Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment Materials
2.2. Experimental Design
2.3. Measurement Indicators
2.4. Principal Component Analysis Salt Tolerance Evaluation
2.5. Membership Function Analysis
2.6. Calculation of the Comprehensive Evaluation Value of Salt Tolerance
2.7. Pearson Correlation Analysis
2.8. Stepwise Regression Analysis
2.9. Hierarchical Cluster Analysis
2.10. Statistical Analysis
3. Results
3.1. Determination of the Best-Fit Salt Concentration
3.2. Comprehensive Evaluation of Salt Tolerance of 114 Rice Germplasm Resources under Optimal Salt Concentrations
3.3. Stepwise Regression Analysis of 114 Rice Germplasm Resources
3.4. Pearson Correlation between Salt Gradient and Germination Index
3.5. Salt Tolerance Classification and Screening for a Reliable Salt Tolerance Indicator
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Yang, A.; Zhang, W. Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J. Exp. Bot. 2016, 67, 6431–6444. [Google Scholar] [CrossRef] [Green Version]
- Long, X.; Liu, L.; Shao, T.; Shao, H.; Liu, Z. Developing and sustainably utilize the coastal mudflat areas in China. Sci. Total Environ. 2016, 56, 1077–1086. [Google Scholar] [CrossRef]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 2012, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Yahia, O.; Ghazi, A.K.; Tawaha, A.; Rahman, A.; Horani, A. Variation in Germination and Ion Uptake in Barley Genotypes under Salinity Conditions. World J. Agric. Sci. 2006, 2, 11–15. [Google Scholar]
- Khan, M.A.; Shirazi, M.U.; Ali, M.; Mumtaz, S.; Ashraf, M.Y. Comparative performance of wheat genotypes growing in saline water. Can. J. Bot. 2006, 38, 1633–1639. [Google Scholar]
- Wang, Y.; Li, T.; Li, Y.; Bjorn, L.O.; Rosendahl, S.; Olsson, P.A.; Li, S.; Fu, X. Community dynamics of arbuscular mycorrhizal fungi in high-input and intensively irrigated rice cultivation systems. Appl. Environ. Microbiol. 2015, 81, 2958–2965. [Google Scholar] [CrossRef] [Green Version]
- Quan, R.; Wang, J.; Hui, J.; Bai, H.; Lyu, X.; Zhu, Y.; Zhang, H.; Zhang, Z.; Li, S.; Huang, R. Improvement of Salt Tolerance Using Wild Rice Genes. Front. Plant Sci. 2017, 8, 2269. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Shannon, M.C. Salinity Effects on Seedling Growth and Yield Components of Rice. Crop Sci. 2000, 40, 996–1003. [Google Scholar] [CrossRef] [Green Version]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt Tolerance in Rice: Focus on Mechanisms and Approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Luo, Y.; Reid, R.; Freese, D.; Li, C.; Watkins, J.; Shi, H.; Zhang, H.; Loraine, A.; Song, B. Salt tolerance response revealed by RNA-Seq in a diploid halophytic wild relative of sweet potato. Sci. Rep. 2017, 7, 9624. [Google Scholar] [CrossRef]
- Maheshwari, P.; Kovalchuk, I. Agrobacterium-Mediated Stable Genetic Transformation of Populus angustifolia and Populus balsamifera. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Xiao, X.; Yang, M.; Gao, Z.; Zang, J.; Fu, X.; Chen, Y. Effects of salt stress on antioxidant defense system in the root of Kandelia candel. Bot. Stud. 2014, 55, 57. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Hui, Z.; Jing, L.; Wen, F. Primary Screening of Salt-tolerant Rice Germplasm in Entire Growth Period. Chin. Agric. Sci. Bull. 2013, 29, 63–68. [Google Scholar]
- Sun, X.; Jiang, Q.; Hu, Z.; Zhang, H.; Xu, C.; Shi, Y.; Han, L.; Zhang, H. Screening and identification of salt-tolerant rice germplasm in whole growth period. Acta Agron. Sin. 2019, 45, 1656–1663. [Google Scholar]
- Wu, H.; Guo, J.; Wang, C.; Li, K.; Zhang, X.; Yang, Z.; Li, M.; Wang, B. An Effective Screening Method and a Reliable Screening Trait for Salt Tolerance of Brassica napus at the Germination Stage. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Hu, J.; Tashpolat, T.; Iiyas, N.; Zhang, F.; Mamat, S. Spatial variability of soil salt content in Yutian oasis. Soils 2017, 49, 162–170. [Google Scholar]
- Qi, D.; Guo, G.; Li, M.; Cao, G.; Zhang, J.; Zhou, Q.; Zhang, S.; Xu, X.; Han, L. Progress of physiology and genetic research on saline-alkaline tolerance in rice. J. Plant Genet. Resour. 2007, 8, 486–493. [Google Scholar]
- Zheng, C.; Zhang, Z.; Zhou, G.; Xie, X. Evaluation on salt tolerance of different rice varieties during germination stage. Shandong Agric. Sci. 2018, 50, 38–42. [Google Scholar]
- Du, Z.; Zhang, Y.; Tian, Z.; Bian, J.; Sun, X. Alkali resistance screening of japonica rice varieties during budding stage in cold region. Heilongjiang Agric. Sci. 2017, 6–10. [Google Scholar] [CrossRef]
- Li, W. Effect of Salt Stress on Seed Germination and Seedling Growth of Rice. Master’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2011. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CMFD&dbname=CMFD2012&filename=1012318229.nh&v=3HNO6lOT58LlgcIa6GcPo%25mmd2B5eLmPFQMTNQGKaL%25mmd2F677CoC8rNLI8ZzMnmCYmrogJIH (accessed on 4 May 2021).
- Agnihotri, R.K.; Palni, L.M.; Spandey, D. Screening of landraces of rice under cultivation in Kumaun Himalaya for salinity stress during germination and early seedling growth. Indian J. Plant Physiol. 2006, 11, 266–272. [Google Scholar]
- Das, D.; Ali, M.A.; Sarkar, T.A.; Ali, M.Y. Germination and seedling growth of indigenous aman rice under salt stress. J. Bangladesh Agric. Univ. 2017, 15, 182. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Niu, X.; Zhang, M.; Wang, C.; Xu, Q.; Feng, Y.; Yang, Y.; Wang, S.; Yuan, X.; Yu, H.; et al. Genome-Wide Association Study of Seed Dormancy and the Genomic Consequences of Improvement Footprints in Rice (Oryza sativa L.). Front. Plant Sci. 2018, 8, 2213. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Lu, X.; Gao, G.; Yang, X. Analysis of the principal components and the subordinate function of alfalfa drought resistance. Acta Agrestia Sin. 2006, 14, 142–146. [Google Scholar]
- Dai, H.; Wu, H.; Amanguli, M.; Wang, L.; Maimaiti, A.; Zhang, J. Analysis of Salt-Tolerance and Determination of Salt-Tolerant Evaluation Indicators in Cotton Seedlings of Different Genotypes. Sci. Agric. Sin. 2014, 47, 1290–1300. [Google Scholar]
- Zhang, S.; Liu, L.; Liu, Z. Research advantages of salt tolerance and breeding in rice. North Rice 2009, 39, 118–121. [Google Scholar]
- Wang, Z.; Wang, J.; Bao, Y.; Wu, Y.; Zhang, H. Quantitative trait loci controlling rice seed germination under salt stress. Euphytica 2010, 178, 297–307. [Google Scholar] [CrossRef]
- Xin, C.; Ma, H.; Zhao, Q.; Wang, Y.; Liu, Q.; Yang, J.; Chen, B.; Lin, X. Effects of salt stress on rice seed germination and seedling growth. Barley Cereal Sci. 2019, 36, 7–10. [Google Scholar]
- Guan, Z.; Chen, S.; Wang, Y.; Chen, F. Screening of salt-tolerance concentration and comparison of salt-tolerance for chrysanthemum and its related taxa. Chin. J. Ecol. 2010, 29, 467–472. [Google Scholar]
- Geng, L.; Ma, X.; Cui, D.; Zhang, Q.; Han, B.; Han, L. Identification and Evaluation Method for Saline Tolerance in Rice During the Whole Growth Stage. J. Plant Genet. Resour. 2019, 20, 267–275. [Google Scholar]
- Zhou, M.; Wang, W. Representative points of student’s tn distribution and their applications in statistical simulation. Acta Math. Appl. Sin. 2016, 39, 620–640. [Google Scholar]
- Guo, Y.; Zhang, W.; Yang, H.; Qu, W. Germination characteristics of rice seed under salt stress and physiological basis of salt-tolerance of rice in seedlings. J. Anhui Agric. Sci. 2006, 34, 1053–1054. [Google Scholar]
- Ruan, S.; Xue, Q. Germination characteristics of seeds under salt stress and physiological basis of salt-tolerance of seedlings in hybrid rice. Chin. J. Rice Sci. 2002, 16, 281–284. [Google Scholar]
- Tian, L.; Chen, Y.; Liu, J.; Ma, X.; Wang, N.; Yang, B.; Li, Y.; Guo, H.; Li, J.; Hu, H. Comprehensive Evaluation and Selection of Rice (Oryza sativa japonica) Germplasm for Saline Tolerance at Germination Stage. Chin. J. Rice Sci. 2017, 31, 75–86. [Google Scholar]
- Feng, Z.; Liu, X.; Jiang, C.; Liang, Z. Comprehensive Evaluation of Rice (Oryza sativa japonica) Germplasm for Alkaline/Saline Tolerance at Germination Stage from Jilin Province, China. Soils Crop 2016, 5, 120–127. [Google Scholar]
- Hu, D.; Wu, D.; You, J.; He, Y.; Qian, W. Principal Component Analysis and Comprehensive Evaluation on Salt Tolerance Related Traits in Brassica napus L. Bot. Res. 2018, 7, 101–112. [Google Scholar]
- Ding, T.; Yang, Z.; Wei, X.; Yuan, F.; Yin, S.; Wang, B. Evaluation of salt-tolerant germplasm and screening of the salt-tolerance traits of sweet sorghum in the germination stage. Funct. Plant Biol. 2018, 45. [Google Scholar] [CrossRef] [PubMed]
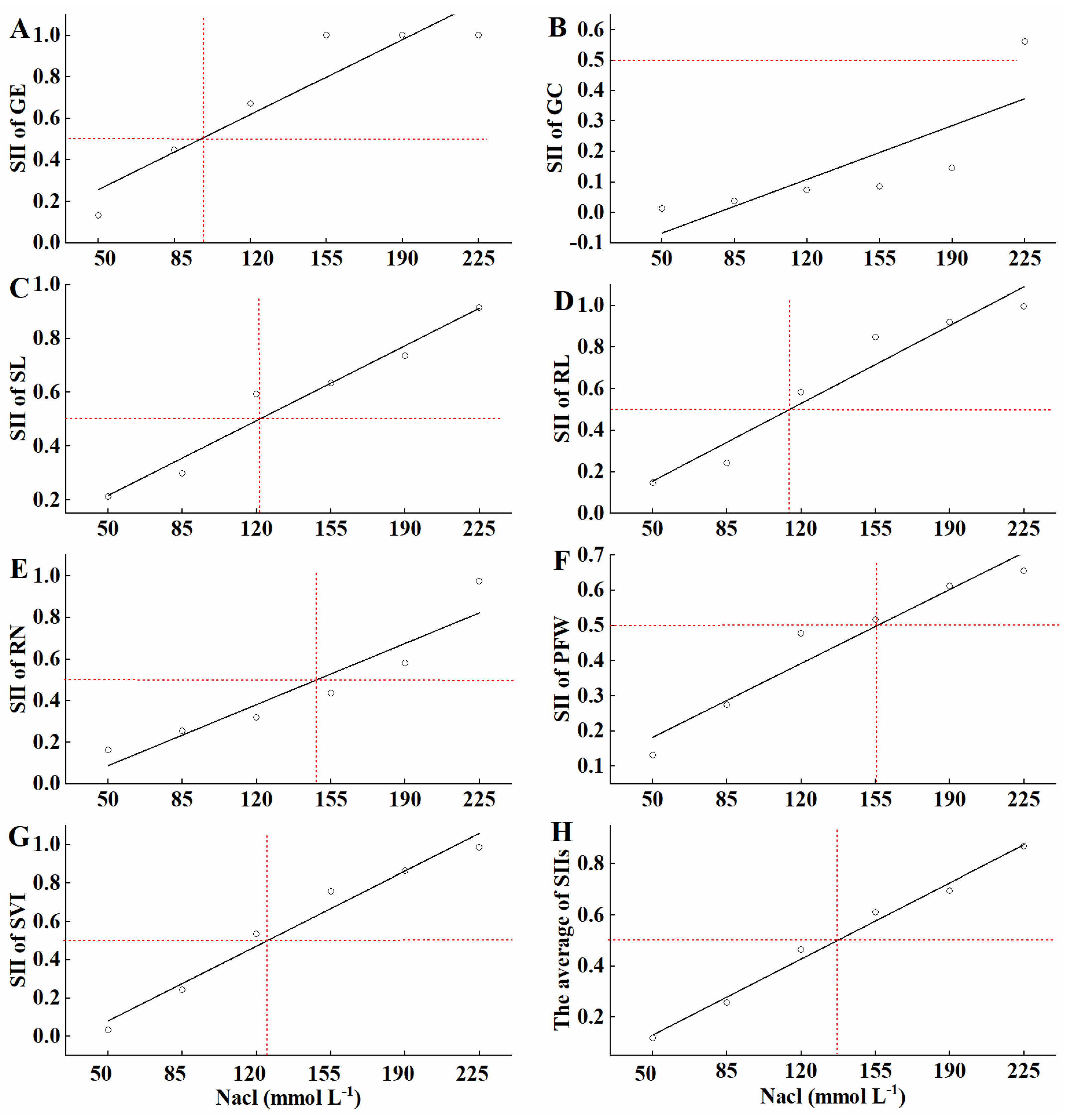
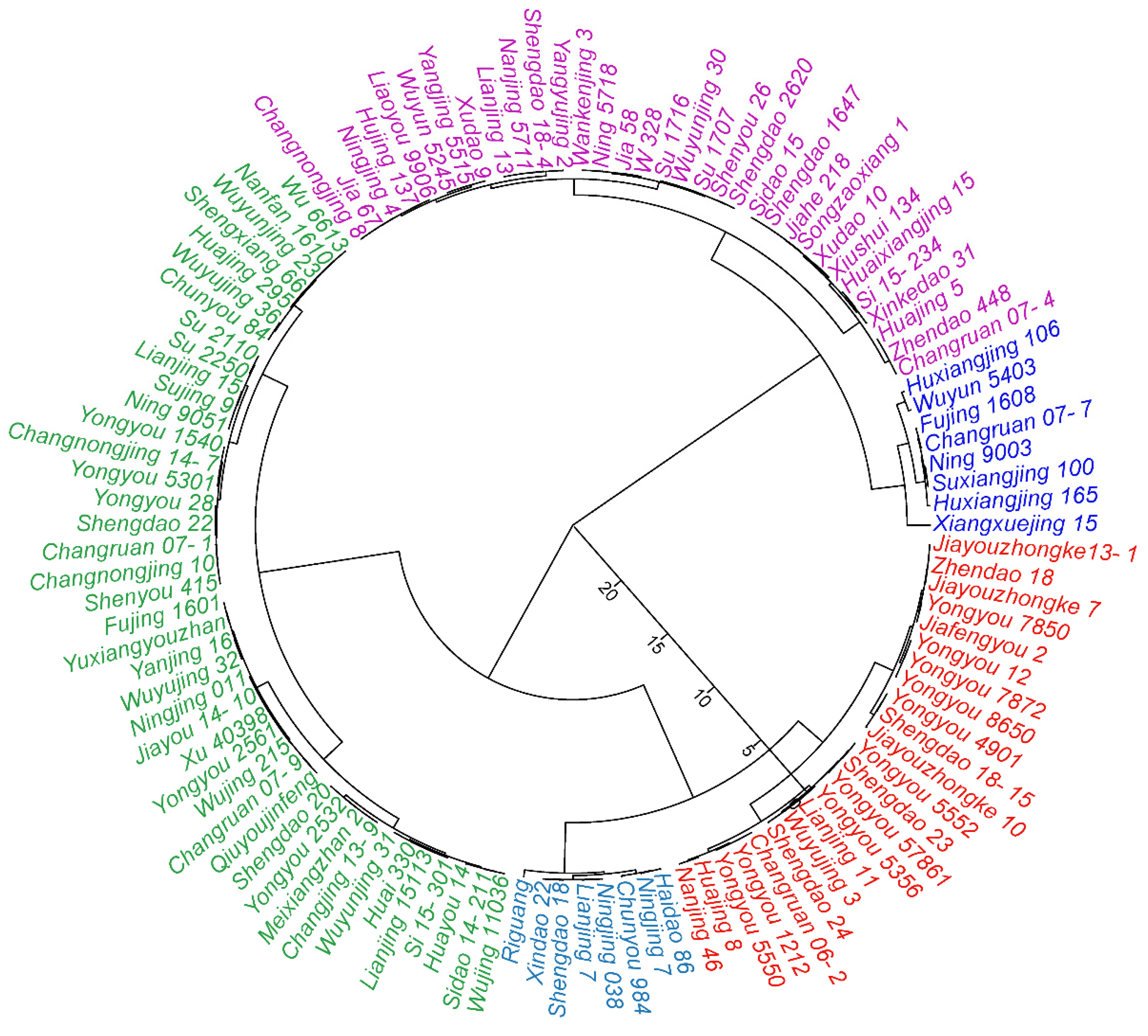
| Trait | Comprehensive Index | ||
|---|---|---|---|
| F1 | F2 | F3 | |
| Germination energy | 0.526 | 0.525 | −0.449 |
| Germination capacity | 0.449 | 0.785 | 0.073 |
| Shoot length | 0.896 | −0.172 | 0.070 |
| Root length | 0.868 | −0.269 | −0.243 |
| Root number | 0.491 | 0.191 | 0.793 |
| Plant fresh weight | 0.845 | −0.293 | 0.142 |
| Seedling vigor index | 0.952 | −0.092 | −0.165 |
| Eigenvalue | 3.897 | 1.125 | 0.948 |
| Weight (%) | 65.292 | 18.841 | 15.874 |
| Contribution ratio (%) | 55.676 | 16.066 | 13.536 |
| Cumulative contribution ratio (%) | 55.676 | 71.741 | 85.278 |
| Variety Name | Principal Components | Membership Function | D-Value | ||||
|---|---|---|---|---|---|---|---|
| F1 | F2 | F3 | μ1 | μ2 | μ3 | ||
| Riguang | 1.97 | 1.08 | 1.54 | 0.85 | 0.78 | 0.81 | 0.83 |
| Ningjing 7 | 1.95 | −0.05 | 1.09 | 0.84 | 0.59 | 0.72 | 0.78 |
| Haidao 86 | 2.75 | −0.86 | −1.45 | 1.00 | 0.45 | 0.19 | 0.76 |
| Chunyou 984 | 2.15 | −0.07 | −0.37 | 0.88 | 0.58 | 0.41 | 0.75 |
| Xindao 22 | 1.82 | 0.53 | −0.25 | 0.82 | 0.69 | 0.44 | 0.73 |
| Shengdao 18 | 0.94 | 2.35 | 1.19 | 0.65 | 1.00 | 0.74 | 0.73 |
| Lianjing 7 | 1.23 | 0.33 | 1.96 | 0.71 | 0.65 | 0.90 | 0.73 |
| Ningjing 038 | 1.35 | −0.72 | 2.46 | 0.73 | 0.47 | 1.00 | 0.72 |
| Shengdao 18-15 | 0.73 | 1.20 | 1.81 | 0.61 | 0.80 | 0.87 | 0.69 |
| Yongyou 4901 | 1.59 | 1.60 | −2.04 | 0.78 | 0.87 | 0.07 | 0.68 |
| Jiayouzhongke 10 | 1.73 | −0.97 | −0.33 | 0.80 | 0.43 | 0.42 | 0.67 |
| Yongyou 7872 | 1.52 | 0.57 | −1.36 | 0.76 | 0.69 | 0.21 | 0.66 |
| Yongyou 12 | 1.66 | −0.20 | −1.15 | 0.79 | 0.56 | 0.25 | 0.66 |
| Yongyou 8650 | 1.91 | −0.46 | −1.94 | 0.84 | 0.52 | 0.09 | 0.66 |
| Jiayouzhongke 7 | 1.77 | −3.46 | 1.29 | 0.81 | 0.00 | 0.76 | 0.65 |
| Zhendao 18 | 0.25 | 1.09 | 2.46 | 0.52 | 0.78 | 1.00 | 0.65 |
| Jiayouzhongke 13-1 | 1.60 | 0.13 | −1.71 | 0.78 | 0.62 | 0.13 | 0.65 |
| Jiafengyou 2 | 1.58 | −0.92 | −0.74 | 0.78 | 0.44 | 0.34 | 0.64 |
| Yongyou 7850 | 1.25 | 0.13 | −0.56 | 0.71 | 0.62 | 0.37 | 0.64 |
| Nanjing 46 | 0.90 | −0.75 | 1.19 | 0.64 | 0.47 | 0.74 | 0.63 |
| Huajing 8 | 0.57 | −0.08 | 1.57 | 0.58 | 0.58 | 0.82 | 0.62 |
| Yongyou 1212 | 1.09 | 0.19 | −0.86 | 0.68 | 0.63 | 0.31 | 0.61 |
| Changruan 06-2 | 0.66 | 0.93 | −0.08 | 0.60 | 0.76 | 0.47 | 0.61 |
| Yongyou 5550 | 0.78 | −0.48 | 0.76 | 0.62 | 0.51 | 0.65 | 0.61 |
| Shengdao 24 | 0.60 | 1.35 | −0.57 | 0.59 | 0.83 | 0.37 | 0.60 |
| Lianjing 11 | 0.45 | −0.46 | 1.71 | 0.56 | 0.52 | 0.84 | 0.60 |
| Yongyou 5356 | 0.59 | 0.40 | 0.32 | 0.59 | 0.66 | 0.56 | 0.60 |
| Wuyujing 3 | 0.57 | 0.30 | 0.44 | 0.58 | 0.65 | 0.58 | 0.59 |
| Shengdao 23 | 0.42 | 1.28 | −0.14 | 0.55 | 0.82 | 0.46 | 0.59 |
| Yongyou 5552 | 0.84 | 0.28 | −0.73 | 0.63 | 0.64 | 0.34 | 0.59 |
| Yongyou 57861 | 1.29 | −1.66 | −0.76 | 0.72 | 0.31 | 0.33 | 0.58 |
| Shengdao 20 | 0.05 | 0.54 | 1.40 | 0.48 | 0.69 | 0.78 | 0.57 |
| Yongyou 2532 | 0.75 | 0.09 | −0.89 | 0.62 | 0.61 | 0.31 | 0.57 |
| Meixiangzhan 2 | 0.34 | 1.31 | −0.60 | 0.54 | 0.82 | 0.36 | 0.56 |
| Changjing 13-9 | 0.19 | −0.52 | 1.74 | 0.51 | 0.51 | 0.85 | 0.56 |
| Sidao 14-211 | 0.06 | 1.92 | −0.33 | 0.48 | 0.93 | 0.42 | 0.56 |
| Wujing 11036 | 0.53 | 0.19 | −0.45 | 0.57 | 0.63 | 0.40 | 0.56 |
| Huayou 14 | 0.53 | −0.51 | 0.08 | 0.57 | 0.51 | 0.51 | 0.55 |
| Lianjing 15113 | 0.59 | −0.58 | −0.24 | 0.59 | 0.50 | 0.44 | 0.55 |
| Si 15-301 | 0.61 | −1.09 | 0.16 | 0.59 | 0.41 | 0.52 | 0.54 |
| Huai 330 | 0.54 | −0.23 | −0.43 | 0.58 | 0.56 | 0.40 | 0.54 |
| Wuyunjing 31 | −0.14 | 0.64 | 1.12 | 0.45 | 0.71 | 0.72 | 0.54 |
| Yanjing 16 | 0.08 | −1.02 | 1.65 | 0.49 | 0.42 | 0.83 | 0.53 |
| Wuyujing 32 | −0.28 | 1.78 | 0.20 | 0.42 | 0.90 | 0.53 | 0.53 |
| Fujing 1601 | 0.60 | −1.47 | −0.03 | 0.59 | 0.34 | 0.48 | 0.52 |
| Yuxiangyouzhan | −0.23 | 0.85 | 0.79 | 0.43 | 0.74 | 0.65 | 0.52 |
| Ningjing 011 | 0.10 | −0.54 | 0.79 | 0.49 | 0.50 | 0.65 | 0.52 |
| Jiayou 14-10 | 0.16 | −1.49 | 1.50 | 0.50 | 0.34 | 0.80 | 0.52 |
| Xu 40398 | −0.10 | 0.69 | 0.34 | 0.45 | 0.71 | 0.56 | 0.52 |
| Yongyou 2561 | 0.83 | −0.81 | −1.69 | 0.63 | 0.46 | 0.14 | 0.52 |
| Wujing 215 | 0.11 | 0.41 | −0.21 | 0.49 | 0.67 | 0.45 | 0.52 |
| Changruan 07-9 | −0.46 | 0.58 | 1.69 | 0.38 | 0.70 | 0.84 | 0.52 |
| Qiuyoujinfeng | −0.05 | 0.69 | −0.01 | 0.46 | 0.71 | 0.49 | 0.51 |
| Su 2110 | 0.43 | 0.03 | −1.35 | 0.56 | 0.60 | 0.21 | 0.51 |
| Su 2250 | −0.20 | 1.85 | −0.78 | 0.44 | 0.91 | 0.33 | 0.51 |
| Sujing 9 | −0.36 | −0.74 | 2.19 | 0.40 | 0.47 | 0.94 | 0.50 |
| Ning 9051 | −0.30 | 1.15 | 0.03 | 0.42 | 0.79 | 0.50 | 0.50 |
| Lianjing 15 | −0.17 | −0.72 | 1.32 | 0.44 | 0.47 | 0.76 | 0.50 |
| Changnongjing 14-7 | 0.06 | 1.06 | −1.39 | 0.48 | 0.78 | 0.20 | 0.49 |
| Yongyou 5301 | 0.12 | −0.24 | −0.40 | 0.50 | 0.55 | 0.41 | 0.49 |
| Yongyou 1540 | 0.30 | −0.42 | −0.98 | 0.53 | 0.52 | 0.29 | 0.49 |
| Yongyou 28 | 0.50 | −0.93 | −1.34 | 0.57 | 0.44 | 0.21 | 0.49 |
| Shengdao 22 | −0.13 | 0.90 | −0.79 | 0.45 | 0.75 | 0.33 | 0.49 |
| Changruan 07−1 | −0.16 | 1.56 | −1.42 | 0.44 | 0.86 | 0.19 | 0.48 |
| Changnongjing 10 | 0.16 | −1.52 | 0.37 | 0.50 | 0.33 | 0.57 | 0.48 |
| Shenyou 415 | −0.21 | −0.07 | 0.31 | 0.43 | 0.58 | 0.55 | 0.48 |
| Wuyujing 36 | −0.51 | 1.00 | 0.19 | 0.38 | 0.77 | 0.53 | 0.47 |
| Chunyou 84 | 0.23 | −1.51 | −0.37 | 0.52 | 0.34 | 0.41 | 0.47 |
| Shengxiang 66 | −0.39 | 1.20 | −0.91 | 0.40 | 0.80 | 0.30 | 0.46 |
| Huajing 295 | −0.67 | 0.31 | 0.94 | 0.35 | 0.65 | 0.68 | 0.46 |
| Wu 6613 | −0.63 | 0.58 | 0.49 | 0.35 | 0.70 | 0.59 | 0.46 |
| Nanfan 1610 | −0.33 | −0.14 | 0.05 | 0.41 | 0.57 | 0.50 | 0.46 |
| Wuyunjing 23 | −0.52 | 0.33 | 0.29 | 0.37 | 0.65 | 0.55 | 0.45 |
| Nanjing 5711 | −0.81 | 0.64 | 0.57 | 0.32 | 0.71 | 0.61 | 0.44 |
| Lianjing 13 | −0.89 | 0.51 | 0.95 | 0.30 | 0.68 | 0.69 | 0.44 |
| Shengdao 18-4 | −0.41 | −0.05 | −0.34 | 0.39 | 0.59 | 0.42 | 0.43 |
| Yangyujing 2 | −0.76 | 0.82 | −0.05 | 0.33 | 0.74 | 0.48 | 0.43 |
| Xudao 9 | −0.80 | 0.78 | −0.11 | 0.32 | 0.73 | 0.47 | 0.42 |
| Yangjing 5515 | −0.69 | 0.05 | 0.15 | 0.34 | 0.60 | 0.52 | 0.42 |
| Wuyun 5245 | −0.52 | 0.52 | −0.97 | 0.37 | 0.68 | 0.29 | 0.42 |
| Ningjing 4 | −0.46 | −0.21 | −0.74 | 0.39 | 0.56 | 0.34 | 0.41 |
| Jia 67 | −0.93 | 0.68 | 0.10 | 0.30 | 0.71 | 0.51 | 0.41 |
| Changnongjing 8 | −1.02 | 0.56 | 0.51 | 0.28 | 0.69 | 0.60 | 0.41 |
| Liaoyou 9906 | −0.42 | −0.07 | −1.16 | 0.39 | 0.58 | 0.25 | 0.41 |
| Hujing 137 | −0.78 | −0.11 | 0.17 | 0.33 | 0.58 | 0.53 | 0.41 |
| Shengdao 2620 | −0.93 | −0.02 | 0.53 | 0.30 | 0.59 | 0.60 | 0.40 |
| Shenyou 26 | −0.65 | 0.68 | −1.23 | 0.35 | 0.71 | 0.24 | 0.40 |
| Wuyunjing 30 | −0.78 | 0.59 | −0.70 | 0.32 | 0.70 | 0.35 | 0.40 |
| Su 1716 | −0.81 | −0.07 | 0.01 | 0.32 | 0.58 | 0.49 | 0.40 |
| Su 1707 | −1.13 | 0.58 | 0.52 | 0.26 | 0.70 | 0.60 | 0.40 |
| W 328 | −0.64 | −1.24 | 0.42 | 0.35 | 0.38 | 0.58 | 0.39 |
| Jia 58 | −0.39 | −1.67 | −0.10 | 0.40 | 0.31 | 0.47 | 0.39 |
| Ning 5718 | −0.99 | 0.82 | −0.42 | 0.28 | 0.74 | 0.40 | 0.39 |
| Wankenjing 3 | −1.05 | 0.13 | 0.41 | 0.27 | 0.62 | 0.57 | 0.39 |
| Si 15-234 | −0.78 | −0.41 | −0.29 | 0.32 | 0.52 | 0.43 | 0.38 |
| Huaixiangjing 15 | −0.95 | −0.09 | 0.03 | 0.29 | 0.58 | 0.50 | 0.38 |
| Xinkedao 31 | −1.23 | 1.11 | −0.29 | 0.24 | 0.79 | 0.43 | 0.37 |
| Songzaoxiang 1 | −1.18 | −0.69 | 1.13 | 0.25 | 0.48 | 0.72 | 0.37 |
| Jiahe 218 | −0.62 | −2.06 | 0.36 | 0.36 | 0.24 | 0.56 | 0.37 |
| Shengdao 1647 | −1.20 | 0.28 | 0.20 | 0.25 | 0.64 | 0.53 | 0.37 |
| Sidao 15 | −0.93 | −0.41 | −0.14 | 0.30 | 0.53 | 0.46 | 0.37 |
| Xiushui 134 | −0.64 | −1.34 | −0.38 | 0.35 | 0.37 | 0.41 | 0.36 |
| Xudao 10 | −0.96 | −0.62 | 0.08 | 0.29 | 0.49 | 0.51 | 0.36 |
| Zhendao 448 | −1.24 | 0.09 | −0.07 | 0.24 | 0.61 | 0.48 | 0.35 |
| Huajing 5 | −1.62 | 0.38 | 1.01 | 0.16 | 0.66 | 0.70 | 0.34 |
| Changruan 07-4 | −1.18 | −0.09 | −0.22 | 0.25 | 0.58 | 0.44 | 0.34 |
| Changruan 07-7 | −0.76 | −2.02 | −0.26 | 0.33 | 0.25 | 0.44 | 0.33 |
| Fujing 1608 | −1.22 | 0.43 | −1.34 | 0.24 | 0.67 | 0.21 | 0.32 |
| Suxiangjing 100 | −1.20 | −0.72 | −0.56 | 0.24 | 0.47 | 0.37 | 0.31 |
| Ning 9003 | −1.62 | 0.89 | −0.69 | 0.16 | 0.75 | 0.35 | 0.30 |
| Huxiangjing 165 | −1.33 | −0.44 | −1.13 | 0.22 | 0.52 | 0.26 | 0.28 |
| Wuyun 5403 | −1.63 | −1.05 | −0.84 | 0.16 | 0.41 | 0.32 | 0.24 |
| Huxiangjing 106 | −1.92 | −1.65 | −0.57 | 0.11 | 0.31 | 0.37 | 0.19 |
| Xiangxuejing 15 | −2.48 | −3.21 | −2.36 | 0.00 | 0.04 | 0.00 | 0.01 |
| Num | Predictive D-Value | Primary Value | Difference | Evaluation Accuracy | Num | Predictive D-Value | Primary Value | Difference | Evaluation Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.523 | 0.540 | −0.018 | 96.74 | 58 | 0.659 | 0.640 | 0.019 | 97.07 |
| 2 | 0.035 | 0.008 | 0.027 | 21.79 | 59 | 0.480 | 0.482 | −0.002 | 99.63 |
| 3 | 0.524 | 0.520 | 0.004 | 99.30 | 60 | 0.511 | 0.519 | −0.008 | 98.54 |
| 4 | 0.614 | 0.605 | 0.009 | 98.60 | 61 | 0.481 | 0.460 | 0.021 | 95.67 |
| 5 | 0.390 | 0.398 | −0.008 | 97.91 | 62 | 0.610 | 0.589 | 0.021 | 96.48 |
| 6 | 0.667 | 0.661 | 0.006 | 99.11 | 63 | 0.500 | 0.509 | −0.009 | 98.16 |
| 7 | 0.654 | 0.660 | −0.007 | 99.01 | 64 | 0.496 | 0.495 | 0.001 | 99.73 |
| 8 | 0.490 | 0.493 | −0.004 | 99.28 | 65 | 0.489 | 0.483 | 0.006 | 98.79 |
| 9 | 0.589 | 0.566 | 0.023 | 96.14 | 66 | 0.746 | 0.764 | −0.018 | 97.66 |
| 10 | 0.681 | 0.681 | 0.000 | 99.99 | 67 | 0.559 | 0.545 | 0.014 | 97.57 |
| 11 | 0.654 | 0.671 | −0.017 | 97.41 | 68 | 0.392 | 0.387 | 0.005 | 98.72 |
| 12 | 0.631 | 0.625 | 0.006 | 99.11 | 69 | 0.526 | 0.531 | −0.005 | 99.12 |
| 13 | 0.624 | 0.612 | 0.012 | 98.15 | 70 | 0.584 | 0.570 | 0.014 | 97.53 |
| 14 | 0.491 | 0.491 | 0.000 | 99.99 | 71 | 0.592 | 0.599 | −0.008 | 98.70 |
| 15 | 0.381 | 0.379 | 0.002 | 99.51 | 72 | 0.361 | 0.373 | −0.012 | 96.76 |
| 16 | 0.611 | 0.649 | −0.038 | 94.17 | 73 | 0.353 | 0.344 | 0.009 | 97.59 |
| 17 | 0.399 | 0.400 | −0.001 | 99.65 | 74 | 0.487 | 0.475 | 0.013 | 97.43 |
| 18 | 0.419 | 0.405 | 0.014 | 96.56 | 75 | 0.363 | 0.368 | −0.005 | 98.63 |
| 19 | 0.325 | 0.330 | −0.005 | 98.44 | 76 | 0.371 | 0.366 | 0.004 | 98.84 |
| 20 | 0.233 | 0.235 | −0.002 | 99.05 | 77 | 0.351 | 0.342 | 0.009 | 97.53 |
| 21 | 0.388 | 0.397 | −0.009 | 97.75 | 78 | 0.375 | 0.366 | 0.009 | 97.68 |
| 22 | 0.671 | 0.645 | 0.025 | 96.21 | 79 | 0.506 | 0.498 | 0.008 | 98.40 |
| 23 | 0.519 | 0.515 | 0.004 | 99.23 | 80 | 0.461 | 0.455 | 0.006 | 98.69 |
| 24 | 0.552 | 0.550 | 0.002 | 99.72 | 81 | 0.568 | 0.558 | 0.010 | 98.27 |
| 25 | 0.357 | 0.363 | −0.007 | 98.11 | 82 | 0.446 | 0.435 | 0.012 | 97.42 |
| 26 | 0.594 | 0.564 | 0.030 | 94.92 | 83 | 0.415 | 0.439 | −0.023 | 94.73 |
| 27 | 0.610 | 0.609 | 0.001 | 99.86 | 84 | 0.576 | 0.594 | −0.017 | 97.06 |
| 28 | 0.772 | 0.751 | 0.021 | 97.28 | 85 | 0.491 | 0.486 | 0.006 | 98.87 |
| 29 | 0.645 | 0.657 | −0.012 | 98.18 | 86 | 0.433 | 0.420 | 0.013 | 97.10 |
| 30 | 0.560 | 0.556 | 0.004 | 99.26 | 87 | 0.401 | 0.390 | 0.012 | 97.08 |
| 31 | 0.506 | 0.520 | −0.014 | 97.38 | 88 | 0.385 | 0.380 | 0.006 | 98.57 |
| 32 | 0.646 | 0.647 | −0.001 | 99.88 | 89 | 0.560 | 0.544 | 0.017 | 96.99 |
| 33 | 0.740 | 0.724 | 0.016 | 97.88 | 90 | 0.365 | 0.363 | 0.002 | 99.35 |
| 34 | 0.353 | 0.346 | 0.007 | 97.88 | 91 | 0.471 | 0.458 | 0.013 | 97.17 |
| 35 | 0.524 | 0.517 | 0.008 | 98.50 | 92 | 0.596 | 0.620 | −0.024 | 96.14 |
| 36 | 0.572 | 0.580 | −0.007 | 98.76 | 93 | 0.522 | 0.525 | −0.003 | 99.39 |
| 37 | 0.589 | 0.596 | −0.007 | 98.86 | 94 | 0.526 | 0.545 | −0.019 | 96.60 |
| 38 | 0.219 | 0.188 | 0.031 | 85.74 | 95 | 0.722 | 0.732 | −0.010 | 98.58 |
| 39 | 0.384 | 0.367 | 0.017 | 95.52 | 96 | 0.264 | 0.282 | −0.018 | 93.57 |
| 40 | 0.415 | 0.393 | 0.022 | 94.60 | 97 | 0.387 | 0.401 | −0.014 | 96.63 |
| 41 | 0.391 | 0.395 | −0.004 | 98.91 | 98 | 0.533 | 0.529 | 0.004 | 99.29 |
| 42 | 0.471 | 0.455 | 0.017 | 96.49 | 99 | 0.837 | 0.829 | 0.007 | 99.14 |
| 43 | 0.295 | 0.307 | −0.012 | 96.00 | 100 | 0.732 | 0.735 | −0.002 | 99.71 |
| 44 | 0.398 | 0.408 | −0.010 | 97.58 | 101 | 0.311 | 0.304 | 0.007 | 97.81 |
| 45 | 0.374 | 0.392 | −0.018 | 95.36 | 102 | 0.414 | 0.407 | 0.007 | 98.30 |
| 46 | 0.619 | 0.641 | −0.022 | 96.51 | 103 | 0.413 | 0.411 | 0.002 | 99.41 |
| 47 | 0.526 | 0.503 | 0.023 | 95.60 | 104 | 0.795 | 0.775 | 0.020 | 97.48 |
| 48 | 0.601 | 0.588 | 0.013 | 97.85 | 105 | 0.503 | 0.520 | −0.017 | 96.71 |
| 49 | 0.435 | 0.419 | 0.016 | 96.37 | 106 | 0.590 | 0.597 | −0.007 | 98.90 |
| 50 | 0.478 | 0.481 | −0.003 | 99.41 | 107 | 0.431 | 0.437 | −0.006 | 98.70 |
| 51 | 0.468 | 0.509 | −0.041 | 92.00 | 108 | 0.290 | 0.317 | −0.027 | 91.40 |
| 52 | 0.499 | 0.501 | −0.002 | 99.58 | 109 | 0.715 | 0.728 | −0.013 | 98.21 |
| 53 | 0.480 | 0.487 | −0.007 | 98.58 | 110 | 0.443 | 0.430 | 0.013 | 97.13 |
| 54 | 0.415 | 0.409 | 0.006 | 98.54 | 111 | 0.392 | 0.422 | −0.030 | 92.78 |
| 55 | 0.468 | 0.465 | 0.003 | 99.35 | 112 | 0.617 | 0.690 | −0.073 | 89.41 |
| 56 | 0.454 | 0.456 | −0.002 | 99.48 | 113 | 0.521 | 0.524 | −0.004 | 99.31 |
| 57 | 0.515 | 0.520 | −0.005 | 99.07 | 114 | 0.581 | 0.565 | 0.017 | 97.13 |
| STI-GE | STI-GC | STI-SL | STI-RL | STI-RN | STI-PFW | STI-SVI | D-Value | |
|---|---|---|---|---|---|---|---|---|
| STI-GE | 1.00 | |||||||
| STI-GC | 0.41 ** | 1.00 | ||||||
| STI-SL | 0.34 ** | 0.29 ** | 1.00 | |||||
| STI-RL | 0.36 ** | 0.19 * | 0.72 ** | 1.00 | ||||
| STI-RN | 0.11 | 0.32 ** | 0.41 ** | 0.22 * | 1.00 | |||
| STI-PFW | 0.29 ** | 0.15 | 0.81 ** | 0.68 ** | 0.43 ** | 1.00 | ||
| STI-SVI | 0.44 ** | 0.40 ** | 0.82 ** | 0.96 ** | 0.31 ** | 0.74 ** | 1.00 | |
| D-value | 0.51 ** | 0.63 ** | 0.81 ** | 0.69 ** | 0.71 ** | 0.75 ** | 0.83 ** | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Hussain, S.; Wang, Y.; Liu, Y.; Li, Q.; Chen, Y.; Wei, H.; Gao, P.; Dai, Q. Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy 2021, 11, 1569. https://doi.org/10.3390/agronomy11081569
Zhang R, Hussain S, Wang Y, Liu Y, Li Q, Chen Y, Wei H, Gao P, Dai Q. Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy. 2021; 11(8):1569. https://doi.org/10.3390/agronomy11081569
Chicago/Turabian StyleZhang, Rui, Shahid Hussain, Yang Wang, Yonghao Liu, Qing Li, Yinglong Chen, Huanhe Wei, Pinglei Gao, and Qigen Dai. 2021. "Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage" Agronomy 11, no. 8: 1569. https://doi.org/10.3390/agronomy11081569
APA StyleZhang, R., Hussain, S., Wang, Y., Liu, Y., Li, Q., Chen, Y., Wei, H., Gao, P., & Dai, Q. (2021). Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy, 11(8), 1569. https://doi.org/10.3390/agronomy11081569






