Laboratory of Analytical Services of Soils, Plants, and Environment (CERZOS-CONICET, Universidad Nacional del Sur, Bahia Blanca, Argentina) to determine the contents of available N forms (NH4+ and NO3−) by semi-micro Kjeldahl methodology, and available phosphorus (Pa) by Bray I [23]. The gravimetric water content (H, % (w/w)) was determined after oven-drying 10 g soil (100 °C, 48 h) per sample.
2.3. Rhizospheric Soil Sampling
Oat plants with their intact root systems and attached soil were randomly collected from each experimental unit by careful excavation (40 cm each side) before (SS) the date of application of suppression methods and 13 days after suppression (R and DQ). The roots were carefully shaken to discard bulk soil, while the soil attached to the roots was collected using a sterile brush [26]. The rhizospheric soil obtained was kept in sterile plastic bags at −80 °C for DNA extraction and molecular analysis, and a subsample was stored at 4 °C for physiological analyses.
2.4. Determination of Potential Nitrification Activity (PNA) and Aerobic Heterotrophic Bacteria
The potential nitrification activity assay was performed as described by Hart et al. [27], with slight modifications described by Allegrini et al. [20]. The PNA was calculated with the formula proposed by Drury et al. [28], reporting results in µg N-NO2− g−1 dw soil h−1.
Culturable aerobic heterotrophic bacteria (AHB) were counted by the plate count method [29]. One gram of rhizospheric soil was suspended in 9 mL sterile 0.85% NaCl containing glass breads (0.2 mm). Soil suspension was shaken for 30 min at 180 rpm and 25 °C to disperse bacteria, serially decimal diluted in 9 mL sterile saline and 100 μL of the 10−4 dilution were plated in triplicate on 0.1% Tryptic Soy Agar (TSA, Laboratorios Britania, Buenos Aires, Argentina). Plates were incubated at 25 °C for 6 days. Plates containing between 30 and 300 colonies were counted. The number of cultivable AHB obtained was expressed as log10 colony forming units (CFU) g−1 soil.
2.5. Rhizospheric Soil DNA Extraction and Quantification
Rhizospheric soil DNA extraction was performed using DNeasy PowerSoil Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The extracted DNA was quantitated with QuantiFluor® dsDNA kit in a Quantus fluorometer (Promega®, Madison, WI, USA). The quality of the DNA was checked by agarose gel electrophoresis and measuring the 260:230 and 260:280 nm absorbance ratios (DS-11 FX spectrophotometer, DeNovix Inc., Wilmington, DE, USA).
2.5.1. Quantification by Real-Time PCR of Nitrifying and Denitrifying Prokaryotes and Total Bacteria and Archaea
Quantitative PCR was used to quantitate the abundance of 16S rRNA gene of Bacteria and Archaea, amoA gene of AOA and AOB, and nitrite reductase genes (nirS and nirK), as indicators of the abundances of these microbial groups. The primers used for molecular analyses are indicated in Supplementary Table S1 [30,31,32,33,34,35,36]. Copy numbers were not converted to cell numbers to avoid introducing biases. The PCR mixture and amplification programs for 16S rRNA gene of Bacteria and Archaea, amoA of AOA and AOB were performed according to Allegrini et al. [13].
The PCR reaction mixture for amplifying the nirS gene contained 7.5 μL PCR iTaq Universal SYBR Green Supermix (2×; Bio-Rad Laboratories, Hercules, CA, USA), 0.75 μL of each primer (10 μM stocks, Invitrogen, Waltham, MA, USA), 5 μL sterilized bi-distilled water, and 1 μL of template DNA (∼1–10 ng μL−1). The following amplification program was used: preincubation at 95 °C for 5 min followed by 40 cycles at 95 °C for 15 s, 60 °C for 20 s, 72 °C for 45 s (amplification), and a melting curve analysis (65–95 °C).
The PCR reaction mixture for amplifying the nirK gene contained 7.5 μL of PCR iTaq Universal SYBR Green Supermix (2×; Bio-Rad Laboratories, Hercules, CA, USA), 0.45 μL of each primer (10 μM stocks, Invitrogen, Waltham, MA, USA), 5.6 μL sterilized bi-distilled water, and 1 μL of template DNA (∼1–10 ng μL−1). The following amplification program was used: preincubation at 95 °C for 5 min followed by 40 cycles at 95 °C for 20 s, 55 °C for 30 s, 72 °C for 45 s (amplification), and a melting curve analysis (65–95 °C).
Standard curves were prepared with decimal dilutions of the respective genes amplified by PCR and cloned in plasmids to serve as references for copy numbers calculations. Equations and efficiency of qPCR standard curves for AOB, AOA, nitrite reductase genes (nirS and nirK), Bacteria, and Archaea are indicated in Supplementary Table S2.
All amplifications were conducted in an ABI 7500 Real−Time System and data processed with 7500 Software v2.0.3 (Applied Biosystems, Foster City, CA, USA).
2.5.2. Metabarcoding of ITS and Bacterial 16S rRNA Gene
The bacterial V4 region of 16S rRNA gene and the fungal internal transcribed spacer (ITS) region were sequenced with Illumina MiSeq (Illumina, Inc., San Diego, CA, USA) using paired-end sequencing, resulting in reads 250 nt in length. The primer sets used for amplification were 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSGGGTATCTAAT-3′) for the bacterial 16S rRNA gene and 7F (5′-GTGARTCATCGAATCTTTG-3′) and 4R (5′-TCCTCCGCTTATTGATATGC) for the fungal ITS region, using the Fluidigm™ protocol at the DNA Service Laboratory, Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign, USA. FASTQ files were generated and demultiplexed by the sequencing service with the bcl2fastq v2.20 Conversion Software (Illumina, San Diego, CA, USA).
2.5.3. Bioinformatics Analysis
The resulting FASTQ files were processed in QIIME2 [37] using the recommended pipelines for paired-end reads based on DADA2 denoising algorithm. Briefly, primers were removed using p-trim-left argument within dada2 denoised-paired script. The trimming argument within dada2 denoised-paired script was set on 283 bp for forward reads and 251 bp for reverse reads for Bacteria and 288 bp for forward reads and 266 bp for reverse reads for Fungi (median Q values > 27 reported by Interactive Quality Plot tool of QIIME2), with a default value of 2 for the expected number of errors. The amplicon sequence variants (ASVs) obtained after removal of primers, trimming, filtering, denoising, and chimera removal steps, were classified with the Ribosomal Database Project (RDP) naïve Bayesian classifier version 2.11 [38].
The ASVs table was rarefied (rarefy function) according to the sample containing the lowest number of bacterial (IVSSR_2; 22,538 reads) and fungi (IVSSR_2; 26,785 reads) reads used in vegan package 2.5–6 [39] and the rarefaction curves were obtained (rarecurve function). Alpha-diversity metrics including Shannon diversity index (H’), reciprocal of Simpson index (1/λ), observed richness (S’ = number of ASVs), Chao-1 index (estimated richness), and Shannon equitability index (EH) were calculated using rarefied data of ASVs table. To calculate beta-diversity metrics for each taxon, the representative sequences of the resulting ASV were aligned using MAFFT v. 7 [40] and the aligned sequences were analyzed with phangorn v2.5.5 package [41] to calculate the distance matrix and a neighbor-joining tree. This phylogenetic tree and the ASV table were loaded in GUniFrac package v1.1 [42] to obtain the generalized UniFrac distances. All analyses were run with R Statistical Software v3.6.1 [43].
2.6. Statistical Analysis
Linear mixed models were fit to each set of soil and microbial parameters, e.g., available phosphorus (Pa), ammonium (NH4+), nitrate (NO3−), qPCR (log10 copy number μg−1 DNA), CFU, PNA, and alpha-diversity metrics using PROC GLIMMIX of SAS software version 9.4 [44] (SAS Institute, Cary, NC, USA). The suppression method was considered a fixed effect and year, block, and their interactions with the treatment were considered random terms in the analyses of variance. Water content was used as a covariate in most of these analyses except for Pa, NH4+ and NO3− whose determination include a moisture correction. When appropriate, least-square means were separated using the lines option of the lsmeans statement, setting the probability of Type I error at (α) 0.05.
For multivariate analysis of beta diversity, the UniFrac distances were used as input in vegan package v2.5–6 [39] through non-metric multidimensional scaling (NMDS, metaMDS function). Additionally, amplicon sequencing data were analyzed using the compositional approach [45,46]. Starting from the initial table of ASVs resulting from the RDP classification platform, which contained 15,868 ASVs of Bacteria and 2193 ASVs of Fungi, a series of ASV filtering and selection steps were carried out. First, ASV reads belonging to the same genera were added, resulting in 1079 and 369 total genera of Bacteria and Fungi, respectively. This set was further reduced as only genera with average relative abundances >0.1% were kept [45], rendering 189 and 109 genera of Bacteria and Fungi, respectively. These genera with their reads were then subjected to further analysis in the JMP® predictor screening platform as a preliminary identification tstep to select the genera responding to treatments, through a bootstrap forest partitioning method [44,47,48]. Overall, 27 Bacteria and 27 Fungi were selected, each contributing at least 1% to the variability of the model algorithms. The resulting ASV table was processed with the package zCompositions [49] in R [43], in order to replace zero values, prior to the centered log-ratio transformation [50], as recommended for compositional data [46].
Principal Component Analyses (PCA) were used as a data reduction technique on the top contributing genera for each taxon to further remove redundancy. FACTOR procedure in SAS software version 9.4 [44] with the default specification of priors = 1 summarized the abundances of each genus into a set of uncorrelated composited variables, or Principal Components (PCs). PCs with eigenvalues ≥1 that also explained at least 5% of the variability in the data set were used as independent variables for further analysis. Genera with a significant correlation with each PC (PC loading value > |0.45|) were considered microbial indicators and used in the description of the PC [48]. Linear models were fit to each PC using the GLIMMIX procedure of SAS, with the suppression method as fixed effects, while the year, block, and their interactions with the treatment were considered random terms in the analyses of variance. Least-square means were separated using the lines option of the lsmeans statement, with the probability of Type I error (α) set at 0.05.
A nonparametric multivariate analysis of variance (NPMANOVA with adonis function in the vegan package; [51]) was deployed using a generalized UniFrac distance and 1000 permutations to compare distances between suppression methods. When appropriate pairwise PERMANOVA were conducted with false discovery rate (FDR) p-value adjustment method in RVAideMemoire package v 0.9–80 [52] (pairwise.perm.manova function without corrections, 1000 permutations).
Software SigmaPlot version 10.0 was used to plot the statistically significant relationships detected between PCs and treatments for genera whose loading was >|0.45|.
Pearson correlation analysis was deployed with the CORR procedure of SAS, to evaluate the relationships among the statistically indicative PCs extracted for Bacteria and Fungi and the abundance of genes linked to the nitrogen cycle (AOA, AOB, nirS, and nirK), with ancillary soil properties.
3. Results
3.1. Bulk Soil Chemical Properties
Table 1 shows the mean treatment values and their standard errors, together with the results of the mean separation procedures for the soil nutrients in response to suppression methods. The contents of NH4+, NO3− and available P did not differ statistically between the suppression methods (Table 1).

Table 1.
Treatment mean values (mean), standard errors of the mean (SEM), number of observations (n), as well as probability values (p-value), and degrees of freedom (df) associated with the ANOVA of available phosphorus (Pa), ammonium (NH4+) and nitrate (NO3−).
3.2. Potential Nitrification Activity
The results of the analysis of variance of the PNA in the rhizospheric soil indicated no statistically significant differences (p = 0.598) between DQ, R, and SS (4.42, 3.75 and 3.47 µg N-NO2− g−1 dw soil h−1, respectively).
3.3. Aerobic Heterotrophic Bacteria Counts and Quantitative PCR of Indicator Genes
In this study, the number of cultivable AHB in the rhizospheric soil of oats was similar (p = 0.598) among DQ, R, and SS (4.42, 3.75 and 3.74 log10 CFU g−1 soil, respectively).
Table 2 summarizes the estimated treatment means, standard errors of the mean (SEM), and the probability values (p-values) of the abundances of indicator genes. Statistical analysis for Bacteria, Archaea, AOA, AOB, nirK, and nirS showed no significant effect of the suppression method (Table 2).

Table 2.
Treatment mean values (mean), standard errors of the mean (SEM), number of observations (n), as well as probability values (p-value), and degrees of freedom (df) associated with the ANOVA of copy number of indicator genes (log10 transformed) from different microbial groups in the rhizosphere of oats.
3.4. Metagenomics Sequencing of DNA from the Rhizosphere of Oats
Metabarcoding analysis comprised 1,663,735 bacterial and 1,809,975 fungal sequences. After filtering, denoising, and removing chimeric sequences, the bacterial sequences were clustered into 15,868 ASVs, whereas the fungal sequences were clustered into 2193 ASVs. Rarefaction curves are included as Supplementary Figures S1 and S2 for Bacteria and Fungi, respectively.
3.4.1. Alpha and Beta Diversity
The alpha-diversity measurements of estimated richness (Chao-1), observed richness (S’), reciprocal of Simpson index (1/λ), Shannon index (H’), and Shannon equitability index (EH) for Bacteria and Fungi revealed no statistical effect of the suppression method (Table 3).

Table 3.
Treatment mean values (mean), standard errors of the mean (SEM), number of observations (n), as well as probability values (p-value), and degrees of freedom (df) associated with the ANOVA of the alpha-diversity metrics of Chao-1 index (estimated richness); S’, observed richness (observed ASVs); 1/λ, reciprocal of Simpson index; H’, Shannon index; and EH, Shannon equitability index.
No significant effect of the suppression method was detected in multivariate statistical analysis through PERMANOVA to investigate the beta diversity of Bacteria and Fungi (F. model = 1.38, p = 0.186 and F. model = 0.994, p = 0.407, respectively). These results were visualized in NMDS analyses based on generalized UniFrac distance, confirming no separation between suppression methods (Figure S3a,b).
3.4.2. The Composition of Communities of Bacteria and Fungi in the Oats Rhizosphere
Bacteria
The PCA on top bacterial indicators resulted in a group of seven uncorrelated PCs with eigenvalue >1 that, together, explained 71% of the variability in the data set (Table S3). PC1 presented positive loadings for Curvibacter, Duganella, Erythrobacter, Flavilitoribacter, Longimicrobium, Massilia, Novosphingobium, Parviterribacter, and Phenylobacterium, and negative loadings for Brevifollis, Chitinispirillum, Lacunisphaera, and Nocardioides. PC2 contained a contrast between two groups of bacteria, those with positive loadings: Abditibacterium, Massilia, and Sediminibacterium; and those with negative loadings: Azoarcus, Gp17, Litorilinea, Parviterribacter, and Thermanaerothrix. PC3 had positive loadings for Geminisphaera, Gemmata, and Methylobacterium, and negative loadings for Stella. The PC4 eigenvector had positive loadings for Azoarcus while including a negative loading for Brevifollis. PC5 showed a positive loading for Longimicrobium and negative loadings for Kineosporia and Thermanaerothrix. The PC6 eigenvectors included positive loadings for Brevundimonas. Notably, PC7 no had genus with PC loadings > |0.45|.
Linear mixed model ANOVAs were deployed to test the effect of the suppression method on the bacterial communities of the oats rhizosphere, using the seven PCs as independent variables. The suppression method effects were significant for PC1 (p = 0.027) and PC3 (p = 0.019), while no statistically significant effects were detected for PC2, PC4, PC5, PC6, or PC7. Figure 3a shows a graph of the PC1 means for each suppression method with their respective standard error bars, together with the means separation results (Table 4). The contribution of each bacterial genus to these results is shown in Figure 3b. Compared to the SS treatment, the group of bacteria with positive loadings significantly decreased with DQ and R treatment, while the opposite behavior was observed for those responsive genera with negative loadings (Figure 3b). Figure 4a shows a graph of the PC3 means for each suppression method with their respective standard error bars, accompanying the means separation results (Table 4). The contribution of each bacterial genus to these results is shown in Figure 4b. Compared to R, the group of bacteria with positive loadings significantly decreased in the rhizosphere of chemically terminated oats (DQ), and the opposite response was observed for those indicators with negative loadings (Figure 4b).
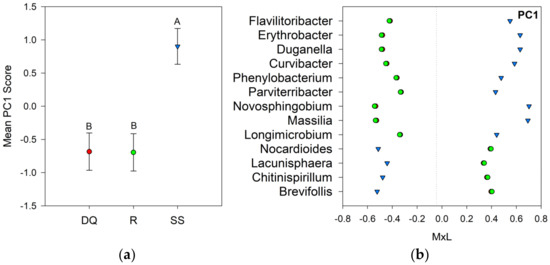
Figure 3.
Visual representation of the combined results of the principal component analyses (PCAs) and their mean separation procedure showing the genera with loadings >|0.45| according to Supplementary Table S3 for PC1: (a) Mean values of the PC1 scores for each suppression method with their standard errors (as error bars). Different capital letters indicate significant differences between suppression methods (p < 0.05); (b) Contribution of each indicator genera to the PC1 mean value (Table 4) for each suppression method (DQ, red circles: chemical suppression; R, green circles: rolling, and SS, blue triangles: no suppression) multiplied by the loading of the specific genera within the PC (Table S3), named M×L.

Table 4.
Mean values and probability values (p-values) associated with the analysis of variance (ANOVA) results for the effects of the suppression method on each group of principal components (PCs) calculated for Bacteria datasets comprised of indicator ASVs.
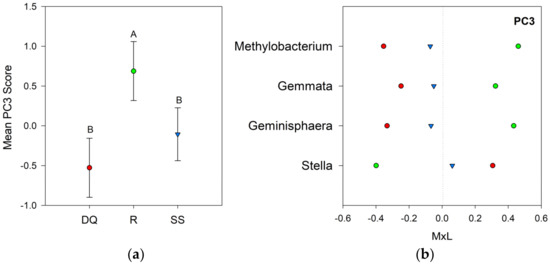
Figure 4.
Visual representation of the combined results of the principal component analyses (PCAs) and their mean separation procedure showing the genera with loadings >|0.45| according to Supplementary Table S3 for PC3: (a) Mean values of the PC3 scores for each suppression method with their standard errors (as error bars). Different capital letters indicate significant differences between suppression methods (p < 0.05); (b) Contribution of each indicator genera to the PC3 mean value (Table 4) for each suppression method (DQ, red circles: chemical suppression; R, green circles: rolling, and SS, blue triangles: no suppression) multiplied by the loading of the specific genera within the PC (Table S3), named M×L.
Fungi
The PCA on top fungal genera returned a set of eight uncorrelated PCs with eigenvalues >1 that, together, explained 73% of the variability in the data set (Table S4). PC1 presented positive loadings for Alternaria, Davidiella, and Podospora, and negative loadings for Dokmaia, Haematonectria, Phialophora, and Spizellomyces. PC2 was a contrast between two groups of microbes, those with positive loadings: Auricularia, Corynascus, Phialocephala, Pseudallescheria, and Exophiala; while the group with negative loadings included Edenia, Elaphocordyceps, Myrmecridium, and Phoma. PC3 had positive loadings for Dokmaia, Lecythophora, and Rhizopycnis, and negative loadings for Corynascus and Mycena. The PC4 eigenvector included positive loadings for Microdochium while including a negative loading for Phaeosphaeria and Powellomyces. PC5 showed a positive loading for Phialocephala and negative loadings for Edenia and Sclerostagonospora. PC6 comprised two genera with positive loadings: Exophiala and Tricladium. The PC7 and PC8 eigenvectors included negative loadings for Myrmecridium and Periconia, respectively.
Linear mixed model ANOVAs assessing the effects of suppression methods on each of the PCs representing the fungal community (Table 5) indicate a statistically significant effect of suppression method only for PC1 (p < 0.001). Figure 5a shows a graph of the fungal PC1 means for each suppression method with their respective standard error bars, accompanying the means comparison results (Table 5). The contribution of each indicator to these results is shown in Figure 5b. Thus, PC1 means statistically decreased within the R and DQ compared to SS. Therefore, fungal indicators with positive loadings on PC1 increased in abundance under SS, while those with negative loadings increased with R and DQ (Figure 5b).

Table 5.
Mean values and probability values (p-values) associated with the analysis of variance (ANOVA) results for the effects of the suppression method on each group of principal components (PCs) calculated for fungi datasets comprised of indicator ASVs.
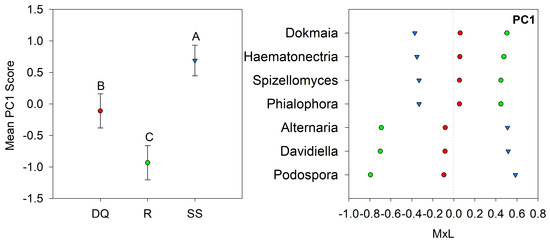
Figure 5.
Visual representation of the combined results of the principal component analyses (PCAs) and their mean separation procedure showing the genera with loadings >|0.45| according to Supplementary Table S4 for PC1: (a) Mean values of the PC1 scores for each suppression method with their standard errors (as error bars). Different capital letters indicate significant differences between suppression method (p < 0.05); (b) Contribution of each indicator genera to the PC1 mean value (Table 5) for each suppression method (DQ, red circles: chemical suppression; R, green circles: rolling, and SS, blue triangles: no suppression) multiplied by the loading of the specific genera within the PC (Table S4), named M×L.
3.5. Pearson’s Correlation among Variables
Supplementary Table S5 shows the matrix of Pearson’s correlation coefficients among NO3−, NH4+, and Pa and the PCs responsive to treatments for each taxon; Bacteria PC1 and PC3 (BPC1 and BPC3), and Fungi PC1 (FPC1). We found one strong (|0.6–0.8|) and four moderate (|0.4–0.6|) associations of statistical significance (p < 0.05). Thus, Bacterial PC1 showed a moderate positive correlation with FPC1 scores (r = 0.53, p = 0.004) and negative correlation with NO3− (r = −0.41, p = 0.047). BPC3 did not have any significant correlation. FPC1 was moderately and negatively correlated with NH4+ (r = −0.41, p = 0.045) and NO3− (r = −0.40, p = 0.012). Among the soil properties examined, NH4+ showed a moderate positive correlation with NO3− (r = 0.68, p = 0.0003).
Supplementary Table S6 shows the matrix of Pearson’s correlation coefficients among NO3−, NH4+, and Pa and the abundance of nitrogen cycle genes (AOA, AOB, nirS, and nirK). Four of the eleven statistically significant correlation coefficients found in the analyses (bolded, Table S6) fell within the “moderate” (|0.4–0.6|) association range, whereas three and six correlations were within the “strong” (|0.6–0.8|) and “very strong” (>|0.8|) association ranges, respectively. AOB was very strongly and positively correlated with the nirK gene (r = 0.91, p < 0.0001) and NH4+ (r = 0.86, p < 0.0001), and moderately associated with NO3− (r = 0.41, p = 0.046), while showing a negative strong correlation to the nirS gene (r = −0.75, p <0.0001). AOA was moderately and positively correlated with NO3− (r = 0.50, p = 0.013). The NirS gene showed a strong negative correlation with the nirK gene (r = −0.84, p < 0.0001), NH4+ (r = −0.78, p < 0.0001), and a moderate negative association to NO3− (r = −0.51, p = 0.011). The NirK gene displayed very strong and moderate positive correlations with NH4+ (r = 0.90, p < 0.0001) and NO3− (r = 0.53, p = 0.007), respectively.
4. Discussion
4.1. The Abundance of Different Microbial Groups
Culturable bacteria represent less than 1% of total bacterial populations, which, however, can very quickly respond to root exudates [53]. In this study, the aerobic heterotrophic bacteria count showed no significant differences among suppression methods. This may be due to the climatic variability, spatial heterogeneity of soil nutrients, soil moisture, and complexity of soil microbial communities that make the detection of treatment effects difficult when working under field conditions [54,55,56]. In contrast, Imparato et al. [57] reported that foliar application of glyphosate doubled culturable bacteria abundance in the rhizosphere of barley (Hordeum vulgare L.) compared to cut or untreated treatments. However, this experiment was carried out in a different grass species, grown for 25 days in a greenhouse, so their results are not strictly comparable with ours. In addition, there are several studies that have shown that the impact of root exudates on the rhizosphere microbiome depends on plant species, genotype, age, and root morphology [58,59,60].
Total bacterial and archaeal abundances were not affected by suppression methods. In agreement with our results, Romdhane et al. [21] did not observe changes in the abundance of bacteria and archaea in soil after CC suppression by different methods, either chemically (glyphosate + 2,4-dichlorophenoxyacetic acid), mechanically (rolling), or naturally by frost (winter-kill) in a field study with mixture CC.
The correlation between genes involved in nitrification and denitrification was significant. Strong negative associations were found among AOB, nirK, and nirS. However, no significant impacts of suppression methods were found on the estimated abundance of nitrifiers and denitrifiers in the oats rhizosphere. The lack of significant differences in the abundance of AOB was consistent with the results obtained by sequencing at the genus level. Here also, Romdhane et al. [21] reported that CC suppression methods did not affect the abundance of AOA and AOB, while the number of copies of nitrite reductase genes (nirK and nirS) decreased in glyphosate-terminated CC compared to rolling and frost treatments. In agreement with our results, Jenkins et al. [61] did not observe any differences in the abundance of the amoA gene of AOB and AOA resulting from treatment with glyphosate in non-glyphosate-resistant (DeKalb DKC65-18) and glyphosate-resistant (DeKalb DKC65-17) corn (Zea mays L.) under reduced tillage. Meanwhile, Zhang et al. [62] observed that the mowing of grasses (Stipa krylovii, Agropyron cristattum, and Cleistogenes squarrosa) did not affect the abundances of amoA, nirS, nirK, and nosZ genes in soil. In contrast, Allegrini et al. [20] observed that the abundance of amoA gene of AOA in the rhizosphere of oats as WCC was higher with mechanical suppression compared to chemical suppression, although this study was done under greenhouse conditions.
Similar to Romdhane et al. [21], we found positive and significant correlations between the abundance of AOB, AOA, and nirK genes and the available N forms (NH4+ and NO3−).
In this study, potential nitrification activity showed no significant differences among the suppression methods. In contrast, Liang et al. [63] reported that flail mowing (cut and chopped) compared to spraying (glyphosate), significantly increased nitrification potential by ~36% in bulk soil, 12 weeks after legume cover crop termination. However, this experiment was carried out in legume WCC over one growing season, so their results are not strictly comparable with ours.
4.2. Bacterial Community Composition
Two groups of bacteria were identified as indicators for suppression method (PC1, Figure 3b). The BPC1 was moderately and negatively associated with soil NO3−. Thus, genera with negative loadings on PC1 were associated positively with soil NO3−, and the opposite occurs for genera with positive loadings on PC1. The group with positive loadings, which responded negatively to chemical suppression and rolling, comprises nine genera: Duganella, Massilia, Erythrobacter, Novosphingobium, Curvibacter, and Phenylobacterium (Proteobacteria), Flavilitoribacter (Bacteroidetes), Longimicrobium (Gemmatimonadetes), and Parviterribacter (Actinobacteria). Some species in the genus Curvibacter are denitrifiers [64] that reduce NO3− to NO2−, and further to gaseous forms of N, which is a metabolism that is widespread among members of the Proteobacteria phylum, and could partially explain the existing correlation between soil NO3− content and PC1.
The genera Duganella and Massilia, representative of the Oxalobacteraceae family, are heterotrophic and non-spore-forming Gram-negative bacteria, commonly found in water, soil, and associated plants [65]. Duganella has been isolated from the rhizosphere of sugarcane (Saccharum officinarum L.) and has plant-growth-promoting capabilities, such as the production of extracellular-polysaccharides [66]. Yin et al. [67] identified Duganella in the rhizosphere of wheat infested with Rhizoctonia solani AG-8, where it could act as a biocontrol agent [68]. Massilia has been found in the rhizosphere of many plant species, such as sugarcane [69], wheat [70], or corn [71]. In addition, this genus exhibited mechanisms of phytopathogen control, including the production of siderophores and extracellular lytic enzymes [72].
The Erythrobacter and Novosphingobium genera belong to the Erythrobacteraceae family, whose members show different application properties as bioremediation of a variety of xenobiotics, production of carotenoids, cytotoxic compounds, among others [73]. Erythrobacter has been isolated from the rhizosphere of Suaeda japonica [74] and Kandelia candel [75]. Bacteria of the genus Erythrobacter presented genes with plant growth promoting traits (as biosynthesis of antibiotics, siderophores production, root colonization, and tolerance to harsh environments) [76]. Novosphingobium was isolated from the rhizosphere soil of grasses as maize [77] and rice (Oryza sativa L.) [78]. This genus also has the potential to act as plant-growth promoting rhizobacteria (PGPR), producing IAA, acetoin, and siderophores.
The genus Phenylobacterium (Alphaaproteobacteria) has been identified in wheat rhizosphere [15]. This bacterium was associated with phenolic compounds degradation in contaminated soil with polycyclic aromatic hydrocarbons (PAHs) [79]. Flavilitoribacter belongs to the order Saprospirales, which has been identified in the rhizosphere of corn and switchgrass (Panicum virgatum L.) [80]. Members of the phylum Gemmatimonadetes have a cosmopolitan distribution in terrestrial systems, although they are generally found in low frequency in soil microbial communities, with relative abundances ranging from 0.2 to 6.5% [81]. Longimicrobium, an oligotrophic bacterium, is the only genus in the family Longimicrobiaceae, and was isolated from soil [82]. Parviterribacter genera are strictly aerobic rods isolated from soil [83]. This genus was favored in contaminated soil with atrazine [84].
The group of four indicator genera favored by the chemical suppression and rolling methods (PC1, Figure 3b) were: Brevifollis and Lacunisphaera (Verrucomicrobia), Chitinispirillum (Fibrobacteres), and Nocardioides (Actinobacteria). Representatives of the Verrucomicrobia phylum are ubiquitous in soil, accounting for 1 to 10% of the bacterial 16S rRNA present in the soil [85]. Verrucomicrobial community structure and abundance are sensitive to changes in soil moisture and fertility, they respond negatively to high soil fertility and low moisture [86,87]. Brevifollis is a Gram-negative, obligately aerobic, chemoorganotrophic bacterium, isolated from an artificial consortium of Chlorella vulgaris (green algae) and bacteria originating from soil [88]. Lacunisphaera has been identified in the corn rhizosphere and showed positive correlations with soil NO3− [89]. The Opitutaceae family, to which Lacunisphaera belongs, exhibited antagonistic activity against soil-borne pathogens in the cotton rhizosphere [90].
Fibrobacteres members are specialists in cellulose degradation, although Chitinispirillum utilizes chitin as the sole growth substrate [91]. This phylum has been identified in the wheat rhizosphere and showed positive correlations with soil NO3− and wheat yield [92]. Nocardioides has been detected in the rhizospheric soil after the continuous cropping of barley [93]. Piutti et al. [94] reported that a strain of the genus Nocardioides, isolated from atrazine-treated bulk- and maize rhizosphere soil, can degrade atrazine. This genus also could act as biocontrol agent [95].
Indicators with a positive response to the rolling method were Gemmata, Geminisphaera, and Methylobacterium (PC3, Figure 4b). The Planctomycetes phylum is ubiquitously distributed in a wide range of aquatic and terrestrial environments and its diversity is sensitive to soil management history [96]. The only representative of this phylum was Gemmata, a genus recently identified in the rhizosphere of corn [97]. Gemmata species have the capacity to carry out heterotrophic nitrification and anaerobic ammonia oxidation (anammox) [98]. Kim et al. [47] reported that this genus decreased in abundance in soil upon introducing cover crops (Secale cereale L. and Vicia villosa Roth.) to corn monoculture. Methylobacterium is a strictly aerobic bacteria belonging to the phylum Proteobacteria, able to grow using compounds containing only one carbon [99]. Members of this genus occupy different habitats, including soil, rhizosphere, water, grains, leaves, nodules, and air [100]. Methylobacterium has been associated with plant growth promotion, biocontrol activity, and bioremediation [101].
Stella is a representative of the phylum Proteobacteria (Alphaproteobacteria) identified as an indicator in the group that responds positively to the DQ and SS methods. It is a Gram-negative bacterium belonging to the Stellacea family, recently proposed by Hördt et al. [102]. Stella was isolated from the soil and associated with organic matter decomposition processes, which improves soil fertility and crop productivity [103].
4.3. Fungal Community Composition
Two groups of fungi were identified as indicators for suppression method (PC1, Figure 5b). The FPC1 was moderately and negatively associated with soil NH4+ and NO3−. Thus, genera with negative loadings on PC1 were positively associated with soil NH4+ and NO3− and the opposite occurs for genera with positive loadings on PC1. The group with negative loadings on PC1, that responded positively to chemical suppression and rolling, comprises 4 genera: Dokmaia, Haematonectria, and Phialophora (Ascomycota), and Spizellomyces (Chytridiomycota).
Representatives of the phylum Ascomycota are diverse and have been detected in different ecosystems such as forests, agricultural lands, and commercial forests [104], and represent the main soil fungal decomposers [105]. Dokmaia and Phialophora were identified in the rhizosphere of Pisum sativum L. [106] and grasses [14,107] and live as saprotrophs in soil, where they usually are non-pathogenic for plants. In agreement with our results, Schlatter et al. [14] found that the application of glyphosate in wheat favored the Phialophora genus in the rhizosphere of this specie. Newsham et al. [108] reported a positive growth response of Vulpia ciliata to inoculation with Phialophora graminicola under controlled conditions. Haematonectria (an anamorph of Fusarium spp.) was found in alfalfa (Medicago sativa L.) rhizosphere and identified as a plant pathogen [109]. Dean et al. [110] reported that Haematonectria was affected by management systems, being more common in conventional soybean (Glycine max L. Merr.) than in organic soybean.
The Chytridiomycota phylum comprises zoosporic fungi that occur in soil as saprophytes growing on organic material [111]. Spizellomyces have been identified in soybean rhizosphere as a plant pathogen infecting soybean during farm cultivation [112]. Schlatter et al. [113] reported that glyphosate, by compromising the plant defense system, promotes the colonization of root pathogens, and can result in a “green bridge” between weeds and crop hosts.
The group of 3 indicator genera that decreased in abundance with chemical suppression and rolling belong to the Ascomycota phylum. Podospora has been associated with N-fertilized soils [47], which could partially explain the correlation among FPC1 and available forms of N (NH4+ and NO3−). Alternaria, Davidiella, and Podospora were identified in wheat rhizosphere and negatively correlated with plant density [114]. Additionally, species of Davidiella and Podospora may reduce wheat yield [114]. The genus Alternaria is widely distributed in soil and organic matter, with a majority of species acting as plant pathogens, saprophytes, and endophytes [115]. A previous study found that Alternaria decreased in abundance in the wheat rhizosphere with glyphosate treatment, indicating that the herbicide can be toxic to some plant pathogens [14]. Podospora, a saprotrophic fungus, has genes potentially involved in lignin degradation and efficient cellulose breakdown [116].
5. Conclusions
The results of this study show that functional genes, aerobic heterotrophic bacteria count, potential nitrification activity, and soil properties were not responsive to the oats suppression methods. Genus-level indicators with high-taxonomic resolution instead showed clear differences between suppression methods. Some bacterial genera, including fungal antagonists and chitin degraders, and two fungi associated with plant potential pathogens, were favored by both suppression methods, yet both methods negatively affected other genera associated with plant growth promotion characteristics.
Therefore, the decaying roots of the suppressed winter cover crop in soil, particularly through chemical desiccation, may act as a “green bridge,” transmitting pathogens to the next crop in the rotation and affecting the ability of plant-growth promoting rhizobacteria to suppress these pathogens. Longer-term studies would be necessary to achieve a better understanding of the effects of these management practices on soil properties and the diversity of rhizospheric microbial communities.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12102246/s1. Table S1: Primer used for qPCR. Table S2: Equations and efficiency of qPCR standard curves for samples processed in 2018 and 2019. The results for ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA), nitrite reductase genes (nirS and nirK), Bacteria, and Archaea are indicated. Table S3: Principal component analysis of bacteria genera showing the eigenvector, the eigenvalue, and the cumulative proportion the of the dataset variability explained by each of the seven principal components (PCs) extracted. Genera with loadings >|0.45| are bolded. Table S4: Principal component analysis of fungal genera showing the eigenvector, the eigenvalue, and the cumulative proportion the of the dataset variability explained by each of the eight principal components (PCs) extracted. Genera with loadings >|0.45| are bolded. Table S5. Matrix of Pearson’s correlation coefficients among microbial indicator groups for each taxon responsive to treatments (Bacteria PC1 and PC3, Fungi PC1) and soil properties of nitrate (NO3−), ammonium (NH4+), and available P (Pa). Table S6. Matrix of Pearson’s correlation coefficients among the abundance of nitrogen cycle genes (AOA, AOB, nirS, and nirK) and soil properties of nitrate (NO3−), ammonium (NH4+), and available P (Pa). Figure S1: Rarefaction curves of the different bacterial samples analyzed through barcoded amplicon-sequencing. Figure S2: Rarefaction curves of the different fungal samples analyzed through barcoded amplicon-sequencing. Figure S3: Non-metric multidimensional scaling of Bacteria (Stress = 0.08) (A) and Fungi (Stress = 0.105) (B) using generalized UniFrac distance.
Author Contributions
Conceptualization, M.C.Z. and M.B.V.; methodology, M.E.M., M.A., G.A.I. and J.B.; resources, M.C.Z. and M.B.V.; formal analysis and data curation, M.E.M., M.A. and M.B.V.; writing—original draft preparation, M.E.M.; writing—review and editing, M.B.V., M.C.Z., M.A., G.A.I. and J.B.; supervision, project administration and funding acquisition, M.C.Z. and M.B.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Argentinean National Agency for Scientific and Technological Promotion (ANPCyT) grant PICT 2015-1556; the Universidad Nacional del Sur grant PGI 24/A250, and the University of Illinois’ Office of International Programs at the College of Agricultural, Consumer and Environmental Sciences (ACES) International Seed Grant, award ISGF2018-MV.
Data Availability Statement
The datasets have been deposited in Sequence Read Archive (SRA) repository under the accession PRJNA861243.
Acknowledgments
The authors acknowledge A.M. Zamponi (CONICET) and M. De Lucía (UNS) for their assistance with the field assay, and CONICET for the fellowships awarded to M. Morales, M. Allegrini and G. Iocoli. We are also grateful to C.L. Wright and M. Band (Roy J. Carver Biotechnology Center, University of Illinois) for their valuable assistance and support in the sequencing service.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Scholberg, J.M.S.; Dogliotti, S.; Leoni, C.; Cherr, C.M.; Zotarelli, L.; Rossing, W.A.H. Cover Crops for Sustainable Agrosystems in the Americas. In Sustainable Agriculture Reviews 4: Genetic Engineering, Biofertilisation, Soil Quality and Organic Farming, 1st ed.; Lichtfouse, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 4, pp. 23–58. [Google Scholar] [CrossRef]
- Blanco-Canqui, H.; Shaver, T.M.; Lindquist, J.L.; Shapiro, C.A.; Elmore, R.W.; Francis, C.A.; Hergert, G.W. Cover Crops and Ecosystem Services: Insights from Studies in Temperate Soils. Agron. J. 2015, 107, 2449–2474. [Google Scholar] [CrossRef]
- Daryanto, S.; Fu, B.; Wang, L.; Jacinthe, P.A.; Zhao, W. Quantitative Synthesis on the Ecosystem Services of Cover Crops. Earth Sci. Rev. 2018, 185, 357–373. [Google Scholar] [CrossRef]
- Kim, N.; Zabaloy, M.C.; Guan, K.; Villamil, M.B. Do Cover Crops Benefit Soil Microbiome? A Meta-Analysis of Current Research. Soil Biol. Biochem. 2020, 142, 107701. [Google Scholar] [CrossRef]
- Morales, M.E.; Iocoli, G.A.; Villamil, M.B.; Zabaloy, M.C. Efecto de Los Cultivos de Cobertura Invernales Sobre El Microbioma Del Suelo: Revisión Sistemática de La Literatura. Rev. Argent. Microbiol. 2021, 54, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Mirsky, S.B.; Curran, W.S.; Mortensen, D.A.; Ryan, M.R.; Shumway, D.L. Control of Cereal Rye with a Roller/Crimper as Influenced by Cover Crop Phenology. Agron. J. 2009, 101, 1589–1596. [Google Scholar] [CrossRef]
- Baigorria, T.; Alvarez, C.; Cazorla, C.; Belluccini, P.; Aimetta, B.; Pegoraro, V.; Boccolini, M.; Conde, B.; Faggioli, V.; Ortiz, J. Impacto Ambiental Y Rolado De Cultivos De Cobertura En Producción De Soja Bajo Siembra Directa. Cienc. Suelo 2019, 37, 355–366. [Google Scholar]
- Benbrook, C.M. Trends in Glyphosate Herbicide Use in the United States and Globally. Environ. Sci. Eur. 2016, 28, 3. [Google Scholar] [CrossRef]
- Van Bruggen, A.H.C.; Finckh, M.R.; He, M.; Ritsema, C.J.; Harkes, P.; Knuth, D.; Geissen, V. Indirect Effects of the Herbicide Glyphosate on Plant, Animal and Human Health Through Its Effects on Microbial Communities. Front. Environ. Sci. 2021, 9, 763917. [Google Scholar] [CrossRef]
- Duke, S.O.; Lydon, J.; Koskinen, W.C.; Moorman, T.B.; Chaney, R.L.; Hammerschmidt, R. Glyphosate Effects on Plant Mineral Nutrition, Crop Rhizosphere Microbiota, and Plant Disease in Glyphosate-Resistant Crops. J. Agric. Food Chem. 2012, 60, 10375–10397. [Google Scholar] [CrossRef]
- Kremer, R.J.; Means, N.E.; Kim, S. Glyphosate Affects Soybean Root Exudation and Rhizosphere Micro-Organisms. Int. J. Environ. Anal. Chem. 2005, 85, 1165–1174. [Google Scholar] [CrossRef]
- Laitinen, P.; Rämö, S.; Siimes, K. Glyphosate Translocation from Plants to Soil—Does This Constitute a Significant Proportion of Residues in Soil? Plant Soil 2007, 300, 51–60. [Google Scholar] [CrossRef]
- Allegrini, M.; Gomez, E.d.V.; Smalla, K.; Zabaloy, M.C. Suppression Treatment Differentially Influences the Microbial Community and the Occurrence of Broad Host Range Plasmids in the Rhizosphere of the Model Cover Crop Avena sativa L. PLoS ONE 2019, 14, e0223600. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Yin, C.; Burke, I.; Hulbert, S.; Paulitz, T. Location, Root Proximity, and Glyphosate-Use History Modulate the Effects of Glyphosate on Fungal Community Networks of Wheat. Microb. Ecol. 2018, 76, 240–257. [Google Scholar] [CrossRef]
- Lupwayi, N.Z.; Fernandez, M.R.; Kanashiro, D.A.; Petri, R.M. Profiles of Wheat Rhizobacterial Communities in Response to Repeated Glyphosate Applications, Crop Rotation, and Tillage. Can. J. Soil Sci. 2021, 101, 157–167. [Google Scholar] [CrossRef]
- Paterson, E.; Sim, A. Effect of Nitrogen Supply and Defoliation on Loss of Organic Compounds from Roots of Festuca Rubra. J. Exp. Bot. 2000, 51, 1449–1457. [Google Scholar] [CrossRef]
- Guo, X.; Zhou, X.; Hale, L.; Yuan, M.; Feng, J.; Ning, D.; Shi, Z.; Qin, Y.; Liu, F.; Wu, L.; et al. Taxonomic and Functional Responses of Soil Microbial Communities to Annual Removal of Aboveground Plant Biomass. Front. Microbiol. 2018, 9, 954. [Google Scholar] [CrossRef]
- Basche, A.D.; Miguez, F.E.; Kaspar, T.C.; Castellano, M.J. Do Cover Crops Increase or Decrease Nitrous Oxide Emissions? A Meta-Analysis. J. Soil Water Conserv. 2014, 69, 471–482. [Google Scholar] [CrossRef]
- Singh, H.; Kandel, T.P.; Gowda, P.H.; Northup, B.K.; Kakani, V.G.; Baath, G.S. Soil N2O Emissions Following Termination of Grass Pea and Oat Cover Crop Residues with Different Maturity Levels. J. Plant Nutr. Soil Sci. 2020, 183, 734–744. [Google Scholar] [CrossRef]
- Allegrini, M.; Morales, M.E.; Villamil, M.B.; Zabaloy, M.C. Ammonia Oxidizing Prokaryotes Respond Differently to Fertilization and Termination Methods in Common Oat’s Rhizosphere. Front. Microbiol. 2021, 12, 746524. [Google Scholar] [CrossRef]
- Romdhane, S.; Spor, A.; Busset, H.; Falchetto, L.; Martin, J.; Bizouard, F.; Bru, D.; Breuil, M.C.; Philippot, L.; Cordeau, S. Cover Crop Management Practices Rather than Composition of Cover Crop Mixtures Affect Bacterial Communities in No-till Agroecosystems. Front. Microbiol. 2019, 10, 1618. [Google Scholar] [CrossRef]
- Soil Survey Staff. Official Soil Series Descriptions. 2019. Available online: https://www.nrcs.usda.gov/wps/portal/nrcs/detail/soils/home/?cid=nrcs142p2_053587 (accessed on 18 September 2019).
- Bray, R.H.; Kurtz, L.T. Determination of Total, Organic, and Available Forms of Phosphorus in Soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A Decimal Code for the Growth Stages of Cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Ashford, D.L.; Reeves, D.W. Use of a Mechanical Roller-Crimper as an Alternative Kill Method for Cover Crops. Am. J. Altern. Agric. 2003, 18, 37–45. [Google Scholar] [CrossRef]
- Yanai, R.D.; Majdi, H.; Park, B.B. Measured and Modelled Differences in Nutrient Concentrations between Rhizosphere and Bulk Soil in a Norway Spruce Stand. Plant Soil 2003, 257, 133–142. [Google Scholar] [CrossRef]
- Hart, S.C.; Stark, J.M.; Davidson, E.A.; Firestone, M.K. Nitrogen Mineralization, Immobilization, and Nitrification. In Methods of Soil Analysis, Part 2—Microbiological and Biochemical Properties; Weaver, R.W., Angle, S., Bottomed, P., Bezdicek, D., Smith, S., Tabatabai, A., Wollum, A., Eds.; Soil Science Society of America: Madison, WI, USA, 1994; pp. 985–1018. [Google Scholar] [CrossRef]
- Drury, C.F.; Hart, S.C.; Yang, X.M. Nitrification Techniques for Soils. In Soil Sampling Methods of Analysis, 2nd ed.; Carter, M.R., Gregorich, E.G., Eds.; Canadian Society Soil Science: Pinawa, MB, Canada; CRC Press Taylor and Francis Group: Boca Raton, FL, USA, 2006; pp. 495–515. [Google Scholar] [CrossRef]
- Bevivino, A.; Paganin, P.; Bacci, G.; Florio, A.; Pellicer, M.S.; Papaleo, M.C.; Mengoni, A.; Ledda, L.; Fani, R.; Benedetti, A.; et al. Soil Bacterial Community Response to Differences in Agricultural Management along with Seasonal Changes in a Mediterranean Region. PLoS ONE 2014, 9, e105515. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of Soil Microbial Community Structure by Use of Taxon-Specific Quantitative PCR Assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef]
- Hoshino, Y.T.; Morimoto, S.; Hayatsu, M.; Nagaoka, K.; Suzuki, C.; Karasawa, T.; Takenaka, M.; Akiyama, H. Effect of Soil Type and Fertilizer Management on Archaeal Community in Upland Field Soils. Microbes Environ. 2011, 26, 307–316. [Google Scholar] [CrossRef]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The Ammonia Monooxygenase Structural Gene Amoa as a Functional Marker: Molecular Fine-Scale Analysis of Natural Ammonia-Oxidizing Populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea Predominate among Ammonia-Oxidizing Prokaryotes in Soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef]
- Schauss, K.; Focks, A.; Leininger, S.; Kotzerke, A.; Heuer, H.; Thiele-Bruhn, S.; Sharma, S.; Wilke, B.M.; Matthies, M.; Smalla, K.; et al. Dynamics and Functional Relevance of Ammonia-Oxidizing Archaea in Two Agricultural Soils. Environ. Microbiol. 2009, 11, 446–456. [Google Scholar] [CrossRef]
- Henry, S.; Baudoin, E.; López-Gutiérrez, J.C.; Martin-Laurent, F.; Brauman, A.; Philippot, L. Quantification of Denitrifying Bacteria in Soils by NirK Gene Targeted Real-Time PCR. J. Microbiol. Methods 2004, 59, 327–335. [Google Scholar] [CrossRef]
- Kandeler, E.; Deiglmayr, K.; Tscherko, D.; Bru, D.; Philippot, L. Abundance of NarG, NirS, NirK, and NosZ Genes of Denitrifying Bacteria during Primary Successions of a Glacier Foreland. Appl. Environ. Microbiol. 2006, 72, 5957–5962. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Henry, H.; Stevens, M.; et al. Vegan: Community Ecology Package; R Package Version 2.5–7. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 15 April 2022).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic Analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, X.; Yang, L. GUniFrac: Generalized UniFrac Distances and Distance-Based Multivariate Analysis of Variance. R Package Version 1.2. Available online: https://cran.r-project.org/web/packages/GUniFrac/GUniFrac.pdf (accessed on 12 May 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: https://www.r-project.org/ (accessed on 17 April 2022).
- SAS Institute Inc. JMP 14 Predictive and Specialized Modeling; SAS Institute: Cary, NC, USA, 2018. [Google Scholar]
- Gloor, G.B.; Reid, G. Compositional Analysis: A Valid Approach to Analyze Microbiome High-Throughput Sequencing Data. Can. J. Microbiol. 2016, 62, 692–703. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Kim, N.; Riggins, C.W.; Zabaloy, C.; Allegrini, M.; Rodriguez-zas, S.L.; Villamil, B. High-Resolution Indicators of Soil Microbial Responses to N Fertilization and Cover Cropping in Corn Monocultures. Agronomy 2022, 12, 954. [Google Scholar] [CrossRef]
- Villamil, M.B.; Kim, N.; Riggins, C.W.; Zabaloy, M.C.; Allegrini, M.; Rodríguez-Zas, S.L. Microbial Signatures in Fertile Soils Under Long-Term N Management. Front. Soil Sci. 2021, 1, 1–22. [Google Scholar] [CrossRef]
- Palarea-Albaladejo, J.; Martín-Fernández, J.A. ZCompositions - R Package for Multivariate Imputation of Left-Censored Data under a Compositional Approach. Chemom. Intell. Lab. Syst. 2015, 143, 85–96. [Google Scholar] [CrossRef]
- Aitchison, J. The Statistical Analysis of Compositional Data. J. R. Stat. Soc. Ser. B Stat. Methodol. 1982, 44, 139–177. [Google Scholar] [CrossRef]
- Anderson, M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Hervé, M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R Package Version 0.9–80. Available online: https://cran.r-project.org/web/packages/RVAideMemoire/RVAideMemoire.pdf (accessed on 21 February 2022).
- Kozdrój, J.; Van Elsas, J.D. Response of the Bacterial Community to Root Exudates in Soil Polluted with Heavy Metals Assessed by Molecular and Cultural Approaches. Soil Biol. Biochem. 2000, 32, 1405–1417. [Google Scholar] [CrossRef]
- Sheng, M.; Hamel, C.; Fernandez, M.R. Cropping Practices Modulate the Impact of Glyphosate on Arbuscular Mycorrhizal Fungi and Rhizosphere Bacteria in Agroecosystems of the Semiarid Prairie. Can. J. Microbiol. 2012, 58, 990–1001. [Google Scholar] [CrossRef]
- Zabaloy, M.C.; Carné, I.; Viassolo, R.; Gómez, M.A.; Gomez, E. Soil Ecotoxicity Assessment of Glyphosate Use under Field Conditions: Microbial Activity and Community Structure of Eubacteria and Ammonia-Oxidising Bacteria. Pest Manag. Sci. 2015, 72, 684–691. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Yin, C.; Hulbert, S.; Burke, I. Impacts of Repeated Glyphosate Use on Wheat-Associated Bacteria Are Small and Depend on Glyphosate Use History. Appl. Environ. Microbiol. 2017, 83, e01354-17. [Google Scholar] [CrossRef]
- Imparato, V.; Santos, S.S.; Johansen, A.; Geisen, S.; Winding, A. Stimulation of Bacteria and Protists in Rhizosphere of Glyphosate-Treated Barley. Appl. Soil Ecol. 2016, 98, 47–55. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Ndour, P.M.S.; Barry, C.M.; Tine, D.; De la Fuente Cantó, C.; Gueye, M.; Barakat, M.; Ortet, P.; Achouak, W.; Ndoye, I.; Sine, B.; et al. Pearl Millet Genotype Impacts Microbial Diversity and Enzymatic Activities in Relation to Root-Adhering Soil Aggregation. Plant Soil 2021, 464, 109–129. [Google Scholar] [CrossRef]
- Lei, S.; Xu, X.; Cheng, Z.; Xiong, J.; Ma, R.; Zhang, L.; Yang, X.; Zhu, Y.; Zhang, B.; Tian, B. Analysis of the Community Composition and Bacterial Diversity of the Rhizosphere Microbiome across Different Plant Taxa. Microbiologyopen 2019, 8, e00762. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.; Locke, M.; Reddy, K.; McChesney, D.S.; Steinriede, R. Glyphosate Applications, Glyphosate Resistant Corn, and Tillage on Nitrification Rates and Distribution of Nitrifying Microbial Communities. Soil Sci. Soc. Am. J. 2017, 81, 1371–1380. [Google Scholar] [CrossRef]
- Zhang, C.J.; Yang, Z.L.; Shen, J.P.; Sun, Y.F.; Wang, J.T.; Han, H.Y.; Wan, S.Q.; Zhang, L.M.; He, J.Z. Impacts of Long-Term Nitrogen Addition, Watering and Mowing on Ammonia Oxidizers, Denitrifiers and Plant Communities in a Temperate Steppe. Appl. Soil Ecol. 2018, 130, 241–250. [Google Scholar] [CrossRef]
- Liang, S.; Grossman, J.; Shi, W. Soil Microbial Responses to Winter Legume Cover Crop Management during Organic Transition. Eur. J. Soil Biol. 2014, 65, 15–22. [Google Scholar] [CrossRef]
- Zielińska, M.; Rusanowska, P.; Jarząbek, J.; Nielsen, J.L. Community Dynamics of Denitrifying Bacteria in Full-Scale Wastewater Treatment Plants. Environ. Technol. 2016, 37, 2358–2367. [Google Scholar] [CrossRef]
- Baldani, J.I.; Rouws, L.; Cruz, L.M.; Olivares, F.L.; Schmid, M.; Hartmann, A. The Family Oxalobacteraceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 919–974. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Saravanan, V.S.; Hari, K.; Lee, K.C.; Lee, J.S. Duganella Sacchari Sp. Nov. and Duganella Radicis Sp. Nov., Two Novel Species Isolated from Rhizosphere of Field-Grown Sugar Cane. Int. J. Syst. Evol. Microbiol. 2013, 63, 1126–1131. [Google Scholar] [CrossRef]
- Yin, C.; Hulbert, S.H.; Schroeder, K.L.; Mavrodi, O.; Mavrodi, D.; Dhingra, A.; Schillinger, W.F.; Paulitz, T.C. Role of Bacterial Communities in the Natural Suppression of Rhizoctonia Solani Bare Patch Disease of Wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 2013, 79, 7428–7438. [Google Scholar] [CrossRef]
- Jiang, P.X.; Wang, H.S.; Zhang, C.; Lou, K.; Xing, X.H. Reconstruction of the Violacein Biosynthetic Pathway from Duganella Sp. B2 in Different Heterologous Hosts. Appl. Microbiol. Biotechnol. 2010, 86, 1077–1088. [Google Scholar] [CrossRef]
- Pisa, G.; Magnani, G.S.; Weber, H.; Souza, E.M.; Faoro, H.; Monteiro, R.A.; Daros, E.; Baura, V.; Bespalhok, J.P.; Pedrosa, F.O.; et al. Diversity of 16S RRNA Genes from Bacteria of Sugarcane Rhizosphere Soil. Braz. J. Med. Biol. Res. 2011, 44, 1215–1221. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Yin, C.; Hulbert, S.; Paulitz, T.C. Core Rhizosphere Microbiomes of Dryland Wheat Are Influenced by Location and Land Use History. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Li, X.; Rui, J.; Mao, Y.; Yannarell, A.; Mackie, R. Dynamics of the Bacterial Community Structure in the Rhizosphere of a Maize Cultivar. Soil Biol. Biochem. 2014, 68, 392–401. [Google Scholar] [CrossRef]
- Ofek, M.; Hadar, Y.; Minz, D. Ecology of Root Colonizing Massilia (Oxalobacteraceae). PLoS ONE 2012, 7, e40117. [Google Scholar] [CrossRef]
- Tonon, L.A.C.; Moreira, A.P.B.; Thompson, F. The Family Erythrobacteraceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 214–235. [Google Scholar] [CrossRef]
- Lee, S.D.; Kim, Y.-J.; Kim, I.S. Erythrobacter Suaedae sp. nov., Isolated from a Rhizosphere Mudflat of a Halophyte (Suaeda japonica). Int. J. Syst. Evol. Microbiol. 2019, 69, 3287–3292. [Google Scholar] [CrossRef]
- Ye, Y.H.; Anwar, N.; Xamxidin, M.; Zhang, R.; Yan, C.; Nie, Y.F.; Zhao, Z.; Sun, C.; Wu, M. Description of Erythrobacter mangrovi sp. nov., an Aerobic Bacterium from Rhizosphere Soil of Mangrove Plant (Kandelia candel). Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2020, 113, 1425–1435. [Google Scholar] [CrossRef]
- Tang, T.; Sun, X.; Dong, Y.; Liu, Q. Erythrobacter aureus sp. nov., a Plant Growth-Promoting Bacterium Isolated from Sediment in the Yellow Sea, China. 3 Biotech 2019, 9, 430. [Google Scholar] [CrossRef]
- Kämpfer, P.; Martin, K.; McInroy, J.A.; Glaeser, S.P. Proposal of Novosphingobium rhizosphaerae sp. nov., Isolated from the Rhizosphere. Int. J. Syst. Evol. Microbiol. 2015, 65, 195–200. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, J.S.; Kim, S.G.; Zhang, C.W.; Jiang, J.Q.; Ma, X.T.; Zhang, J.; Zhang, X.X. Novosphingobium oryzae sp. nov., a Potential Plant-Promoting Endophytic Bacterium Isolated from Rice Roots. Int. J. Syst. Evol. Microbiol. 2016, 66, 302–307. [Google Scholar] [CrossRef]
- Rodgers-Vieira, E.A.; Zhang, Z.; Adrion, A.C.; Gold, A.; Aitken, M.D. Identification of Anthraquinone-Degrading Bacteria in Soil Contaminated with Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2015, 81, 3775–3781. [Google Scholar] [CrossRef]
- Hargreaves, S.K.; Williams, R.J.; Hofmockel, K.S. Environmental Filtering of Microbial Communities in Agricultural Soil Shifts with Crop Growth. PLoS ONE 2015, 10, e0134345. [Google Scholar] [CrossRef]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global Biogeography and Quantitative Seasonal Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef]
- Pascual, J.; García-López, M.; Bills, G.F.; Genilloud, O. Longimicrobium Terrae gen. nov., sp. nov., an Oligotrophic Bacterium of the under-Represented Phylum Gemmatimonadetes Isolated through a System of Miniaturized Diffusion Chambers. Int. J. Syst. Evol. Microbiol. 2016, 66, 1976–1985. [Google Scholar] [CrossRef]
- Foesel, B.U.; Geppert, A.; Rohde, M.; Overmann, J. Parviterribacter kavangonensis gen. nov., sp. nov. and Parviterribacter multiflagellatus sp. nov., novel members of Parviterribacteraceae fam. nov. within the order Solirubrobacterales, and emended descriptions of the classes Thermoleophilia and Rubrobact and their orders and families. Int. J. Syst. Evol. Microbiol. 2016, 66, 652–665. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, X.; Zhang, T.; He, W.; Song, F. Effects of the Long-Term Application of Atrazine on Soil Enzyme Activity and Bacterial Community Structure in Farmlands in China. Environ. Pollut. 2020, 262, 114264. [Google Scholar] [CrossRef]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The Under-Recognized Dominance of Verrucomicrobia in Soil Bacterial Communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef]
- Buckley, D.H.; Schmidt, T.M. Environmental Factors Influencing the Distribution of rRNA from Verrucomicrobia in Soil. FEMS Microbiol. Ecol. 2001, 35, 105–112. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Soares, T.; Rossetto, R.; van Veen, J.A.; Tsai, S.M.; Kuramae, E.E. Verrucomicrobial Community Structure and Abundance as Indicators for Changes in Chemical Factors Linked to Soil Fertility. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2015, 108, 741–752. [Google Scholar] [CrossRef]
- Otsuka, S.; Suenaga, T.; Vu, H.T.; Ueda, H.; Yokota, A.; Senoo, K. Brevifollis gellanilyticus gen. nov., sp. nov., a Gellan-Gum-Degrading Bacterium of the Phylum Verrucomicrobia. Int. J. Syst. Evol. Microbiol. 2013, 63, 3075–3078. [Google Scholar] [CrossRef]
- Vasconcellos, R.L.F.; Romagnoli, E.M.; Taketani, R.G.; Santos, S.N.; Zucchi, T.D.; Melo, I.S. Impact of Inoculation with Pseudomonas eestus CMAA 1215T on the Non-Target Resident Bacterial Community in a Saline Rhizosphere Soil. Curr. Microbiol. 2021, 78, 218–228. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Ding, C.; Jia, Z.; He, Z.; Zhang, T.; Wang, X. Declined Soil Suppressiveness to Fusarium oxysporum by Rhizosphere Microflora of Cotton in Soil Sickness. Biol. Fertil. Soils 2015, 51, 935–946. [Google Scholar] [CrossRef]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An Important Phylum of Cellulose-Degrading Bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Ren, N.; Wang, Y.; Ye, Y.; Zhao, Y.; Huang, Y.; Fu, W.; Chu, X. Effects of Continuous Nitrogen Fertilizer Application on the Diversity and Composition of Rhizosphere Soil Bacteria. Front. Microbiol. 2020, 11, 1948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yao, Y.; Xu, H.; Xie, Z.; Guo, J.; Qi, Z.; Jiang, H. Soil Metabolomics and Bacterial Functional Traits Revealed the Responses of Rhizosphere Soil Bacterial Community to Long-Term Continuous Cropping of Tibetan Barley. PeerJ 2022, 10, e13254. [Google Scholar] [CrossRef] [PubMed]
- Piutti, S.; Semon, E.; Landry, D.; Hartmann, A.; Dousset, S.; Lichtfouse, E.; Topp, E.; Soulas, G.; Martin-Laurent, F. Isolation and Characterisation of Nocardioides sp. SP12, an Atrazine-Degrading Bacterial Strain Possessing the Gene trzN from Bulk- and Maize Rhizosphere Soil. FEMS Microbiol. Lett. 2003, 221, 111–117. [Google Scholar] [CrossRef]
- Hou, J.; Liu, W.; Wang, B.; Wang, Q.; Luo, Y.; Franks, A.E. PGPR Enhanced Phytoremediation of Petroleum Contaminated Soil and Rhizosphere Microbial Community Response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef]
- Buckley, D.H.; Huangyutitham, V.; Nelson, T.A.; Rumberger, A.; Thies, J.E. Diversity of Planctomycetes in Soil in Relation to Soil History and Environmental Heterogeneity. Appl. Environ. Microbiol. 2006, 72, 4522–4531. [Google Scholar] [CrossRef]
- Akinola, S.A.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity. Microorganisms 2021, 9, 1419. [Google Scholar] [CrossRef]
- Xia, Z.; Wang, Q.; She, Z.; Gao, M.; Zhao, Y.; Guo, L.; Jin, C. Nitrogen Removal Pathway and Dynamics of Microbial Community with the Increase of Salinity in Simultaneous Nitrification and Denitrification Process. Sci. Total Environ. 2019, 697, 134047. [Google Scholar] [CrossRef]
- Dourado, M.N.; Camargo Neves, A.A.; Santos, D.S.; Araújo, W.L. Biotechnological and Agronomic Potential of Endophytic Pink-Pigmented Methylotrophic Methylobacterium spp. BioMed Res. Int. 2015, 2015, 909016. [Google Scholar] [CrossRef]
- Green, P.N. Methylobacterium. In Bergey’s Manual Systematics of Archaea and Bacteria; In Association with Bergey’s Manual Trust; Whitman, W.B., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Jinal, H.N.; Amaresan, N.; Sankaranarayanan, A. Methylobacterium. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Senthil Kumar, M., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 509–519. [Google Scholar] [CrossRef]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Vasilyeva, L.V. Stella, a New Genus of Soil Prosthecobacteria, with Proposals for Stella humosa sp. Nov. and Stella vacuolata sp. nov. Int. J. Syst. Bacteriol. 1985, 35, 518–521. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q. Outline of Ascomycota. In Encyclopedia of Mycology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 1, pp. 246–254. [Google Scholar] [CrossRef]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota Members Dominate Fungal Communities during Straw Residue Decomposition in Arable Soil. PLoS ONE 2013, 8, e66146. [Google Scholar] [CrossRef]
- Xu, L.; Ravnskov, S.; Larsen, J.; Nicolaisen, M. Linking Fungal Communities in Roots, Rhizosphere, and Soil to the Health Status of Pisum Sativum. FEMS Microbiol. Ecol. 2012, 82, 736–745. [Google Scholar] [CrossRef]
- Miura, T.; Niswati, A.; Swibawa, I.G.; Haryani, S.; Gunito, H.; Shimano, S.; Fujie, K.; Kaneko, N. Diversity of Fungi on Decomposing Leaf Litter in a Sugarcane Plantation and Their Response to Tillage Practice and Bagasse Mulching: Implications for Management Effects on Litter Decomposition. Microb. Ecol. 2015, 70, 646–658. [Google Scholar] [CrossRef]
- Newsham, K.K. Phialophora Graminicola, a Dark Septate Fungus, Is a Beneficial Associate of the Grass Vulpia ciliata Ssp. ambigua. New Phytol. 1999, 144, 517–524. [Google Scholar] [CrossRef]
- Yao, Q.; Xu, Y.; Liu, X.; Liu, J.; Huang, X.; Yang, W.; Yang, Z.; Lan, L.; Zhou, J.; Wang, G. Dynamics of Soil Properties and Fungal Community Structure in Continuous-Cropped Alfalfa Fields in Northeast China. PeerJ 2019, 2019, e7127. [Google Scholar] [CrossRef]
- Dean, S.L.; Billingsley Tobias, T.; Phippen, W.B.; Clayton, A.W.; Gruver, J.; Porras-Alfaro, A. A Study of Glycine max (Soybean) Fungal Communities under Different Agricultural Practices. Plant Gene 2017, 11, 8–16. [Google Scholar] [CrossRef]
- Digby, A.L.; Gleason, F.H.; McGee, P.A. Some Fungi in the Chytridiomycota Can Assimilate Both Inorganic and Organic Sources of Nitrogen. Fungal Ecol. 2010, 3, 261–266. [Google Scholar] [CrossRef]
- Tian, L.; Shi, S.; Ma, L.; Tran, L.S.P.; Tian, C. Community Structures of the Rhizomicrobiomes of Cultivated and Wild Soybeans in Their Continuous Cropping. Microbiol. Res. 2020, 232, 126390. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Burke, I.; Paulitz, T.C. Succession of Fungal and Oomycete Communities in Glyphosate-Killed Wheat Roots. Phytopathology 2018, 108, 582–594. [Google Scholar] [CrossRef]
- Borrell, A.N.; Shi, Y.; Gan, Y.; Bainard, L.D.; Germida, J.J.; Hamel, C. Fungal Diversity Associated with Pulses and Its Influence on the Subsequent Wheat Crop in the Canadian Prairies. Plant Soil 2017, 414, 13–31. [Google Scholar] [CrossRef]
- Lou, J.; Fu, L.; Peng, Y.; Zhou, L. Metabolites from Alternaria fungi and Their Bioactivities. Molecules 2013, 18, 5891–5935. [Google Scholar] [CrossRef]
- Espagne, E.; Lespinet, O.; Malagnac, F.; Da Silva, C.; Jaillon, O.; Porcel, B.M.; Couloux, A.; Aury, J.M.; Ségurens, B.; Poulain, J.; et al. The Genome Sequence of the Model Ascomycete Fungus Podospora anserina. Genome Biol. 2008, 9, R77. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).