Mycobiome Composition and Diversity under the Long-Term Application of Spent Mushroom Substrate and Chicken Manure
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sample Collection
2.2. Analysis of the Soil’s Chemical Properties
2.3. Extraction of the Total Genomic DNA from the Soil Samples
2.4. Analysis of the Fungal Abundance by qPCR
2.5. Analysis of the Fungal Community Using the DGGE Approach
2.6. Analysis of the Soil Fungal Community Using a t-RFLP Approach
2.7. Analysis of the Soil Fungal Community Composition Using Illumina MiSeq Platform Sequencing
2.8. Statistical Analysis
3. Results
3.1. Soil Properties
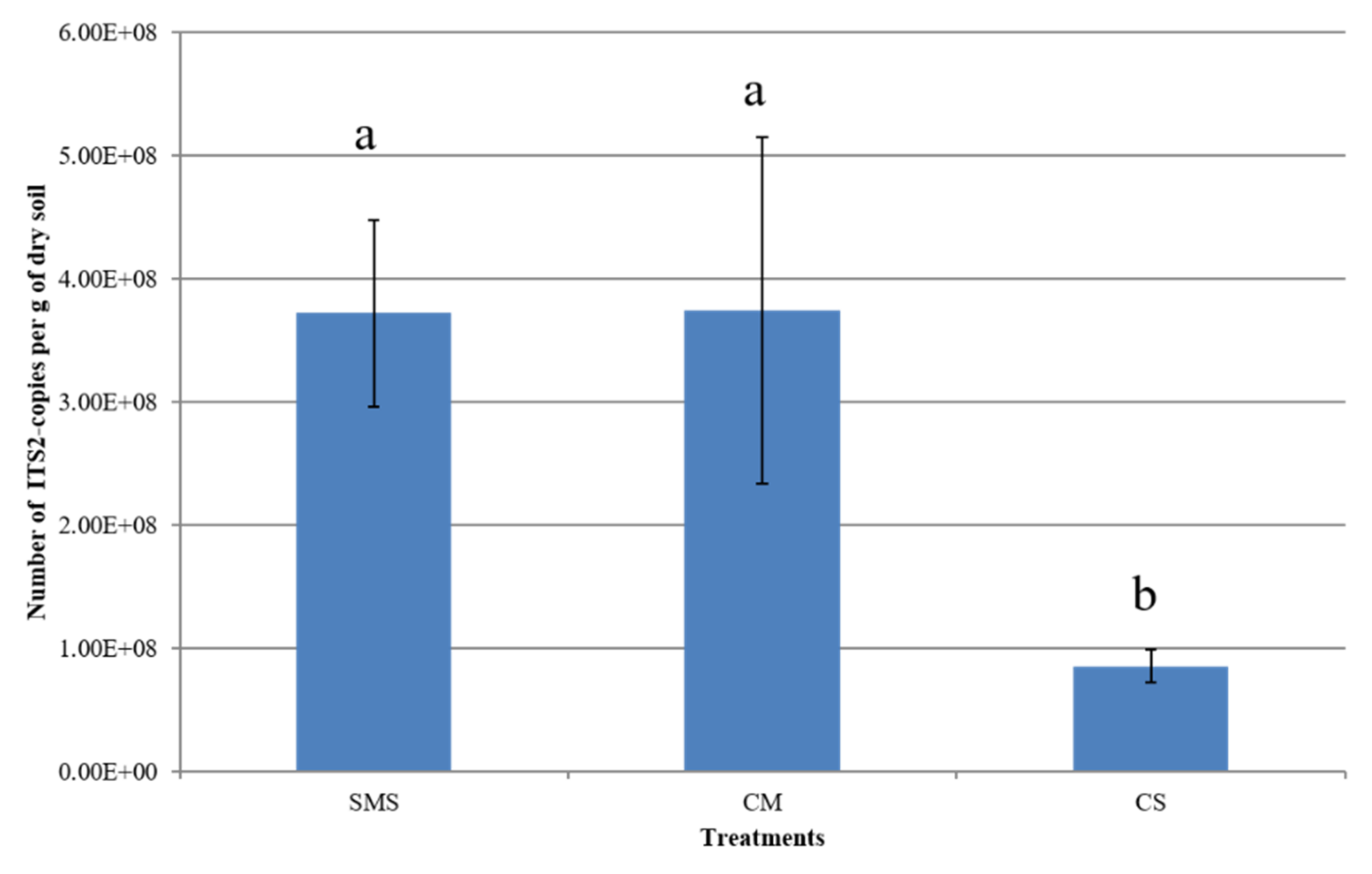
3.2. Effect of Organic Fertilization Treatments on Soil Fungal Abundance
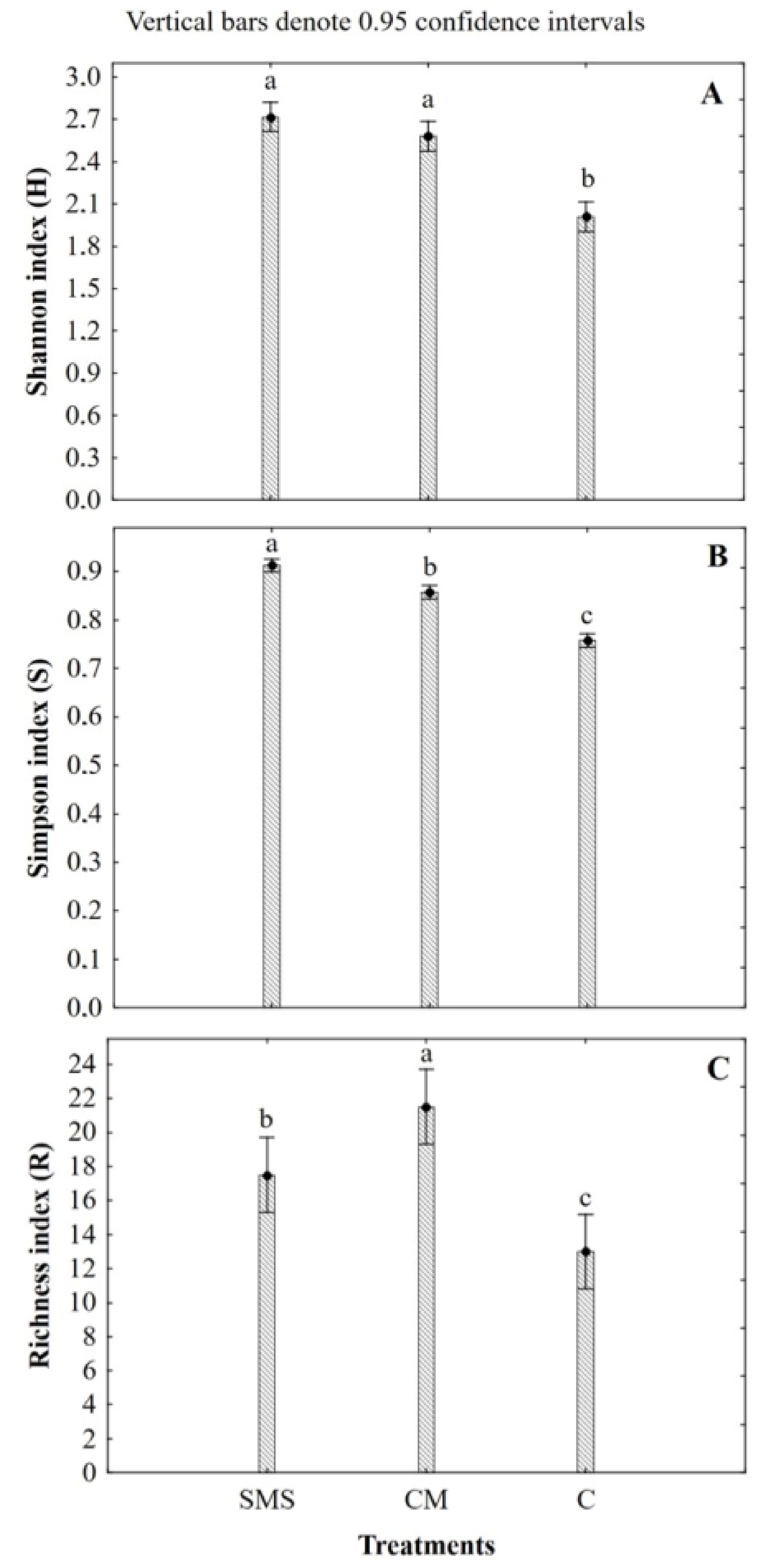
3.3. Effect of the Organic Fertilization Treatments on the Soil Fungal Diversity
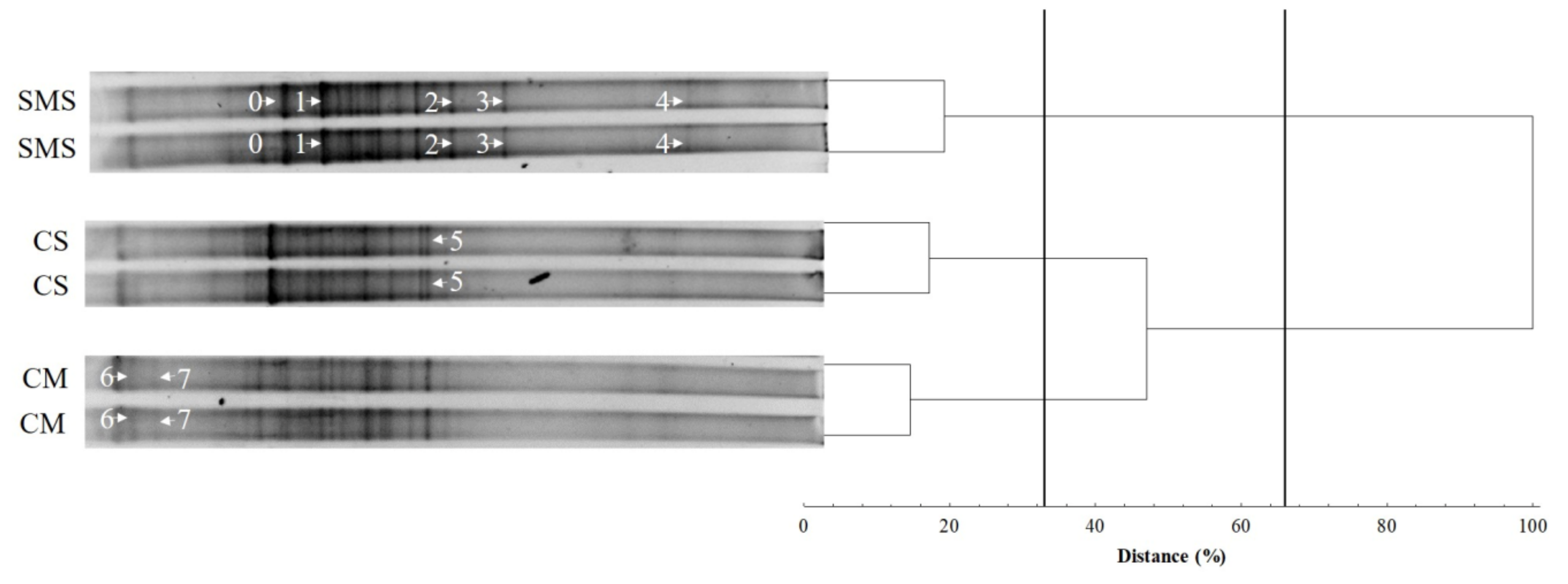
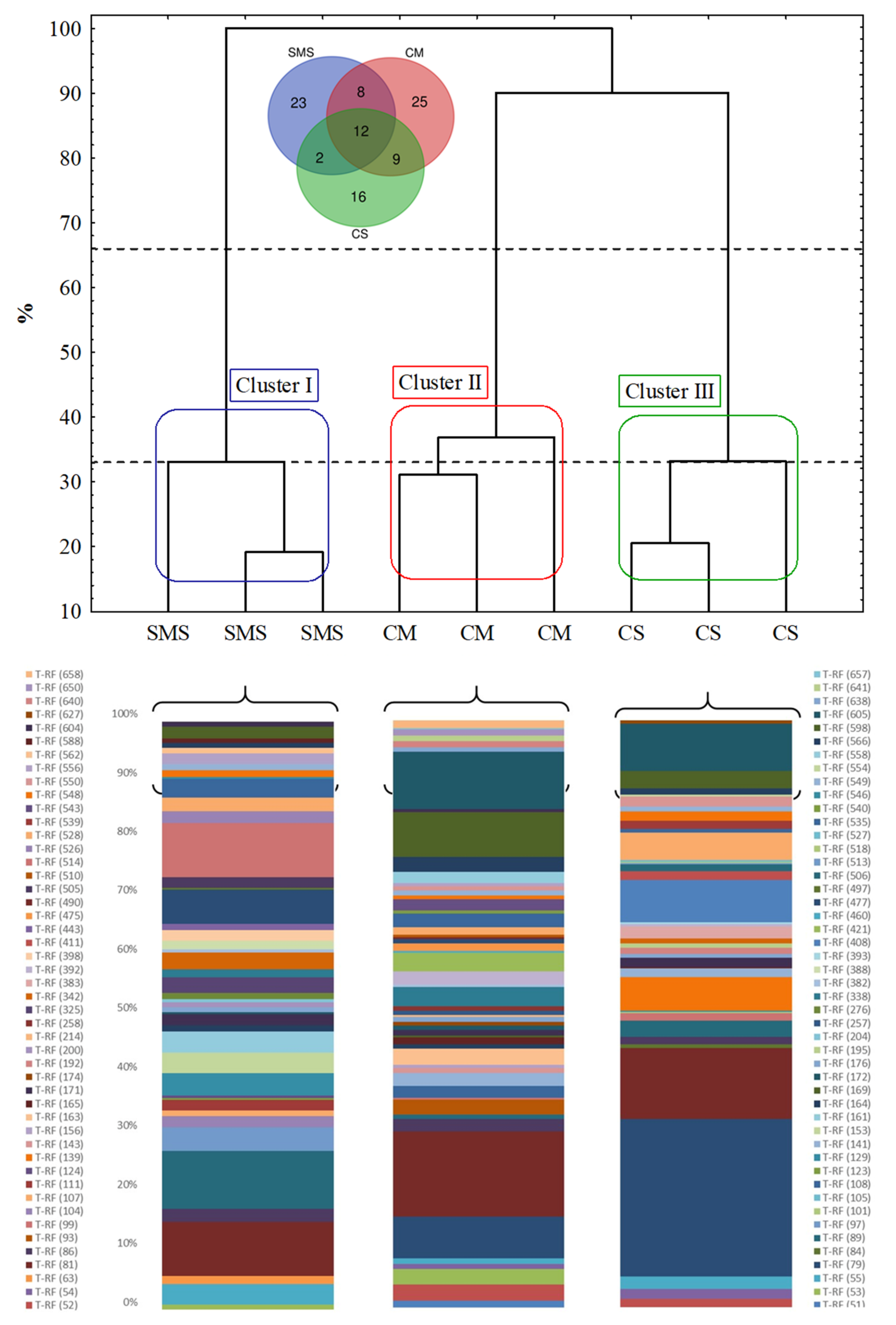
3.4. The Fungal Community, as Determined by t-RFLP
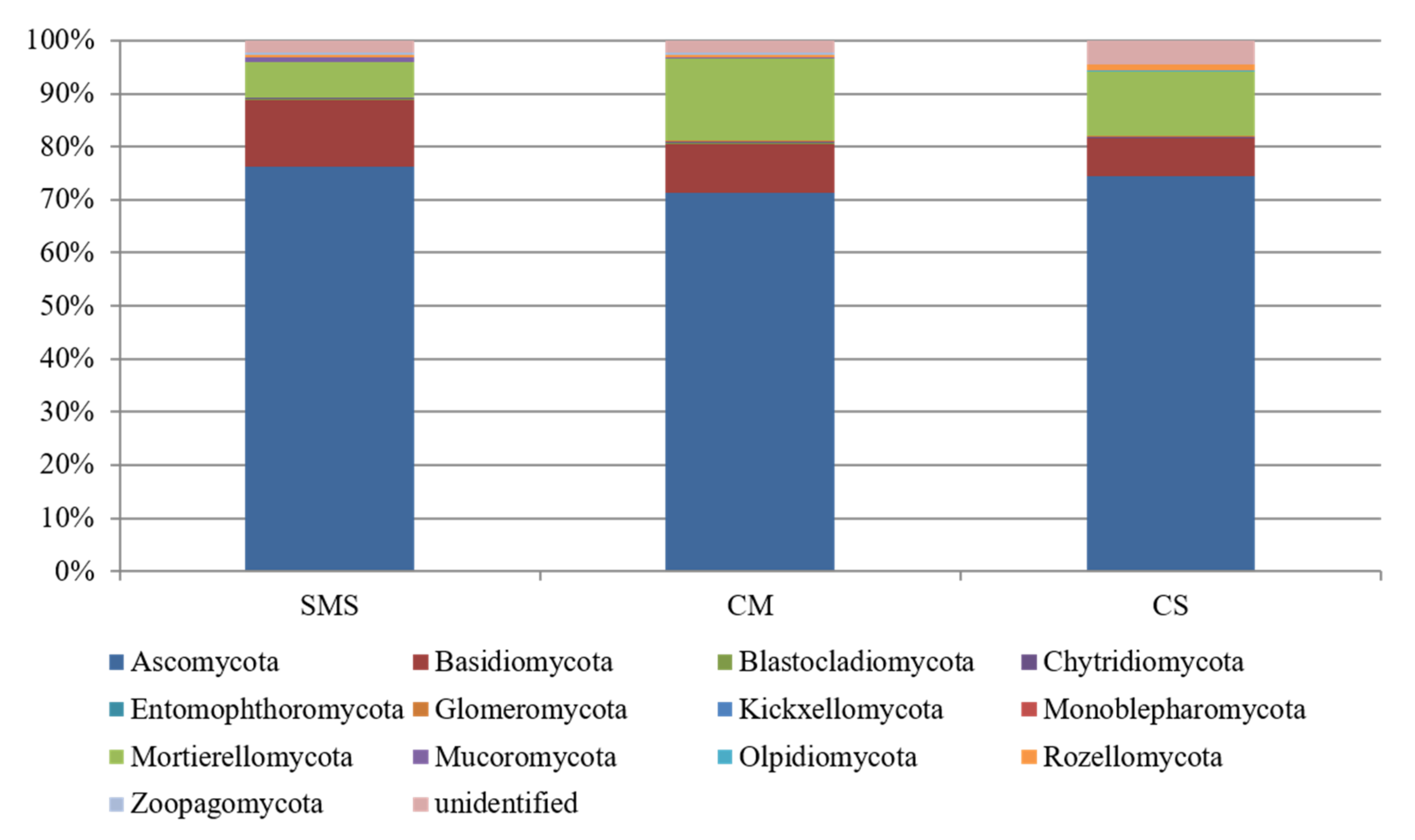
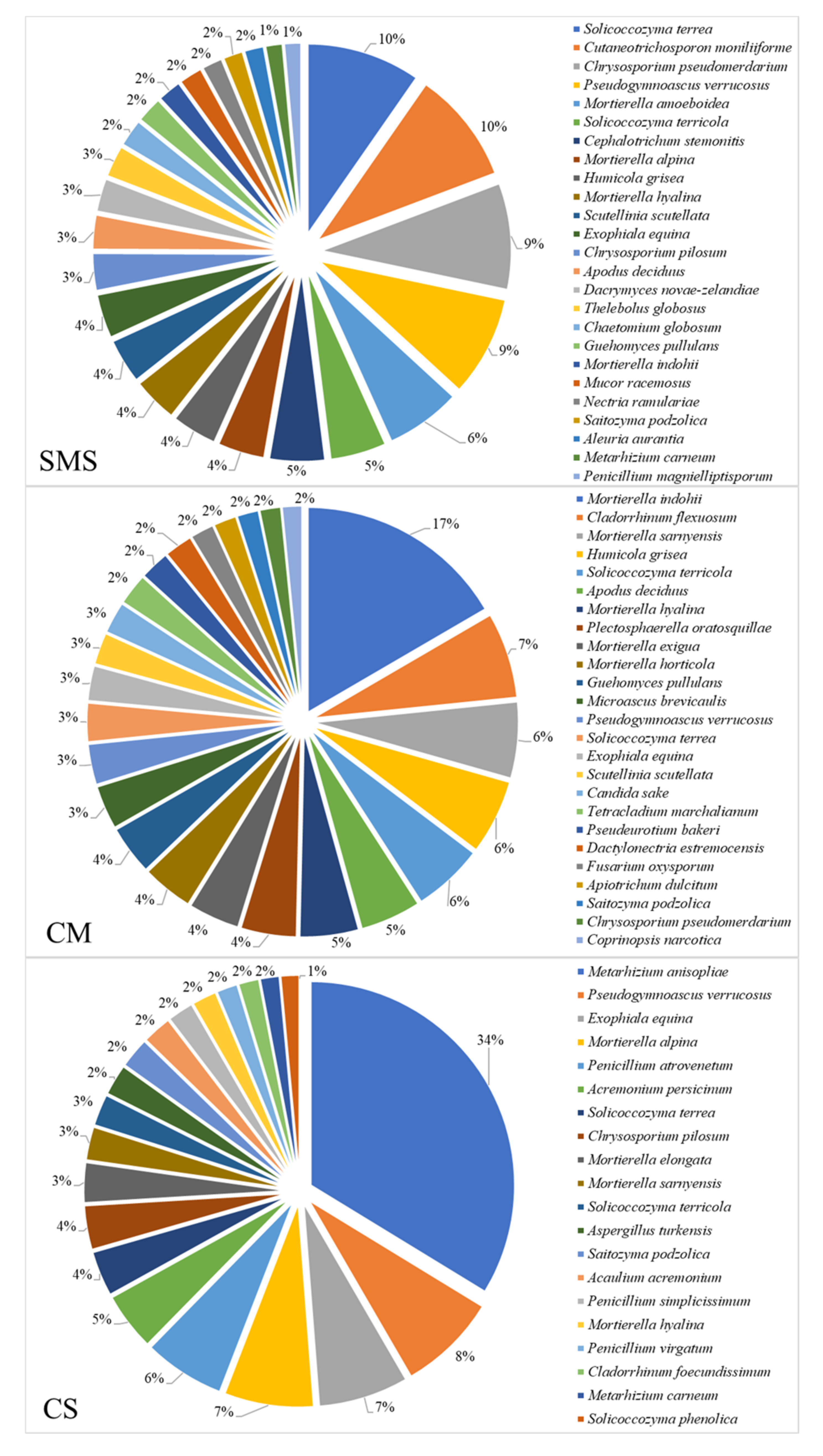
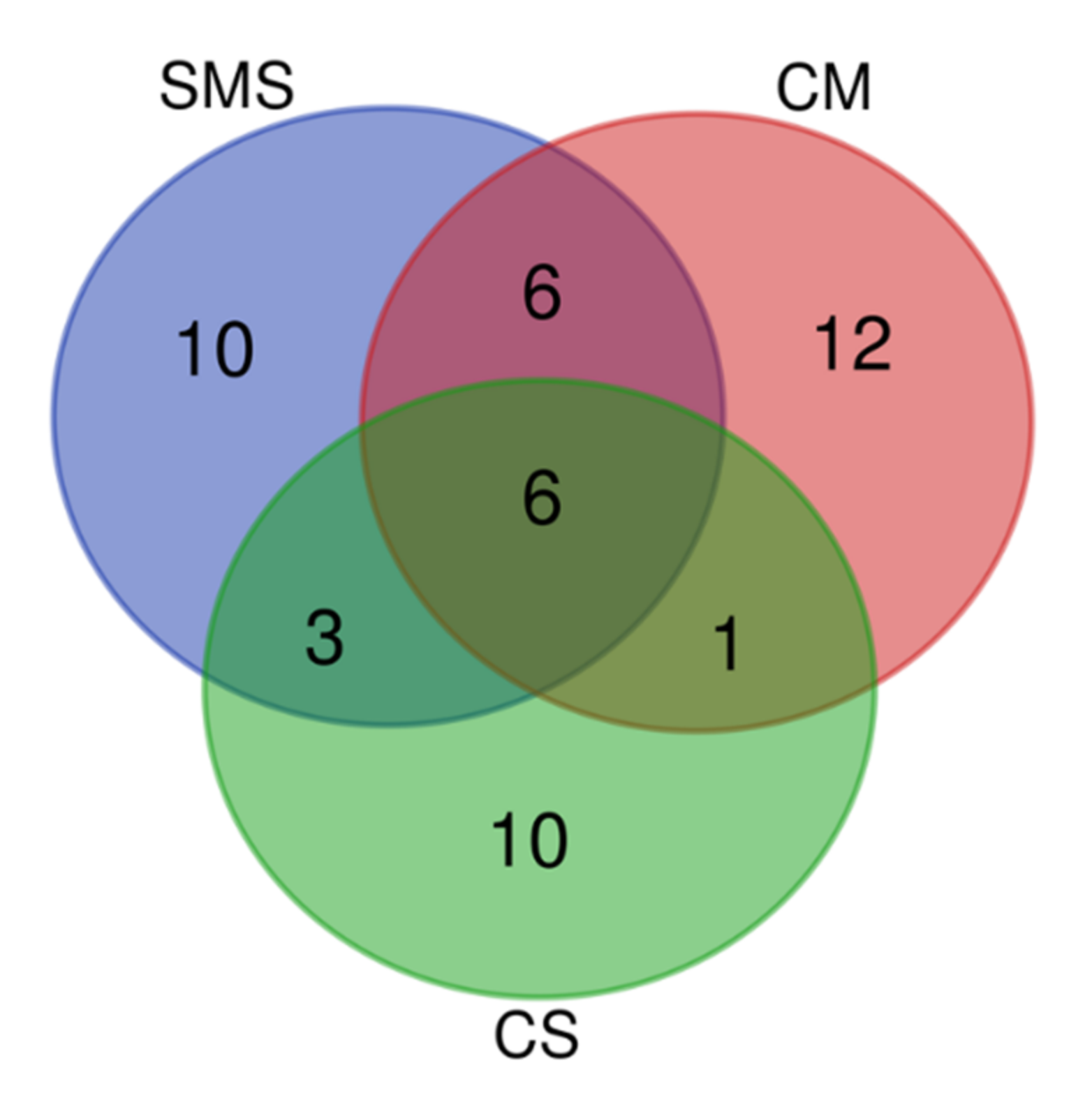
3.5. Relative Abundance of the Fungi and Their Taxonomic Classification, as Determined by NGS
4. Discussion
4.1. Effects of Additives on Soil Properties
4.2. Effects of Organic Additives on Soil Fungal Abundance
4.3. Effects of the Organic Additives on the Soil Fungal Community Structure
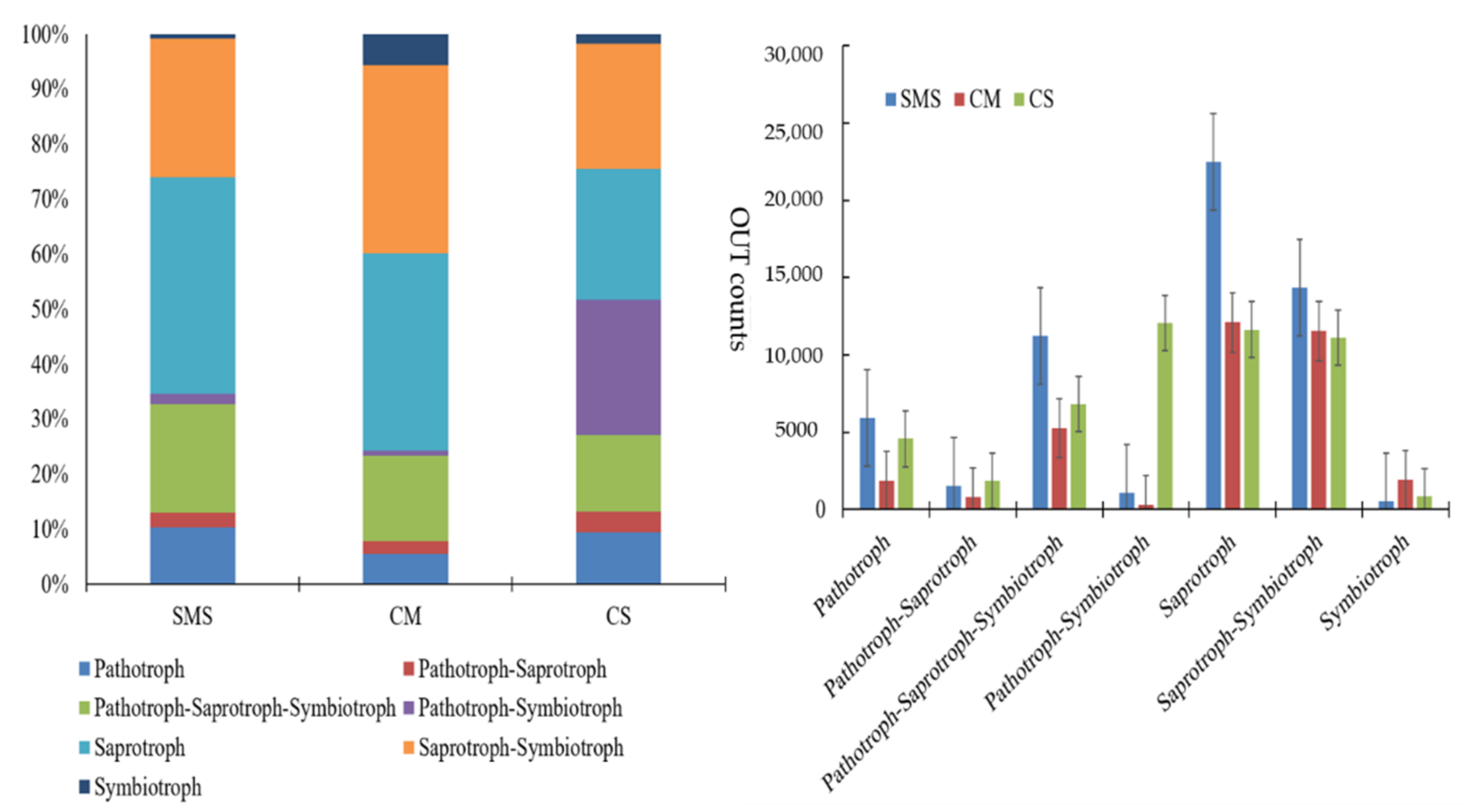
4.4. Effects of Organic Additives on Soil-Borne Pathogens and Functional Guild Groups
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frąc, M.; Ziemiński, K. Methane fermentation process for utilization of organic waste. Int. Agrophys. 2012, 26, 317–330. [Google Scholar] [CrossRef]
- Jezierska-Tys, S.; Frąc, M. Impact of dairy sewage sludge on enzymatic activity and inorganic nitrogen concentrations in the soils. Int. Agrophys. 2009, 23, 31–37. [Google Scholar]
- Marín-Benito, J.M.; Sánchez-Martín, J.M.; Rodríguez-Cruz, M.S. Impact of spent mushroom substrates on the fate of pesticides in soil, and their use for preventing and/or controlling soil and water contamination: A review. Toxics 2016, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Becher, M. Chemical composition of spent mushroom substrate after cultivation of Agaricus bisporus as a waste organic material. Ekon. Sr. 2013, 4, 207–213. (In Polish) [Google Scholar]
- Ahlawat, O.P.; Sagar, M.P. Management of Spent Mushroom Substrate Technical Bulletin; National Research Centre for Mushroom, Indian Council of Agricultural Research: Solan, India, 2007. [Google Scholar]
- Becher, M.; Banach-Szott, M.; Godlewska, A. Organic Matter Properties of Spent Button Mushroom Substrate in the Context of Soil Organic Matter Reproduction. Agronomy 2021, 11, 204. [Google Scholar] [CrossRef]
- Owaid, M.N.; Abed, I.A.; Al-Saeedi, S.S.S. Applicable properties of the bio-fertilizer spent mushroom substrate in organic systems as a byproduct from the cultivation of Pleurotus spp. Inform. Process. Agric. 2017, 4, 78–82. [Google Scholar] [CrossRef]
- Marques, E.L.S.; Martos, E.T.; Souza, R.J.; Silva, R.; Zied, D.C.; Souza Dias, E. Spent mushroom compost as a substrate for the production of lettuce seedlings. J. Agric. Sci. 2014, 6, 138–143. [Google Scholar] [CrossRef]
- Remya, J.S.; Beena, S. Antagonistic potential of fungal microflora from spent mushroom substrate against soil borne pathogens of ginger. Int. J. Appl. Pur. Sci. Agric. 2015, 1, 93–101. [Google Scholar]
- Medina, E.; Paredes, C.; Bustamante, M.A.; Moral, R.; Moreno-Caselles, J. Relationships between soil physico-chemical, chemical and biological properties in a soil amended with spent mushroom substrate. Geoderma 2012, 173, 152–161. [Google Scholar] [CrossRef]
- Chen, Z.; Jiang, X. Microbiological safety of chicken litter or chicken litter-based organic fertilizers: A review. Agriculture 2014, 4, 1–29. [Google Scholar] [CrossRef]
- Quang, N.H. Long-Term Effects of Land Application of Poultry Manure on Crop Production, and Soil and Water Quality Under a Corn-Soybean Rotation System in Iowa. Master’s Thesis, Iowa State University, Ames, IA, USA, 2010; pp. 1–95. [Google Scholar]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, X.; Zhou, B.; Zhao, B.; Ma, M.; Guan, D.; Li, J.; Chen, S.; Cao, F.; Shen, D.; et al. Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast China. Soil Biol. Biochem. 2016, 95, 135–143. [Google Scholar] [CrossRef]
- Klaubauf, S.; Inselsbacher, E.; Zechmeister-Boltenstern, S.; Wanek, W.; Gottsberger, R.; Strauss, J.; Gorfer, M. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fung. Div. 2010, 44, 65–75. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Ijaz, U.Z.; Saunders, A.M.; Theuerl, S. Terminal restriction fragment length polymorphism is an “old school” reliable technique for swift microbial community screening in anaerobic digestion. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Junier, P.; Junier, T.; Witzel, K.P. TRiFLe, a Program for in silico terminal restriction fragment length polymorphism analysis with user-defined sequence sets. Appl. Environ. Microbiol. 2008, 74, 6452–6456. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rincon-Florez, V.A.; Carvalhais, L.C.; Schenk, P.M. Culture-Independent Molecular Tools for Soil and Rhizosphere Microbiology. Diversity 2013, 5, 581–612. [Google Scholar] [CrossRef]
- WRB IUSS Working Group. World Reference Base for Soil Resources 2014, update 2015. International soil classification system for naming soils and creating legends for soil maps. In World Soil Resources Reports; FAO: Rome, Italy, 2015. [Google Scholar]
- Usowicz, B.; Hajnos, M.; Sokołowska, Z.; Józefaciuk, G.; Bowanko, G.; Kossowski, J. Spatial variability of physical and chemical soil properties in a field and commune scale. Acta Agrophysica Monogr. 2004, 3, 90. (In Polish) [Google Scholar]
- Kalembasa, D.; Majchrowska-Safaryan, A. Fraction of heavy metals in the beds after the cultivation mushroom from mushroom factor. Ochr. Sr. Zasobów Nat. 2009, 41, 1–6. (In Polish) [Google Scholar]
- Polish Standard. Determination of Available Phosphorus in Mineral Soils; PN-R-04023:1996; Polish Committee for Standardization: Warsaw, Poland, 1996.
- Polish Standard. Determination of Available Potassium in Mineral Soils; PN-R-04022:1996/Az1:2002; Polish Committee for Standardization: Warsaw, Poland, 1996.
- Polish Standard. Determination of Available Magnesium in Soils; PN-R-04020:1994/Az1:2004; Polish Committee for Standardization: Warsaw, Poland, 1994.
- Ostrowska, A.; Gawliński, S.; Szczubiałka, Z. Analyses and Evaluation Methods of Soil and Plants; Institute of Environmental Protection: Warsaw, Poland, 1991; p. 334. (In Polish).
- Oszust, K.; Frąc, M.; Gryta, A.; Bilińska, N. The influence of ecological and conventional plant production systems on soil microbial quality under hops (Humulus lupulus). Int. J. Mol. Sci. 2014, 15, 9907–9923. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Korabecna, M. The variability in the fungal ribosomal DNA (ITS1, ITS2, and 5.8 S rRNA gene): Its biological meaning and application in medical mycology. Com. Cur. Res. Edu. Top. Trend. Appl. Microbiol. 2007, 108, 783–787. [Google Scholar] [CrossRef]
- Manter, D.K.; Vivanco, J.M. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Met. 2007, 71, 7–14. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov (accessed on 21 January 2021).
- Schmidt, P.A.; Bálint, M.; Greshake, B.; Bandowa, C.; Römbke, J.; Schmitt, I. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013, 65, 128–132. [Google Scholar] [CrossRef]
- Vilgalys Mycology Lab—Duke University. Available online: https://sites.duke.edu/vilgalyslab/rdna_primers_for_fungi (accessed on 21 January 2021).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Aronesty, E. ea-utils: Command-Line Tools for Processing Biological Sequencing Data. 2011. Available online: http://code.google.com/p/ea-utils (accessed on 21 January 2021).
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Islam, M.A.; Islam, S.; Akter, A.; Rahman, M.H.; Nandwani, D. Effect of organic and inorganic fertilizers on soil properties and the growth, yield and quality of tomato in Mymensingh, Bangladesh. Agriculture 2017, 7, 31. [Google Scholar] [CrossRef]
- Oszust, K.; Frąc, M.; Lipiec, J. Soil microbial functionality in response to dairy sewage sludge and mineral fertilisers application under winter rape. Int. J. Environ. Sci. Technol. 2015, 12, 3675–3684. [Google Scholar] [CrossRef]
- Lehmann, J.; Rillig, M.C.; Thies, J.; Masiello, C.A.; Hockaday, W.C.; Crowley, D. Biochar effects on soil biota—a review. Soil Biol. Biochem. 2011, 43, 1812–1836. [Google Scholar] [CrossRef]
- Yao, Q.; Liu, J.; Yu, Z.; Li, Y.; Jin, J.; Liu, X.; Wang, G. Three years of biochar amendment alters soil physiochemical properties and fungal community composition in a black soil of northeast China. Soil Biol. Biochem. 2017, 110, 56–67. [Google Scholar] [CrossRef]
- Ding, J.; Jiang, X.; Guan, D.; Zhao, B.; Ma, M.; Zhou, B.; Cao, F.; Yang, X.; Li, L.; Li, J. Influence of inorganic fertilizer and organic manure application on fungal communities in a long-term field experiment of Chinese Mollisols. Appl. Soil Ecol. 2017, 111, 114–122. [Google Scholar] [CrossRef]
- Bastida, F.; Torres, I.F.; Hernandez, T.; García, C. The impacts of organic amendments: Do they confer stability against drought on the soil microbial community? Soil Biol. Biochem. 2017, 113, 173–183. [Google Scholar] [CrossRef]
- Gul, S.; Whalen, J.K.; Thomas, B.W.; Sachdeva, V.; Deng, H.Y. Physico-chemical properties and microbial responses in biochar-amended soils: Mechanisms and future directions. Agric. Ecosyst. Environ. 2015, 206, 46–59. [Google Scholar] [CrossRef]
- Gul, S.; Whalen, J.K. Biochemical cycling of nitrogen and phosphorus in biochar-amended soils. Soil Biol. Biochem. 2016, 103, 1–15. [Google Scholar] [CrossRef]
- Smith, C.J.; Osborn, A.M. Advantages and limitations of quantitative PCR(Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Gryta, A.; Frąc, M.; Oszust, K. Community shift in structure and functions across soil profile in response to organic waste and mineral fertilization strategies. Appl. Soil Ecol. 2019, 143, 55–60. [Google Scholar] [CrossRef]
- Hannula, S.E.; Ma, H.; Pérez-Jaramillo, J.E.; Pineda, A.; Bezemer, T.M. Structure and ecological function of the soil microbiome affecting plant–soil feedbacks in the presence of a soil-borne pathogen. Environ. Microbiol. 2020, 22, 660–676. [Google Scholar] [CrossRef]
- Thomson, B.C.; Tisserant, E.; Plassart, P.; Uroz, S.; Griffiths, R.I.; Hannula, S.E.; Buee, M.; Mougel, C.; Ranjard, L.; Van Veen, J.A.; et al. Soil conditions and land use intensification effects on soil microbial communities across a range of European field sites. Soil Biol. Biochem. 2015, 88, 403–413. [Google Scholar] [CrossRef]
- Wang, J.; Song, Y.; Ma, T.; Raza, W.; Li, J.; Howland, J.G.; Huang, Q.; Shen, Q. Impacts of inorganic and organic fertilization treatments on bacterial and fungal communities in a paddy soil. Appl. Soil Ecol. 2017, 112, 42–50. [Google Scholar] [CrossRef]
- Quilliam, R.S.; Glanville, H.C.; Wade, S.C.; Jones, D.L. Life in the ‘charosphere’—does biochar in agricultural soil provide a significant habitat for microorganisms? Soil Biol. Biochem. 2013, 65, 287–293. [Google Scholar] [CrossRef]
- Lipiec, J.; Brzezinska, M.; Turski, M.; Szarlip, P.; Frąc, M. Wettability and biogeochemical properties of the drilosphere and casts of endogeic earthworms in pear orchard. Soil Till. Res. 2015, 145, 55–61. [Google Scholar] [CrossRef]
- Król, A.; Lipiec, J.; Frąc, M. The effect of dairy sewage sludge amendment on repellency and hydraulic conductivity of soil aggregates from two depths of Eutric Cambisol. J. Plant Nutr. Soil Sci. 2015, 178, 270–277. [Google Scholar] [CrossRef]
- Frąc, M.; Oszust, K.; Lipiec, J.; Jezierska-Tys, S.; Nwaichi, E.O. Soil microbial functional and fungal diversity as influenced by municipal sewage sludge accumulation. Int. J. Environ. Res. Public Health 2014, 11, 8891–8908. [Google Scholar] [CrossRef] [PubMed]
- Ameloot, N.; Graber, E.R.; Verheijen, F.G.A.; De Neve, S. Interactions between biochar stability and soil organisms: Review and research needs. Eur. J. Soil Sci. 2013, 64, 379–390. [Google Scholar] [CrossRef]
- Narendrula-Kotha, R.; Nkongolo, K.K. Bacterial and fungal community structure and diversity in a mining region under long-term metal exposure revealed by metagenomics sequencing. Ecol. Genet. Genom. 2017, 2, 13–24. [Google Scholar] [CrossRef]
- Brown, S.P.; Jumpponen, A. Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Mol. Ecol. 2014, 23, 481–497. [Google Scholar] [CrossRef]
- Feng, Y.; Yu, Y.; Tang, H.; Zu, Q.; Zhu, J.; Lin, X. The contrasting responses of soil microorganisms in two rice cultivars to elevated ground-level ozone. Environ. Pollut. 2015, 197, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Lou, J.; Brookes, P.C.; Luo, Y.; He, Y.; Xu, J. Taxon-specific responses of soil microbial communities to different soil priming effects induced by addition of plant residues and their biochars. J. Soils Sediments 2017, 17, 674–684. [Google Scholar] [CrossRef]
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef] [PubMed]
- Frąc, M.; Hannula, S.E.; Belka, M.; Jȩdryczka, M. Fungal biodiversity and their role in soil health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef]
- Millanes, A.M.; Diederich, P.; Ekman, S.; Wedin, M. Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi). Mol. Phylogen. Evol. 2011, 61, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Ozimek, E.; Hanaka, A. Mortierella Species as the Plant Growth-Promoting Fungi Present in the Agricultural Soils. Agriculture 2021, 11, 7. [Google Scholar] [CrossRef]
- Fracetto, G.G.M.; Azevedo, L.C.B.; Fracetto, F.J.C.; Andreote, F.D.; Lambais, M.R.; Pfenning, L.H. Impact of Amazon land use on the community of soil fungi. Sci. Agric. 2013, 70, 59–67. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Wu, H.; Wang, J.; Wu, Y.; Lin, S.; Khan, M.U.; Zhang, Z.; Lin, W. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Brachmann, A.; Parniske, M. The Most Widespread Symbiosis on Earth. PLoS Biol. 2006, 4, e239. [Google Scholar] [CrossRef]
- Hart, M.M.; Aleklett, K.; Chagnon, P.-L.; Egan, C.; Ghignone, S.; Helgasson, T.; Lekberg, Y.; Öpik, M.; Pickles, B.J.; Waller, L. Navigating the labyrinth: A guide to sequence-based, community ecology of arbuscular mycorrhizal fungi. New Phytol. 2015, 207, 235–247. [Google Scholar] [CrossRef] [PubMed]
- George, P.B.L.; Creer, S.; Griffiths, R.I.; Emmett, B.A.; Robinson, D.A.; Jones, D.L. Primer and Database Choice Affect Fungal Functional but Not Biological Diversity Findings in a National Soil Survey. Front. Environ. Sci. 2019, 7, 173. [Google Scholar] [CrossRef]
- Tedersoo, L.; Arnold, A.E.; Sen, K.H. Novel aspects in the life cycle and biotrophic interactions in Pezizomycetes (Ascomycota, Fungi). Mol. Ecol. 2013, 22, 1488–1493. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol. Biochem. 2015, 83, 29–39. [Google Scholar] [CrossRef]
- Mueller, R.C.; Belnap, J.; Kuske, C.R. Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in an arid shrubland. Front. Microbiol. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Liu, X.Z.; Wang, Q.M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef]
- Berbee, M.L. The phylogeny of plant and animal pathogens in the Ascomycota. Phys. Mol. Plant Pathol. 2001, 59, 165–187. [Google Scholar] [CrossRef]
- Wagner, L.; Stielow, B.; Hoffmann, K.; Petkovits, T.; Papp, T.; Vágvölgyi, C.; de Hoog, G.S.; Verkley, G.; Voigt, K. A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Persoonia 2013, 30, 77–93. [Google Scholar] [CrossRef]
- Mącik, M.; Gryta, A.; Sas-Paszt, L.; Frąc, M. The Status of Soil Microbiome as Affected by the Application of Phosphorus Biofertilizer: Fertilizer Enriched with Beneficial Bacterial Strains. Int. J. Mol. Sci. 2020, 21, 8003. [Google Scholar] [CrossRef]
- Goodwin, S.B.; Kema, G.H. Gearing up for comparative genomics: Analyses of the fungal class Dothideomycetes. New Phytol. 2009, 183, 250–254. [Google Scholar] [CrossRef]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ohm, R.A.; Feau, N.; Henrissat, B.; Schoch, C.L.; Horwitz, B.A.; Barry, K.W.; Condon, B.J.; Copeland, A.C.; Dhillon, B.; Glaser, F.; et al. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 2012, 8, e1003037. [Google Scholar] [CrossRef] [PubMed]
- Sachs, J.L.; Simms, E.L. Pathways to mutualism breakdown. Trends Ecol. Evol. 2006, 21, 585–592. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Tiago, P.V.; de Oliveira, N.T.; de Luna, E.Á.; Lima, A. Biological insect control using Metarhizium anisopliae: Morphological, molecular, and ecological aspects. Ciên. Rur. St. Maria 2014, 44, 645–651. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Gueidan, C.; Schroers, H.J.; de Hoog, G.S.; Starink, M.; Rosete, A.Y.; Guarro, J.; Scott, J.A. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011, 68, 139–162. [Google Scholar] [CrossRef] [PubMed]
- Summerell, B.A.; Laurence, M.H.; Liew, E.C.Y.; Leslie, J.F. Biogeography and phylogeography of Fusarium: A review. Fungal Divers 2010, 44, 3–13. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, J.; Singh, R.S. Nonpathogenic Fusarium as a Biological Control Agent. Plant Pathol. J. 2010, 9, 79–91. [Google Scholar] [CrossRef]
- Bilal, L.; Asaf, S.; Hamayun, M.; Gul, H.; Iqbal, A.; Ullah, I.; Lee, I.J.; Hussain, A. Plant growth promoting endophytic fungi Asprgillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis 2018, 76, 117–127. [Google Scholar] [CrossRef]

| Treatments | pHKCl | SWC (%) | TOC (%) | N (g/kg) | P (mg/100 g) | K (mg/100 g) | Mg (mg/100 g) |
|---|---|---|---|---|---|---|---|
| SMS | 7.08 | 11.43 | 2.73 | 2.69 | 50 | 30 | 13 |
| CM | 6.68 | 11.43 | 2.03 | 0.87 | 49 | 18 | 17 |
| CS | 4.95 | 1.38 | 0.50 | 0.59 | 24 | 9 | 4 |
| Treatments | Number of OTUs (Richness Index) | Shannon Index | Simpson Index | Total Number of Reds | Total Number of Genus |
|---|---|---|---|---|---|
| SMS | 796 | 5.95 | 0.94 | 140400 | 241 |
| CM | 848 | 6.96 | 0.97 | 60377 | 245 |
| CS | 972 | 6.86 | 0.98 | 82296 | 276 |
| Phylum | Class | Order | Family | Genus | Relative Abundance (%) | ||
|---|---|---|---|---|---|---|---|
| SMS | CM | CS | |||||
| Ascomycota | Dothideomycetes | Pleosporales | Phaeosphaeriaceae | Ophiosphaerella | 0.02 | 0.11 | 1.01 |
| Eurotiomycetes | Chaetothyriales | Herpotrichiellaceae | Exophiala | 1.08 | 2.77 | 0.66 | |
| Eurotiales | Trichocomaceae | Aspergillus | 0.18 | 1.07 | 0.13 | ||
| Penicillium | 1.07 | 4.76 | 0.35 | ||||
| Onygenales | Onygenaceae | Chrysosporium | 4.18 | 2.26 | 1.22 | ||
| unidentified | unidentified | 1.77 | 1.21 | 2.19 | |||
| Leotiomycetes | Helotiales | Helotiales | Cadophora | 0.03 | 0.13 | 2.24 | |
| Thelebolales | Pseudeurotiaceae | Pseudogymnoascus | 2.45 | 3.02 | 0.73 | ||
| Pezizomycetes | Pezizales | Ascobolaceae | Ascobolus | 0.79 | 1.25 | 1.99 | |
| Pyronemataceae | Pseudaleuria | 0.19 | 0.77 | 1.44 | |||
| Scutellinia | 1.09 | 0.04 | 0.58 | ||||
| unidentified | 1.92 | 0.93 | 2.56 | ||||
| Sordariomycetes | Glomerellales | Plectosphaerellaceae | Plectosphaerella | 0.06 | 0.09 | 1.00 | |
| Hypocreales | Clavicipitaceae | Metarhizium | 0.63 | 13.96 | 0.40 | ||
| Hypocreaceae | Trichoderma | 0.35 | 0.66 | 1.08 | |||
| Hypocreales | Acremonium | 0.06 | 1.87 | 0.18 | |||
| Nectriaceae | Fusarium | 0.86 | 1.81 | 1.09 | |||
| unidentified | 4.21 | 1.10 | 1.86 | ||||
| unidentified | unidentified | 0.27 | 1.04 | 0.48 | |||
| Microascales | Microascaceae | Cephalotrichum | 1.34 | 0.12 | 0.09 | ||
| unidentified | unidentified | 7.07 | 0.17 | 0.02 | |||
| Sordariales | Chaetomiaceae | Humicola | 1.10 | 0.76 | 1.34 | ||
| Lasiosphaeriaceae | Apodus | 0.84 | 0.20 | 1.09 | |||
| Cladorrhinum | 0.16 | 0.79 | 1.54 | ||||
| Podospora | 0.17 | 1.37 | 0.92 | ||||
| unidentified | 5.49 | 1.49 | 3.69 | ||||
| Sordariaceae | unidentified | 0.03 | 0.26 | 1.39 | |||
| unidentified | unidentified | unidentified | 0.58 | 3.37 | 8.52 | ||
| unidentified | unidentified | unidentified | unidentified | 6.85 | 9.02 | 12.35 | |
| Basidiomycota | Agaricomycetes | unidentified | unidentified | unidentified | 22.57 | 3.93 | 5.72 |
| Tremellomycetes | Filobasidiales | Piskurozymaceae | Solicoccozyma | 0.01 | 0.10 | 1.04 | |
| Trichosporonales | Trichosporonaceae | Cutaneotrichosporon | 4.50 | 2.90 | 2.00 | ||
| Mortierellomycota | Mortierellomycetes | Mortierellales | Mortierellaceae | Mortierella | 2.74 | 0.05 | 0.00 |
| unidentified | unidentified | unidentified | unidentified | unidentified | 6.52 | 11.85 | 14.91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frąc, M.; Pertile, G.; Panek, J.; Gryta, A.; Oszust, K.; Lipiec, J.; Usowicz, B. Mycobiome Composition and Diversity under the Long-Term Application of Spent Mushroom Substrate and Chicken Manure. Agronomy 2021, 11, 410. https://doi.org/10.3390/agronomy11030410
Frąc M, Pertile G, Panek J, Gryta A, Oszust K, Lipiec J, Usowicz B. Mycobiome Composition and Diversity under the Long-Term Application of Spent Mushroom Substrate and Chicken Manure. Agronomy. 2021; 11(3):410. https://doi.org/10.3390/agronomy11030410
Chicago/Turabian StyleFrąc, Magdalena, Giorgia Pertile, Jacek Panek, Agata Gryta, Karolina Oszust, Jerzy Lipiec, and Bogusław Usowicz. 2021. "Mycobiome Composition and Diversity under the Long-Term Application of Spent Mushroom Substrate and Chicken Manure" Agronomy 11, no. 3: 410. https://doi.org/10.3390/agronomy11030410
APA StyleFrąc, M., Pertile, G., Panek, J., Gryta, A., Oszust, K., Lipiec, J., & Usowicz, B. (2021). Mycobiome Composition and Diversity under the Long-Term Application of Spent Mushroom Substrate and Chicken Manure. Agronomy, 11(3), 410. https://doi.org/10.3390/agronomy11030410







