Discovery of Anthocyanin Biosynthetic Pathway in Cosmos caudatus Kunth. Using Omics Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Cultivation and Maintenance
2.2. RNA Extraction for Transcriptomic Analysis
2.3. cDNA Library Construction and Illumina Sequencing
2.4. De Novo Transcriptome Assembly
2.5. Gene Annotation and Classification
2.6. Metabolite Extraction
2.7. LC-MS Analysis
2.8. Statistical and Multivariate Analysis
2.9. Anatomy Studies
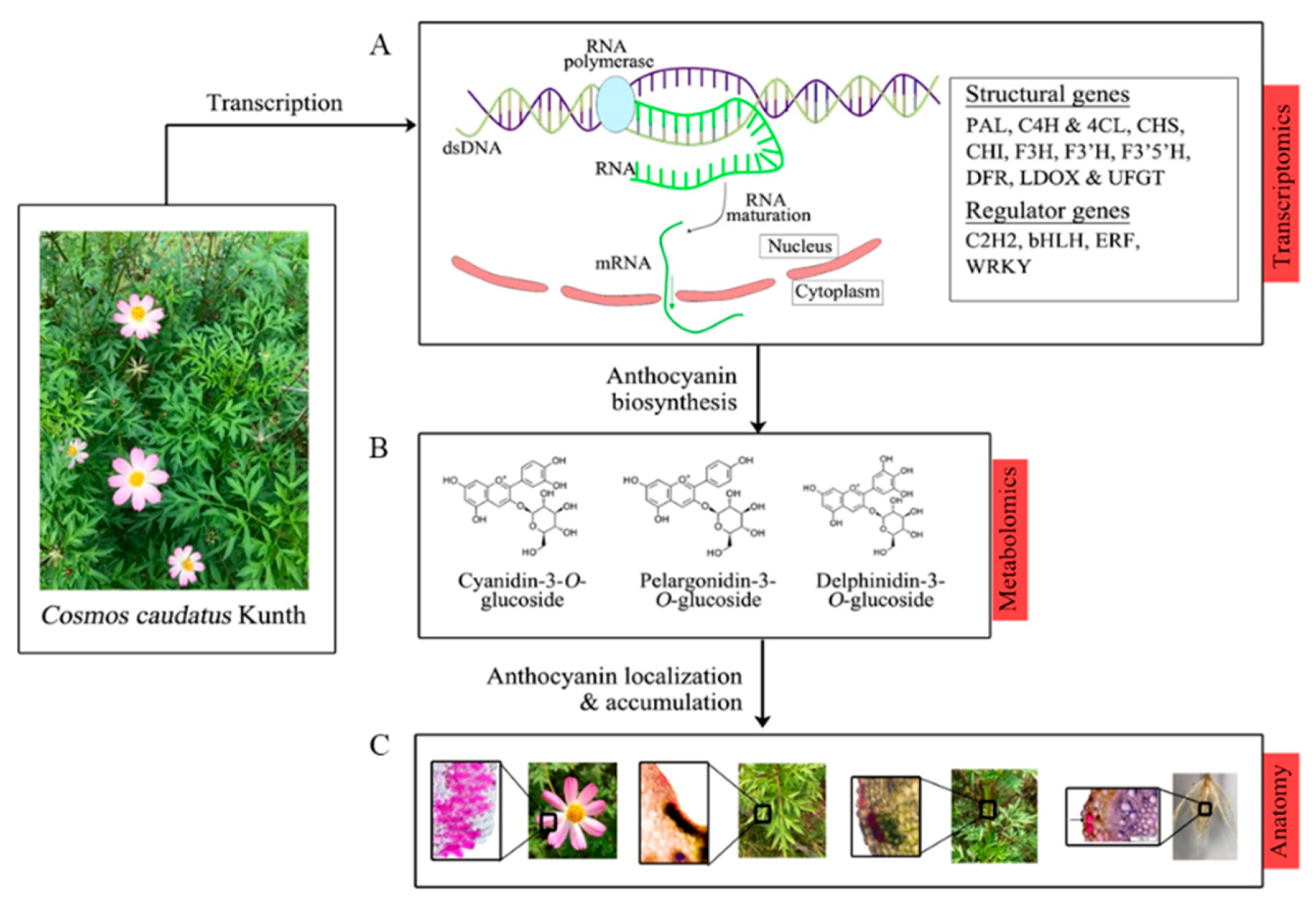
3. Results
3.1. Transcriptome Analysis
3.1.1. High-Throughput Sequencing and De Novo Sequence Assembly of C. caudatus Transcriptome
3.1.2. Functional Annotation of Unigenes
3.1.3. Candidate Genes for Anthocyanin Biosynthesis
3.1.4. Candidate Transcription Factors (TFs) Involved in Anthocyanin Biosynthesis and Transport
3.2. Metabolomic Analysis
3.3. Anatomy Data
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ha, U.; Bae, W.J.; Kim, S.J.; Yoon, B., II; Hong, S.H.; Lee, J.Y.; Hwang, T.-K.; Hwang, S.Y.; Wang, Z.; Kim, S.W. Anthocyanin induces apoptosis of DU-145 cells in vitro and inhibits xenograft growth of prostate cancer. Yonsei Med. J. 2015, 56, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kruger, M.J.; Davies, N.; Myburgh, K.H.; Lecour, S. Proanthocyanidins, anthocyanins and cardiovascular diseases. Food Res. Int. 2014, 59, 41–52. [Google Scholar] [CrossRef]
- Wallace, T.C. Anthocyanins in cardiovascular disease. Adv. Nutr. 2011, 2, 1–7. [Google Scholar] [CrossRef]
- Sancho, R.A.S.; Pastore, G.M. Evaluation of the effects of anthocyanins in type 2 diabetes. Food Res. Int. 2012, 46, 378–386. [Google Scholar] [CrossRef]
- Peiffer, D.S.; Zimmerman, N.P.; Wang, L.-S.; Ransom, B.W.S.; Carmella, S.G.; Kuo, C.-T.; Siddiqui, J.; Chen, J.-H.; Oshima, K.; Huang, Y.-W. Chemoprevention of esophageal cancer with black raspberries, their component anthocyanins, and a major anthocyanin metabolite, protocatechuic acid. Cancer Prev. Res. 2014, 7, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shen, X.; Chen, Y. Effect of pH, temperature and iron on the stability of anthocyanins from black-skinned peanuts (Arachis hypogaea L.). Afr. J. Agric. Res. 2013, 8, 2044–2047. [Google Scholar]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, R.N.; Santos, D.T.; Meireles, M.A.A. Non-thermal stabilization mechanisms of anthocyanins in model and food systems—An overview. Food Res. Int. 2011, 44, 499–509. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef]
- Lim, S.-H.; Ha, S.-H. Marker development for the identification of rice seed color. Plant Biotechnol. Rep. 2013, 7, 391–398. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Andarwulan, N.; Batari, R.; Sandrasari, D.A.; Bolling, B.; Wijaya, H. Flavonoid content and antioxidant activity of vegetables from Indonesia. Food Chem. 2010, 121, 1231–1235. [Google Scholar] [CrossRef]
- Sumazian, Y.; Syahida, A.; Hakiman, M.; Maziah, M. Antioxidant activities, flavonoids, ascorbic acid and phenolic content of Malaysian vegetables. J. Med. Plants Res. 2010, 4, 881–890. [Google Scholar]
- Shipp, J.; Abdel-Aal, E.-S.M. Food applications and physiological effects of anthocyanins as functional food ingredients. Open Food Sci. J. 2010, 4, 1. [Google Scholar] [CrossRef]
- Bunawan, H.; Baharum, S.N.; Bunawan, S.N.; Amin, N.M.; Noor, N.M. Cosmos caudatus Kunth.: A traditional medicinal herb. Glob. J. Pharmacol. 2014, 8, 420–426. [Google Scholar]
- Mohamed, N.; Gwee Sian Khee, S.; Shuid, A.N.; Muhammad, N.; Suhaimi, F.; Othman, F.; Babji, A.S.; Soelaiman, I.-N. The Effects of Cosmos caudatus on Structural Bone Histomorphometry in Ovariectomized Rats. Evidence-Based Complement. Altern. Med. 2012, 2012, 1–6. [Google Scholar] [CrossRef]
- Mohamed, N.; Sahhugi, Z.; Ramli, E.S.M.; Muhammad, N. The effects of Cosmos caudatus (ulam raja) on dynamic and cellular bone histomorphometry in ovariectomized rats. BMC Res. Notes 2013, 6, 239. [Google Scholar] [CrossRef]
- Amalia, L.; Anggadiredja, K.; Sukrasno, I.F.; Inggriani, R. Antihypertensive Potency of Wild Cosmos Caudatus Leaf Extract.Pdf. J. Pharmacol. Toxicol. 2012, 7, 359–368. [Google Scholar] [CrossRef][Green Version]
- Shui, G.; Leong, L.P.; Shih, P.W. Rapid screening and characterisation of antioxidants of Cosmos caudatus using liquid chromatography coupled with mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 827, 127–138. [Google Scholar] [CrossRef]
- Wong, S.P.; Leong, L.P.; Koh, J.H.W.; William Koh, J.H. Antioxidant activities of aqueous extracts of selected plants. Food Chem. 2006, 99, 775–783. [Google Scholar] [CrossRef]
- Reihani, S.F.S.; Azhar, M.E. Antioxidant activity and total phenolic content in aqueous extracts of selected traditional Malay salads (Ulam). Int. Food Res. J. 2012, 19, 4. [Google Scholar]
- Rasdi, N.H.M.; Samah, O.A.; Sule, A.; Ahmed, Q.U. Antimicrobial studies of Cosmos caudatus Kunth. (compositae). J. Med. Plants Res. 2010, 4, 669–673. [Google Scholar]
- Fuzzati, N.; Dyatmiko, W.; Rahman, A.; Hostettmann, K. Phenylpropane derivatives from roots of Cosmos caudatus. Phytochemistry 1995, 39, 409–412. [Google Scholar] [CrossRef]
- Andarwulan, N.; Kurniasih, D.; Apriady, R.A.; Rahmat, H.; Roto, A.V.; Bolling, B.W. Polyphenols, carotenoids, and ascorbic acid in underutilized medicinal vegetables. J. Funct. Foods 2012, 4, 339–347. [Google Scholar] [CrossRef]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef]
- Ward, J.A.; Ponnala, L.; Weber, C.A. Strategies for transcriptome analysis in nonmodel plants. Am. J. Bot. 2012, 99, 267–276. [Google Scholar] [CrossRef]
- Loke, K.-K.; Rahnamaie-Tajadod, R.; Yeoh, C.-C.; Goh, H.-H.; Mohamed-Hussein, Z.-A.; Zainal, Z.; Ismail, I.; Noor, N.M. Transcriptome analysis of Polygonum minus reveals candidate genes involved in important secondary metabolic pathways of phenylpropanoids and flavonoids. PeerJ 2017, 5, e2938. [Google Scholar] [CrossRef] [PubMed]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778. [Google Scholar] [CrossRef]
- Naoumkina, M.; Farag, M.A.; Sumner, L.W.; Tang, Y.; Liu, C.-J.; Dixon, R.A. Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2007, 104, 17909–17915. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; von Roepenack-Lahaye, E.; Schmidt, J.; Schmotz, C.; Neumann, S.; Scheel, D.; Clemens, S. Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 2008, 147, 2107–2120. [Google Scholar] [CrossRef]
- Farag, M.A.; Huhman, D.V.; Dixon, R.A.; Sumner, L.W. Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 2008, 146, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Hirai, M.Y.; Sasaki, E.; Akiyama, K.; Yonekura-Sakakibara, K.; Provart, N.J.; Sakurai, T.; Shimada, Y.; Saito, K. AtMetExpress development: A phytochemical atlas of Arabidopsis development. Plant Physiol. 2010, 152, 566–578. [Google Scholar] [CrossRef]
- Matsuda, F.; Yonekura-Sakakibara, K.; Niida, R.; Kuromori, T.; Shinozaki, K.; Saito, K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009, 57, 555–577. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Recent advances of metabolomics in plant biotechnology. Plant Biotechnol. Rep. 2012, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Kamide, Y.; Hirai, M.Y.; Saito, K. Plant lipidomics based on hydrophilic interaction chromatography coupled to ion trap time-of-flight mass spectrometry. Metabolomics 2013, 9, 121–131. [Google Scholar] [CrossRef]
- Shinozaki, K.; Sakakibara, H. Omics and bioinformatics: An essential toolbox for systems analyses of plant functions beyond 2010 Plant Cell Physiol. Plant Cell Physiol. 2009, 50, 1177–1780. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gunasekaran, D.; Bunawan, H.; Ismail, I.; Noor, N.M. Data on Fourier transform-infrared of Cosmos caudatus Kunth. tissues analyzed with chemometric analysis. Data Br. 2018, 19, 1423–1427. [Google Scholar] [CrossRef]
- Mediani, A.; Abas, F.; Khatib, A.; Maulidiani, H.; Shaari, K.; Choi, Y.H.; Lajis, N.H. 1H-NMR-based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res. Int. 2012, 49, 763–770. [Google Scholar] [CrossRef]
- Javadi, N.; Abas, F.; Hamid, A.A.; Simoh, S.; Shaari, K.; Ismail, I.S.; Mediani, A.; Khatib, A. GC-MS-Based Metabolite Profiling of Cosmos caudatus Leaves Possessing Alpha-Glucosidase Inhibitory Activity. J. Food Sci. 2014, 79, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rahman, A.; Goh, H.H.; Loke, K.K.; Noor, N.M.; Aizat, W.M. RNA-seq analysis of mangosteen (Garcinia mangostana L.) fruit ripening. Genom. Data 2017, 12, 159–160. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Garzón, G.A.; Narváez, C.E.; Riedl, K.M.; Schwartz, S.J. Chemical composition, anthocyanins, non-anthocyanin phenolics and antioxidant activity of wild bilberry (Vaccinium meridionale Swartz) from Colombia. Food Chem. 2010, 122, 980–986. [Google Scholar] [CrossRef]
- Rosli, M.A.F.; Azizan, K.A.; Baharum, S.N.; Goh, H.H. Mass spectrometry data of metabolomics analysis of Nepenthes pitchers. Data Br. 2017, 14, 295–297. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Rohs, R.; Jin, X.; West, S.M.; Joshi, R.; Honig, B.; Mann, R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010, 79, 233–269. [Google Scholar] [CrossRef]
- Hudson, W.H.; Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15, 749–760. [Google Scholar] [CrossRef]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, R.; Jaiswal, A.K. Enzymes in bioconversion and food processing. In Enzymes in Food Technology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 19–40. [Google Scholar]
- Dahl, S.G.; Sylte, I.; Ravna, A.W. Structures and models of transporter proteins. J. Pharmacol. Exp. Ther. 2004, 309, 853–860. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Choi, S.R.; Chhapekar, S.S.; Kim, M.-S.; Singh, S.; Yi, S.Y.; Oh, S.H.; Kim, H.; Lee, C.Y.; Oh, M.-H. Red Chinese cabbage transcriptome analysis reveals structural genes and multiple transcription factors regulating reddish purple color. Int. J. Mol. Sci. 2020, 21, 2901. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Mihaleva, V.V.; Vorst, O.; Maliepaard, C.; Verhoeven, H.A.; de Vos, R.C.H.; Hall, R.D.; van Ham, R.C.H.J. Accurate mass error correction in liquid chromatography time-of-flight mass spectrometry based metabolomics. Metabolomics 2008, 4, 171–182. [Google Scholar] [CrossRef]
- Guo, B.; Wang, M.; Liu, Y.; Zhou, J.; Dai, H.; Huang, Z.; Shen, L.; Zhang, Q.; Chen, B. Wide-scope screening of illegal adulterants in dietary and herbal supplements via rapid polarity-switching and multistage accurate mass confirmation using an LC-IT/TOF hybrid instrument. J. Agric. Food Chem. 2015, 63, 6954–6967. [Google Scholar] [CrossRef] [PubMed]
- Stroh, J.G.; Petucci, C.J.; Brecker, S.J.; Huang, N.; Lau, J.M. Automated sub-ppm mass accuracy on an ESI-TOF for use with drug discovery compound libraries. J. Am. Soc. Mass Spectrom. 2007, 18, 1612–1616. [Google Scholar] [CrossRef]
- V. González de Peredo, A.; Vázquez-Espinosa, M.; Espada-Bellido, E.; Jiménez-Cantizano, A.; Ferreiro-González, M.; Amores-Arrocha, A.; Palma, M.; G. Barroso, C.; F. Barbero, G. Development of new analytical microwave-assisted extraction methods for bioactive compounds from myrtle (myrtus communis L.). Molecules 2018, 23, 2992. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Zaima, N.; Moriyama, T.; Kawamura, Y. Different localization patterns of anthocyanin species in the pericarp of black rice revealed by imaging mass spectrometry. PLoS ONE 2012, 7, e31285. [Google Scholar] [CrossRef]
- Yang, H.; Kim, H.W.; Kwon, Y.S.; Kim, H.K.; Sung, S.H. Fast and Simple Discriminative Analysis of Anthocyanins-Containing Berries Using LC/MS Spectral Data. Phytochem. Anal. 2017, 28, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Cooney, J.M.; Jensen, D.J.; McGhie, T.K. LC-MS identification of anthocyanins in boysenberry extract and anthocyanin metabolites in human urine following dosing. J. Sci. Food Agric. 2004, 84, 237–245. [Google Scholar] [CrossRef]
- Di Stefano, V.; Pitonzo, R.; Novara, M.E.; Bongiorno, D.; Indelicato, S.; Gentile, C.; Avellone, G.; Bognanni, R.; Scandurra, S.; Melilli, M.G. Antioxidant activity and phenolic composition in pomegranate (Punica granatum L.) genotypes from south Italy by UHPLC–Orbitrap-MS approach. J. Sci. Food Agric. 2019, 99, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Slimestad, R.; Solheim, H. Anthocyanins from black currants (Ribes nigrum L.). J. Agric. Food Chem. 2002, 50, 3228–3231. [Google Scholar] [CrossRef]
- Diretto, G.; Jin, X.; Capell, T.; Zhu, C.; Gomez-Gomez, L. Differential accumulation of pelargonidin glycosides in petals at three different developmental stages of the orange-flowered gentian (Gentiana lutea L. var. aurantiaca). PLoS ONE 2019, 14, e0212062. [Google Scholar] [CrossRef]
- Longo, L.; Vasapollo, G. Anthocyanins from bay (Laurus nobilis L.) berries. J. Agric. Food Chem. 2005, 53, 8063–8067. [Google Scholar] [CrossRef] [PubMed]
- Stashenko, E.; Mejía, J.; Sierra, L.J.; Bernal, D.C.; Gualteros, L.; Carrillo, K.; Martínez, J.R. Flower metabolomics: Volatile compound profile, antioxidant capacity and lc-ms-tof identification of flavonoids in various tropical flowers. Planta Med. 2015, 81, PW_201. [Google Scholar] [CrossRef]
- Banaszewski, K.; Park, E.; Edirisinghe, I.; Cappozzo, J.C.; Burton-Freeman, B.M. A pilot study to investigate bioavailability of strawberry anthocyanins and characterize postprandial plasma polyphenols absorption patterns by Q-TOF LC/MS in humans. J. Berry Res. 2013, 3, 113–126. [Google Scholar] [CrossRef]
- Iwashina, T. Flavonoid properties of five families newly incorporated into the order Caryophyllales. Bull. Natl. Mus. Nat. Sci. 2013, 39, 25–51. [Google Scholar]
- Cabrita, L.; Frøystein, N.Å.; Andersen, Ø.M. Anthocyanin trisaccharides in blue berries of Vaccinium padifolium. Food Chem. 2000, 69, 33–36. [Google Scholar] [CrossRef]
- Giordano, L.; Coletta, W.; Rapisarda, P.; Donati, M.B.; Rotilio, D. Development and validation of an LC-MS/MS analysis for simultaneous determination of delphinidin-3-glucoside, cyanidin-3-glucoside and cyanidin-3-(6-malonylglucoside) in human plasma and urine after blood orange juice administration. J. Sep. Sci. 2007, 30, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Stein-Chisholm, R.E.; Beaulieu, J.C.; Grimm, C.C.; Lloyd, S.W. LC–MS/MS and UPLC–UV evaluation of anthocyanins and anthocyanidins during rabbiteye blueberry juice processing. Beverages 2017, 3, 56. [Google Scholar] [CrossRef]
- Li, D.; Meng, X.; Li, B. Profiling of anthocyanins from blueberries produced in China using HPLC-DAD-MS and exploratory analysis by principal component analysis. J. Food Compos. Anal. 2016, 47, 1–7. [Google Scholar] [CrossRef]
- Šavikin, K.; Zdunić, G.; Janković, T.; Gođevac, D.; Stanojković, T.; Pljevljakušić, D. Berry fruit teas: Phenolic composition and cytotoxic activity. Food Res. Int. 2014, 62, 677–683. [Google Scholar] [CrossRef]
- Mertz, C.; Gancel, A.-L.; Gunata, Z.; Alter, P.; Dhuique-Mayer, C.; Vaillant, F.; Perez, A.M.; Ruales, J.; Brat, P. Phenolic compounds, carotenoids and antioxidant activity of three tropical fruits. J. Food Compos. Anal. 2009, 22, 381–387. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, S.Z.; Cheng, Y.W.; Ya, H.Y.; Han, J.M. Transcriptome analysis and anthocyanin- related genes in red leaf lettuce. Genet Mol Res. 2016, 15, 1–11. [Google Scholar] [CrossRef]
- Tu, Y.; Liu, F.; Guo, D.; Fan, L.; Zhu, Z.; Xue, Y.; Gao, Y.; Guo, M. Molecular characterization of flavanone 3-hydroxylase gene and flavonoid accumulation in two chemotyped safflower lines in response to methyl jasmonate stimulation. BMC Plant Biol. 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khumkarjorn, N.; Thanonkeo, S.; Yamada, M.; Thanonkeo, P. Cloning and expression analysis of a flavanone 3-hydroxylase gene in Ascocenda orchid. J. Plant Biochem. Biotechnol. 2017, 26, 179–190. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, S.; Yadav, S.K.; Ahuja, P.S. Characterization of dihydroflavonol 4-reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze]. Plant Biotechnol. Rep. 2009, 3, 95–101. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.-I.; Jung, H.-J.; Yang, T.-J.; Hur, Y.; Nou, I.-S. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 2014, 550, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Kobayashi, M.; Gong, Z.; Tanaka, Y.; Yamazaki, M. Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: Molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 1999, 17, 181–189. [Google Scholar] [CrossRef]
- Chuan-Li, Z.; Peng, C.; Yue-Ming, Z.; Chang-Yuan, Z.; Dan-Hong, S.; Fei, J. Full Length cDNA Cloning and Expression Analysis of UFGT Gene from Ginkgo biloba. Acta Hortic. Sin. 2012, 39, 1903. [Google Scholar]
- Wu, X.; Gong, Q.; Ni, X.; Zhou, Y.; Gao, Z. UFGT: The key enzyme associated with the petals variegation in Japanese apricot. Front. Plant Sci. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Davies, C.; Robinson, S.P. Anthocyanin composition and anthocyanin pathway gene expression in grapevine sports differing in berry skin colour. Aust. J. Grape Wine Res. 1996, 2, 163–170. [Google Scholar] [CrossRef]
- Boss, P.K.; Davies, C.; Robinson, S.P. Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 1996, 32, 565–569. [Google Scholar] [CrossRef]
- Hu, C.; Gong, Y.; Jin, S.; Zhu, Q. Molecular analysis of a UDP-glucose: Flavonoid 3-O-glucosyltransferase (UFGT) gene from purple potato (Solanum tuberosum). Mol. Biol. Rep. 2011, 38, 561–567. [Google Scholar] [CrossRef]
- Stommel, J.R.; Lightbourn, G.J.; Winkel, B.S.; Griesbach, R.J. Transcription factor families regulate the anthocyanin biosynthetic pathway in Capsicum annuum. J. Am. Soc. Hortic. Sci. 2009, 134, 244–251. [Google Scholar] [CrossRef]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN 1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.E.; Teeri, T.H. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant. Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant. J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Chiu, L.-W.; Zhou, X.; Burke, S.; Wu, X.; Prior, R.L.; Li, L. The purple cauliflower arises from activation of a MYB transcription factor. Plant. Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Hosokawa, M.; Hoshino, A.; Kitamura, Y.; Morita, Y.; Park, K.-I.; Nakashima, A.; Deguchi, A.; Tatsuzawa, F.; Doi, M. A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). J. Exp. Bot. 2011, 62, 5105–5116. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant. Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yang, Y.; Gao, J.; Ma, C.; Bi, Y. A comparative transcriptome analysis of a wild purple potato and its red mutant provides insight into the mechanism of anthocyanin transformation. PLoS ONE 2018, 13, e0191406. [Google Scholar] [CrossRef]
- Jaakola, L.; Poole, M.; Jones, M.O.; Kämäräinen-Karppinen, T.; Koskimäki, J.J.; Hohtola, A.; Häggman, H.; Fraser, P.D.; Manning, K.; King, G.J. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant. Physiol. 2010, 153, 1619–1629. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, H.; Wang, N.; Jiang, S.; Fang, H.; Zhang, Z.; Yang, G.; Wang, Y.; Su, M.; Xu, L. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant. Mol. Biol. 2018, 98, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S. Map-based cloning of the pear gene MYB 114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant. J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- An, J.; Zhang, X.; You, C.; Bi, S.; Wang, X.; Hao, Y. Md WRKY 40 promotes wounding-induced anthocyanin biosynthesis in association with Md MYB 1 and undergoes Md BT 2-mediated degradation. New Phytol. 2019, 224, 380–395. [Google Scholar] [CrossRef]
- Cyboran-Mikołajczyk, S.; Jurkiewicz, P.; Hof, M.; Kleszczyńska, H. The Impact of O-Glycosylation on Cyanidin Interaction with POPC Membranes: Structure-Activity Relationship. Molecules 2018, 23, 2771. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Ren, J.; Liu, Z.; Chen, W.; Xu, H.; Feng, H. Anthocyanin degrading and chlorophyll accumulation lead to the formation of bicolor leaf in ornamental kale. Int. J. Mol. Sci. 2019, 20, 603. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Collins, T.M. Phylogenetic and ontogenetic influences on the distribution of anthocyanins and betacyanins in leaves of tropical plants. Int. J. Plant. Sci. 2001, 162, 1141–1153. [Google Scholar] [CrossRef]
- Aras, A.; Cevahir, G.; Yentür, S.; Eryılmaz, F.; Sarsağ, M.; Çağ, S. Investigation of anthocyanin localization in various parts of Impatiens balsamina L. Biotechnol. Biotechnol. Equip. 2007, 21, 69–73. [Google Scholar] [CrossRef][Green Version]
- Zhu, H.; Zhang, T.J.; Zhang, P.; Peng, C.L. Pigment patterns and photoprotection of anthocyanins in the young leaves of four dominant subtropical forest tree species in two successional stages under contrasting light conditions. Tree Physiol. 2016, 36, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.-J.; Chow, W.S.; Liu, X.-T.; Zhang, P.; Liu, N.; Peng, C.-L. A magic red coat on the surface of young leaves: Anthocyanins distributed in trichome layer protect Castanopsis fissa leaves from photoinhibition. Tree Physiol. 2016, 36, 1296–1306. [Google Scholar] [CrossRef]
- Hong, Y.; Tang, X.; Huang, H.; Zhang, Y.; Dai, S. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Suzuki, T.; Nakatsuka, T.; Dohra, H.; Yamagishi, M.; Matsuyama, K.; Matsuura, H. RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genom. 2016, 17, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Li, S.; Feng, C.; Hong, Y.; Huang, H.; Wang, J.; Wang, L.; Dai, S. Metabolite and gene expression analysis reveal the molecular mechanism for petal colour variation in six Centaurea cyanus cultivars. Plant. Physiol. Biochem. 2019, 142, 22–33. [Google Scholar] [CrossRef]
- Seyedreihani, S.F.; Tan, T.-C.; Alkarkhi, A.F.M.; Easa, A.M. Total phenolic content and antioxidant activity of Ulam raja (Cosmos caudatus) and quantification of its selected marker compounds: Effect of extraction. Int. J. Food Prop. 2017, 20, 260–270. [Google Scholar] [CrossRef]
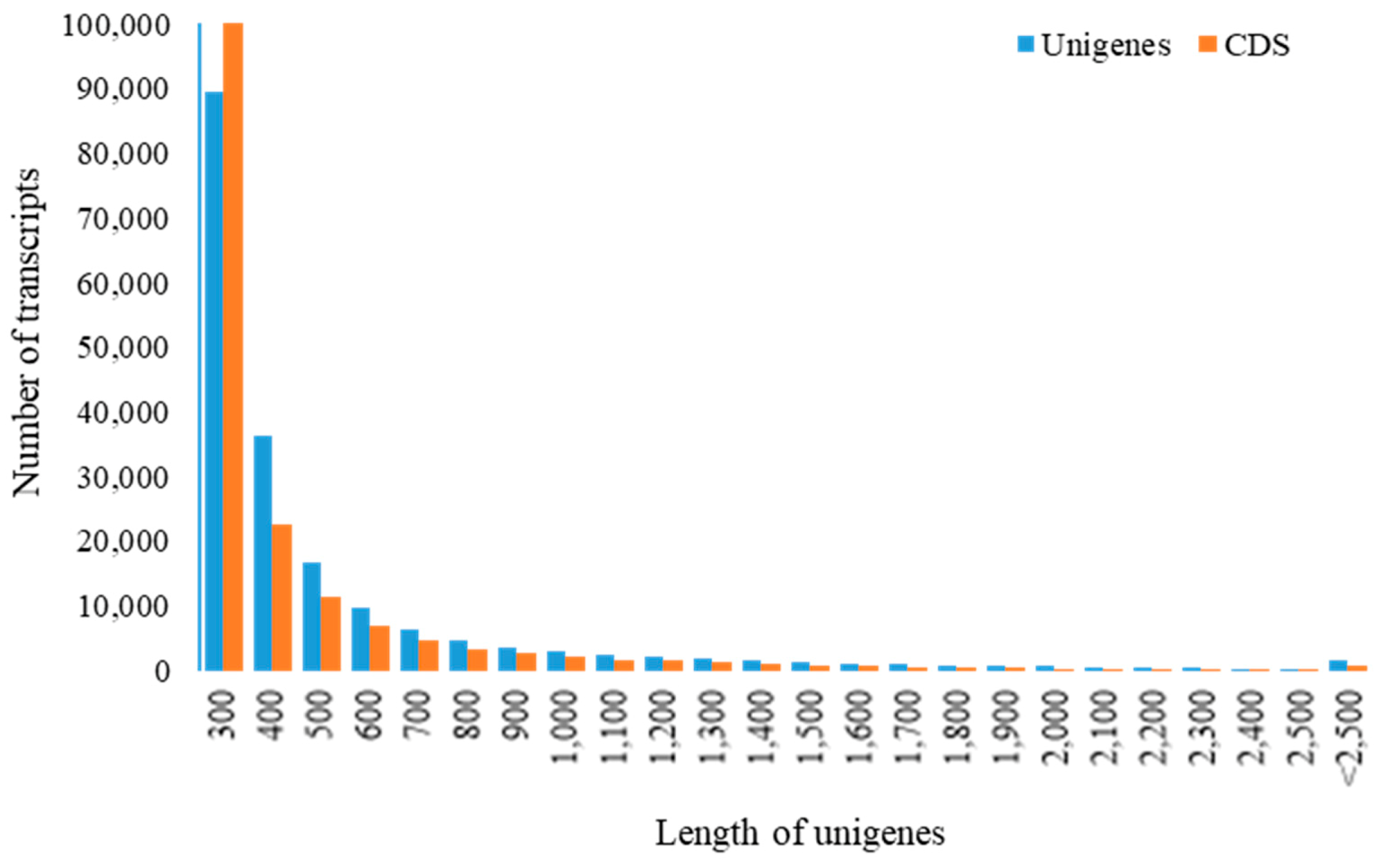
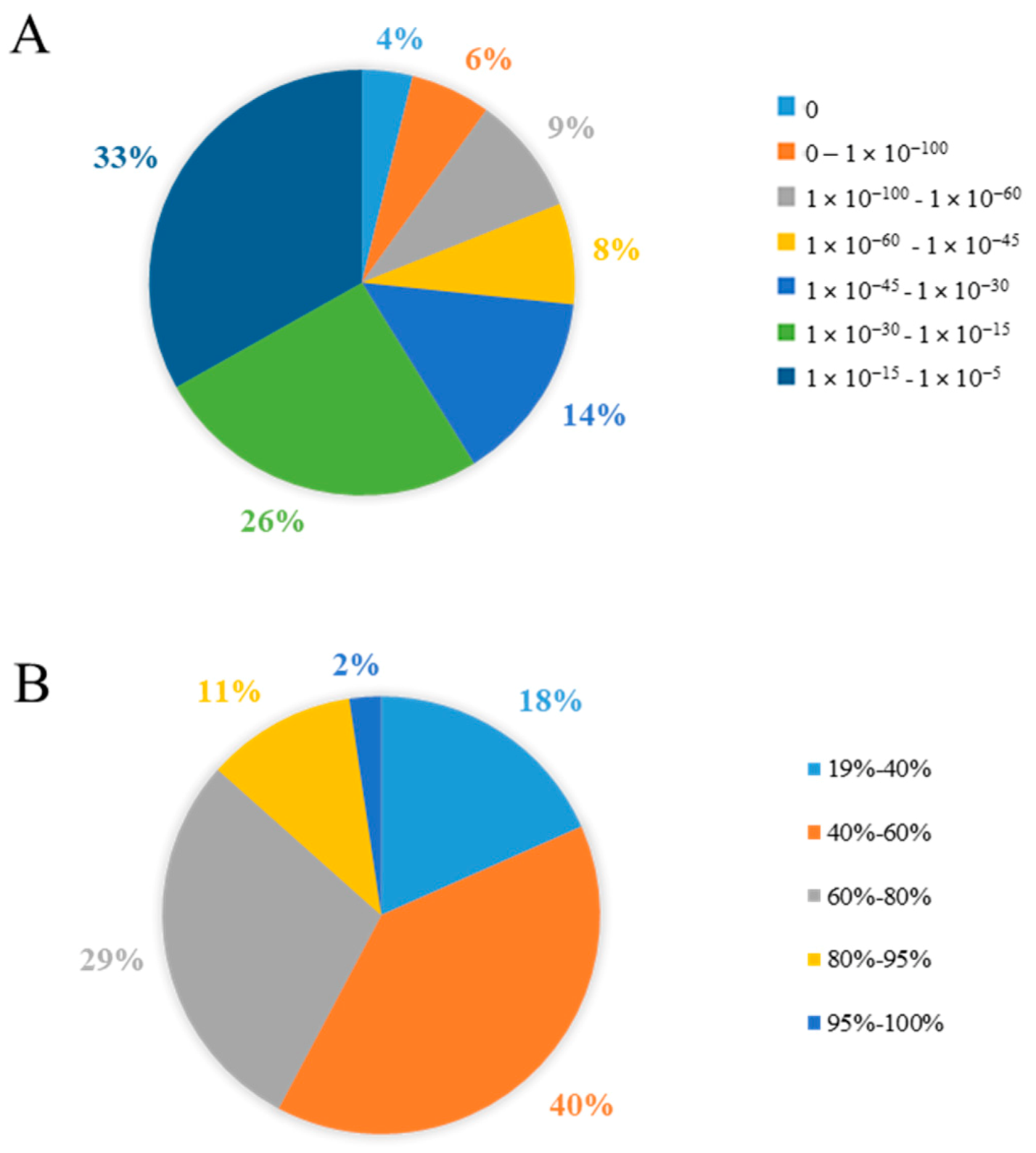
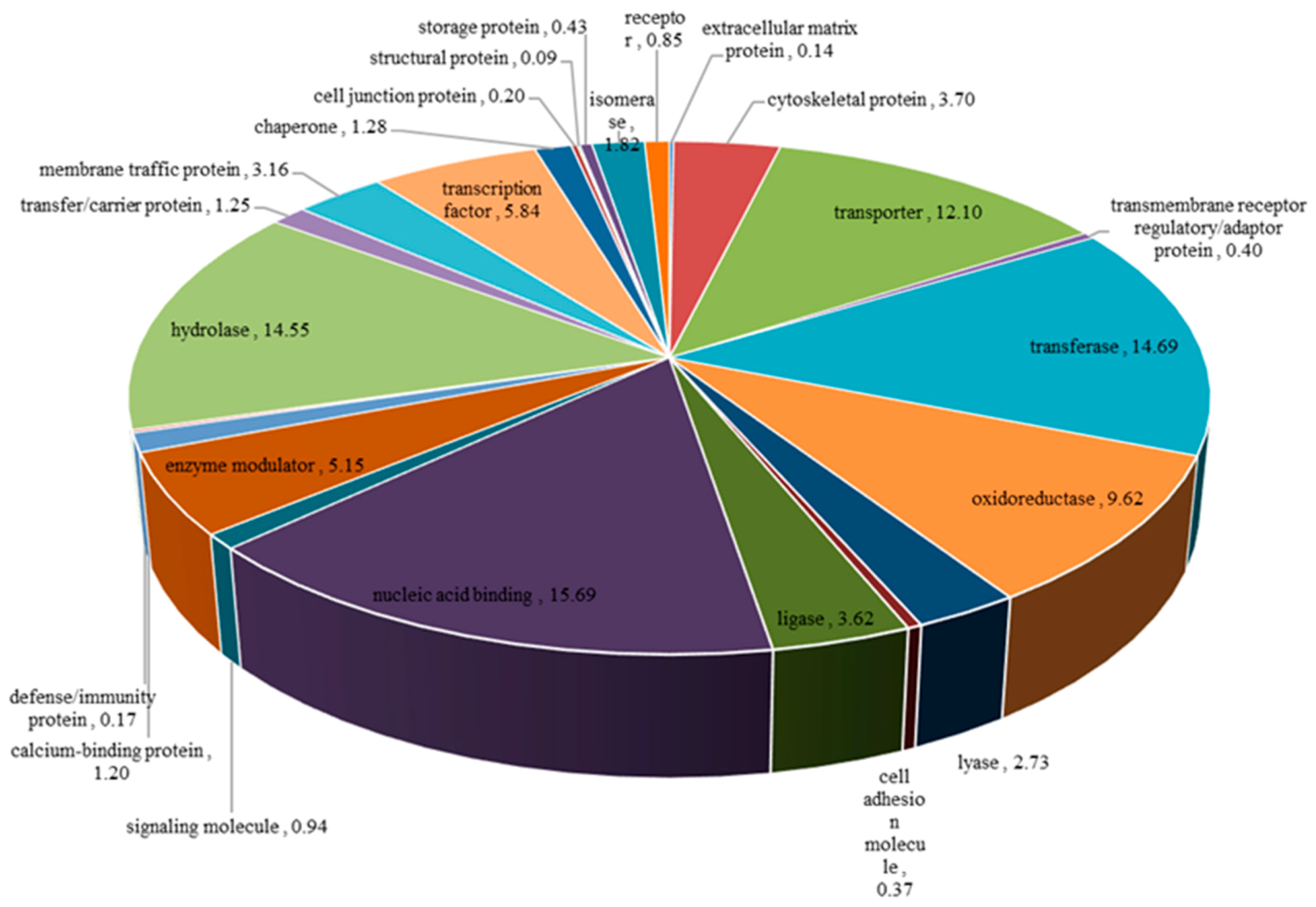
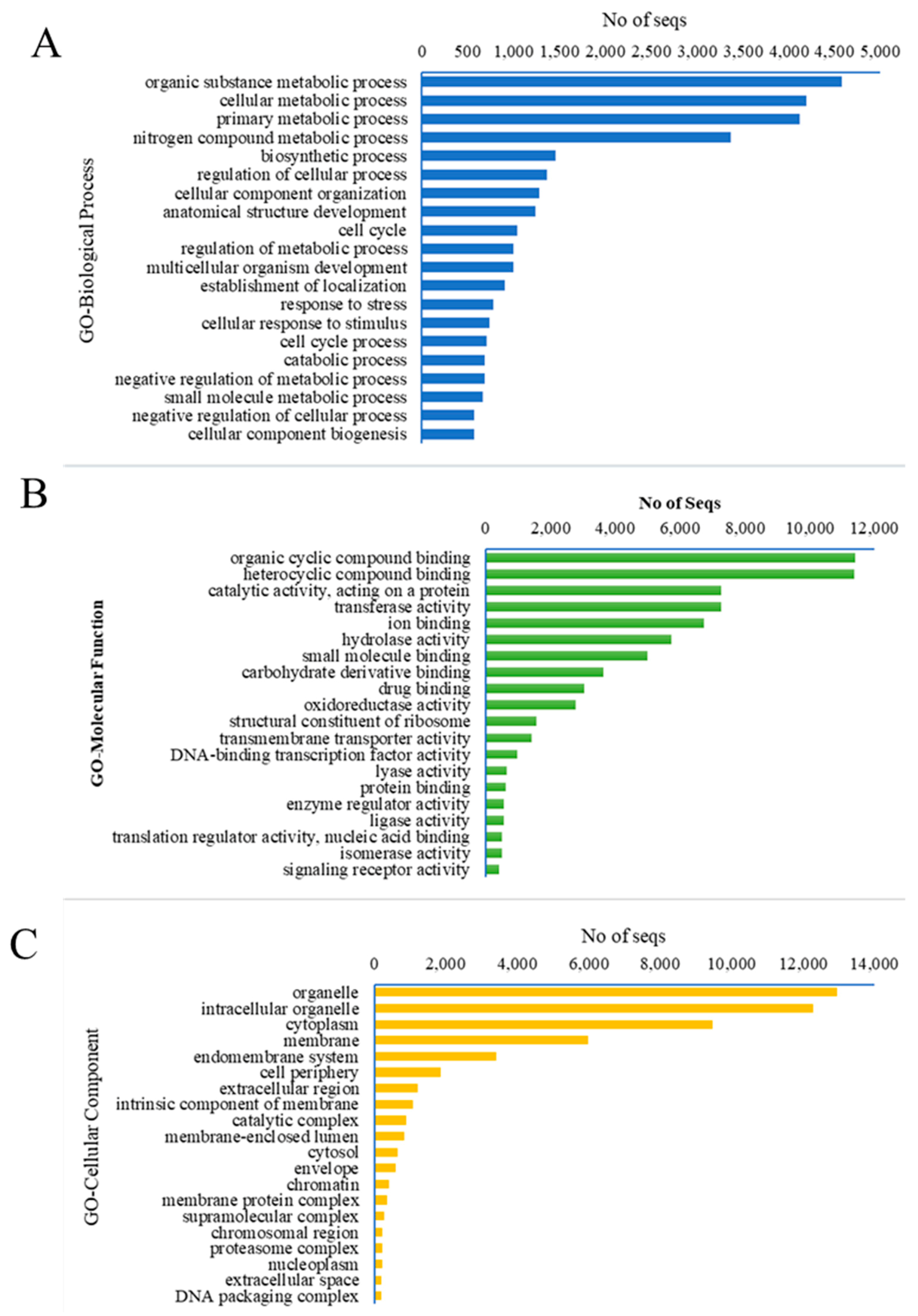
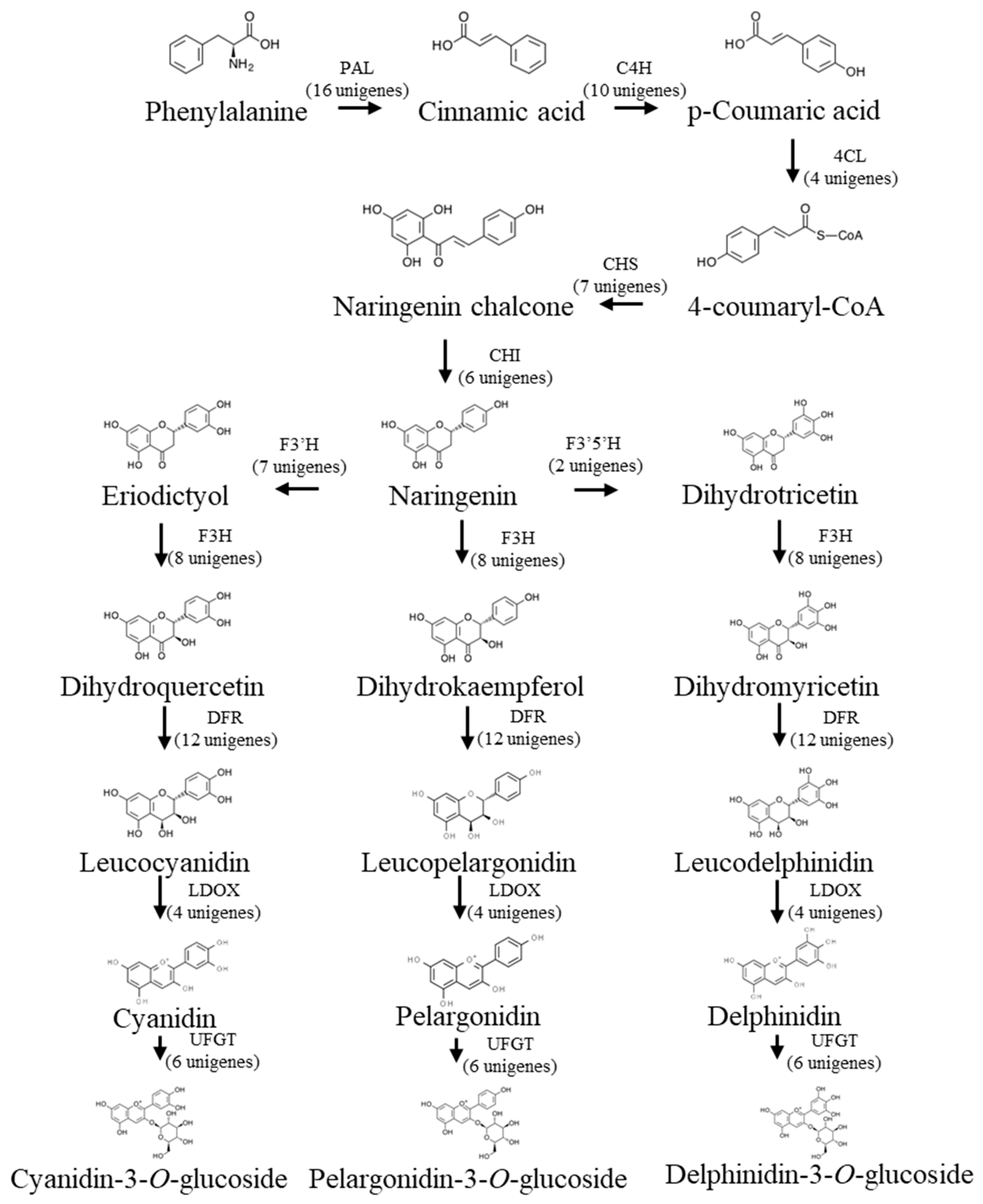
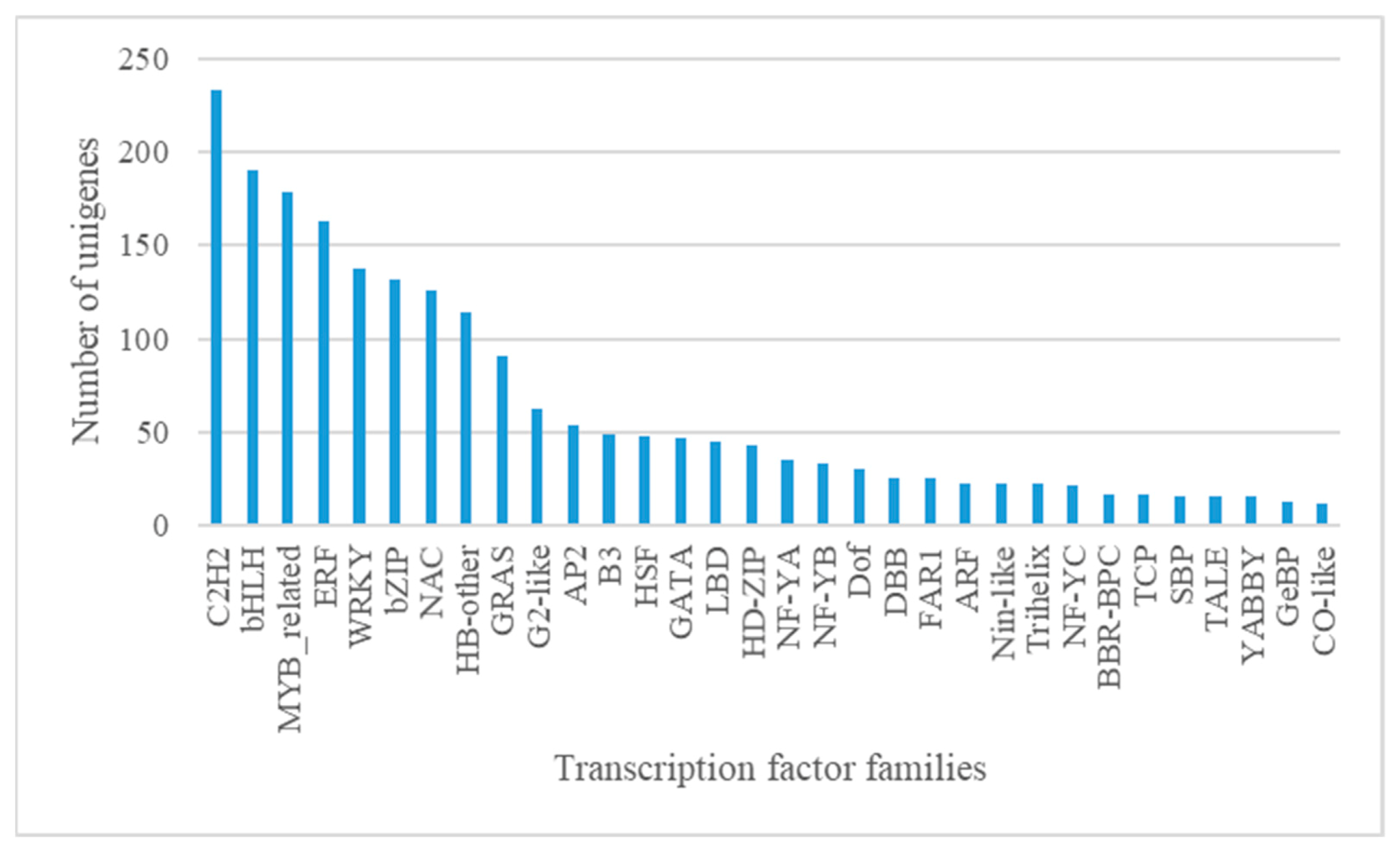
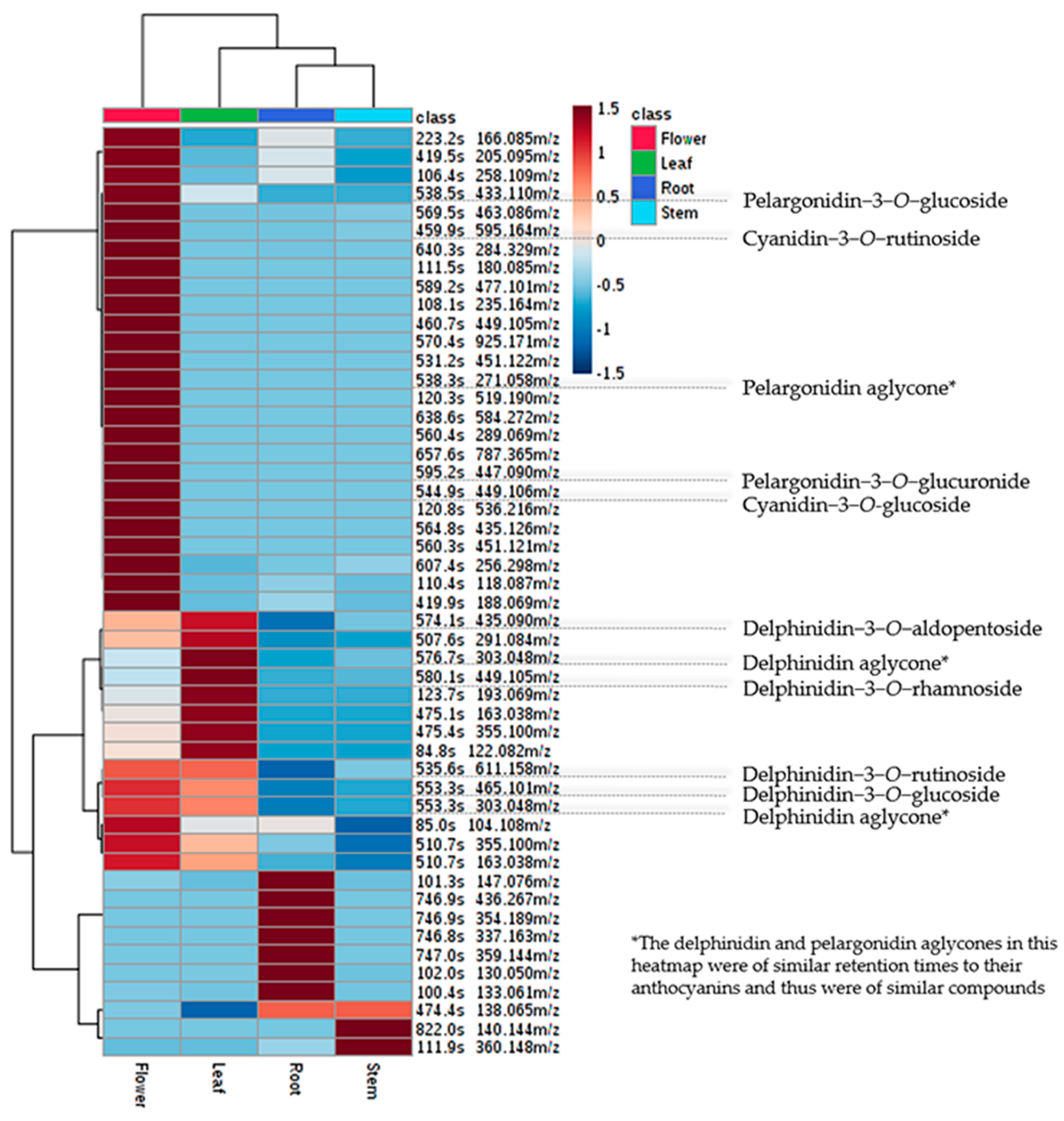
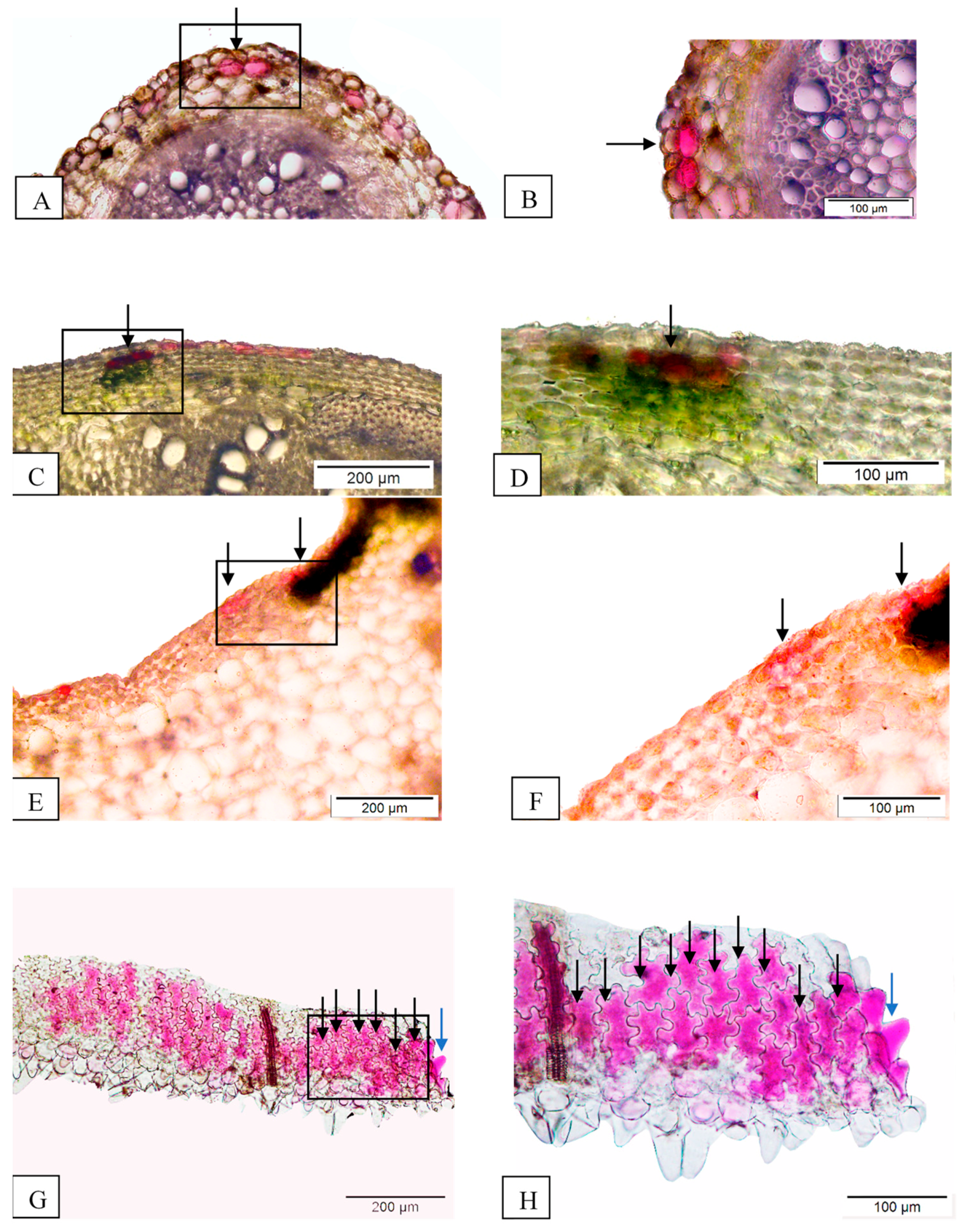
| Attributes | Unigenes | CDS |
|---|---|---|
| Total unigenes | 187,206 | 167,252 |
| GC% | 41.89 | 43.84 |
| Contig N50 | 551 | 468 |
| Median contig length | 308 | 258 |
| Average contig length | 476.71 | |
| Total assembled bases | 89,243,338 |
| Database | Number of Unigenes | Percentages of Unigenes |
|---|---|---|
| NR | 95,943 | 51.25 |
| Swiss-Prot | 71,607 | 38.25 |
| Interpro | 75,629 | 40.40 |
| KEGG | 49,984 | 26.7 |
| GO | 70,065 | 37.43 |
| Genes | Domain Hit (InterProScan) |
|---|---|
| F3H | IPR005123: Oxoglu/Fe-dep_dioxygenase PS51471: FE2OG_OXY PF03171: 2OG-FeII_Oxy IPR026992: DIOX_N PF14226: DIOX_N |
| DFR | IPR001509: Epimerase_deHydtase PF01370: Epimerase |
| LDOX | IPR005123: Oxoglu/Fe-dep_dioxygenase PF03171: 2OG-FeII_Oxy PS51471: FE2OG_OXY |
| UFGT | IPR035595: UDP_glycos_trans_CS PS00375: UDPGT |
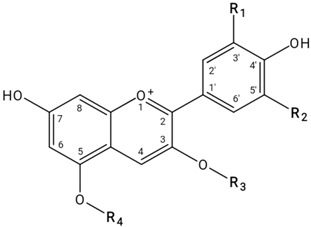 | |||||
|---|---|---|---|---|---|
| Anthocyanin/Anthocyanidin | Molecular Weight (g/mol) | R1 | R2 | R3 | R4 |
| Cyanidin | 287.24 | OH | H | H | H |
| Pelargonidin | 271.24 | H | H | H | H |
| Delphinidin | 303.24 | OH | OH | H | H |
| Cyanidin-3-O-pentoside | 419.35 | OH | H | Pentose | H |
| Cyanidin-3-O-rutinoside (rhamnohexoside) | 595.53 | OH | H | Rutinose | H |
| Cyanidin-3-O-hexoside | 449.38 | OH | H | Hexose | H |
| Pelargonidin-3-O-hexoside | 433.38 | H | H | Hexose | H |
| Pelargonidin-3,5-O-dihexoside | 595.53 | H | H | Hexose | Hexose |
| Pelargonidin-3-O-(6-p-coumaroyl)hexoside | 579.53 | H | H | p-Coumaroylhexose | H |
| Pelargonidin-3-O-glucuronoside | 447.37 | H | H | Glucuronide | H |
| Delphinidin-3-O-rhamnoside | 449.38 | OH | OH | Rhamnose | H |
| Delphinidin-3-O-hexoside | 465.38 | OH | OH | Hexose | H |
| Delphinidin-3-O-aldopentoside | 435.35 | OH | OH | Aldopentose | H |
| Delphinidin-3-O-rutinoside | 611.52 | OH | OH | Rutinose | H |
| Anthocyanin | Calculated Molecular Ion [M]+, (m/z) | Observed [M]+, m/z (Accurate Mass Error) | Retention Time, tR (min) | Molecular Formula | p-Value | References |
|---|---|---|---|---|---|---|
| Cyanidin-3-O-pentoside | 419.0973 | 419.0967 (0.6 mDa, 1.4 ppm) | 9.99 min | C20H19O10+ | 0.049875 | [61,62] |
| Cyanidin-3-O-rutinoside (rhamnoglucoside) | 595.1657 | 595.1656 (0.1 mDa, 0.2 ppm) | 7.64 min | C27H31O15+ | 2.91 × 10−7 | [62,63,64] |
| Cyanidin-3-O-glucoside | 449.1078 | 449.1085 (0.7 mDa, 1.5 ppm) | 8.98 min | C21H21O11+ | 6.48 × 10−9 | [62,65] |
| Pelargonidin-3-O-glucoside | 433.1129 | 433.1133 (0.4 mDa, 1.0 ppm) | 8.98 min | C21H21O10+ | 1.08 × 10−8 | [62,65] |
| Pelargonidin-3,5-O-dihexoside | 595.1657 | 595.1655 (0.2 mDa, 0.4 ppm) | 8.16 min | C27H31O15+ | 1.89 × 10−5 | [65,66] |
| Pelargonidin-3-O-(6-p-coumaroyl) hexoside | 579.1497 | 579.1491 (0.6 mDa, 1.0 ppm) | 8.19 min | C30H27O12+ | 7.74 × 10−9 | [67,68] |
| Pelargonidin-3-O-glucuronide | 447.0922 | 447.0917 (0.5 mDa, 1.0 ppm) | 9.93 min | C21H19O11+ | 4.57 × 10−8 | [69,70] |
| Delphinidin-3-O-rhamnoside | 449.1078 | 449.1078 (0.0 mDa, 0.0 ppm) | 9.69 min | C21H21O11+ | 6.74 × 10−9 | [71,72] |
| Delphinidin-3-O-glucoside | 465.1028 | 465.1027 (0.1 mDa, 0.2 ppm) | 9.28 min | C21H21O12+ | 1.47 × 10−8 | [62,73] |
| Delphinidin-3-O-aldopentoside | 435.0922 | 435.0916 (0.5 mDa, 1.2 ppm) | 9.61 min | C20H19O11+ | 9.57 × 10−9 | [74,75] |
| Delphinidin-3-O-rutinoside | 611.1607 | 611.1606 (0.1 mDa, 0.1 ppm) | 8.94 min | C27H31O16+ | 6.63 × 10−8 | [76,77] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gunasekaran, D.; Tahir, N.I.; Akbar, M.A.; Basir, S.; Ismail, I.; Talip, N.; Ramzi, A.B.; Baharum, S.N.; Noor, N.M.; Bunawan, H. Discovery of Anthocyanin Biosynthetic Pathway in Cosmos caudatus Kunth. Using Omics Analysis. Agronomy 2021, 11, 661. https://doi.org/10.3390/agronomy11040661
Gunasekaran D, Tahir NI, Akbar MA, Basir S, Ismail I, Talip N, Ramzi AB, Baharum SN, Noor NM, Bunawan H. Discovery of Anthocyanin Biosynthetic Pathway in Cosmos caudatus Kunth. Using Omics Analysis. Agronomy. 2021; 11(4):661. https://doi.org/10.3390/agronomy11040661
Chicago/Turabian StyleGunasekaran, Darvien, Noor Idayu Tahir, Muhamad Afiq Akbar, Syazwani Basir, Ismanizan Ismail, Noraini Talip, Ahmad Bazli Ramzi, Syarul Nataqain Baharum, Normah Mohd Noor, and Hamidun Bunawan. 2021. "Discovery of Anthocyanin Biosynthetic Pathway in Cosmos caudatus Kunth. Using Omics Analysis" Agronomy 11, no. 4: 661. https://doi.org/10.3390/agronomy11040661
APA StyleGunasekaran, D., Tahir, N. I., Akbar, M. A., Basir, S., Ismail, I., Talip, N., Ramzi, A. B., Baharum, S. N., Noor, N. M., & Bunawan, H. (2021). Discovery of Anthocyanin Biosynthetic Pathway in Cosmos caudatus Kunth. Using Omics Analysis. Agronomy, 11(4), 661. https://doi.org/10.3390/agronomy11040661






