1. Introduction
Grape is one of the most favorite fruits in the world, which contains a variety of vitamins, carotenoids, and polyphenols which have numerous benefits for human health such as anti-cancer, anti-oxidation, and photoprotective [
1,
2]. Italy, France, Spain, the United States, and China are the main producers of grapes. According to the survey data of the Food and Agriculture Organization of the United Nations, grape disease is the main reason for the decrease in global grape production. However, most grape diseases start from the leaves and then spread to the entire plant. Therefore, a method which could identify grape leaf diseases with high accuracy will help to improve the management of grape production and provide a good growth environment.
Conventional expert diagnosis of grape leaf disease has the disadvantage of high cost and large risk of error. With the development of computer vision (CV), machine learning (ML), and deep learning (DL), technology has been widely applied to crop disease detection [
3,
4]. Conventional machine vision methods segment crop diseases spots using handcraft features such as color, texture, or shape. However, the characteristics of different diseases’ symptoms are highly similar. As a result, it is difficult to judge the types of diseases, and the accuracy of disease recognition is poor, especially in a complex natural environment. Compared to the conventional machine learning method, deep learning can often achieve better performance. A convolutional neural network (CNN) is a high-performance deep learning network which provides end-to-end pipelines to automatically learn the expressed hierarchical features hidden in the images [
5,
6,
7]. Plant disease diagnosis based on deep networks is not only more effective but also avoids the tedious features selection procedure.
Nowadays, CNN-based models have been widely applied for early disease detection in crops and subsequent disease management. Atila et al. [
8] adopted EfficientNet to realize plant disease diagnosis, with the help of transfer learning. A dataset with 38 categories of diseased leaves was used to train the networks. Finally, the highest accuracy of 99.97% was obtained on the B4 model. Long et al. [
9] trained AlexNet and GoogleNet networks combined with a transfer learning strategy for Camellia oleifera diseases identification. Manpreet et al. [
7] used a pre-trained ResNet network to classify 7 tomato diseases and an accuracy of 98.8% was achieved on the test dataset. A deep detection model structure for tomato leaf diseases was proposed by Karthik et al. [
10]. The residual network was optimized and improved by the team to make it learn disease features more effectively. Yang et al. [
11] used the saliency analysis of the image to locate pests in tea gardens. AlexNet was optimized using some tricks such as reducing the number of network layers and convolution kernels to improve the performance. The optimized model was effective against 23 pests in tea gardens, and an average recognition accuracy of 88.1% was obtained. In [
12], a powerful neural network for identification of three different legume species based on the morphological patterns of the leaf veins was proposed by Grinblat et al. In summary, the above-mentioned literature provides a lot of references for the diagnosis of grape leaf disease in the real agriculture environment.
The above studies show that better performance can often be achieved by using a CNN network. However, a large dataset and huge computational resources are indispensable to train a deep network from scratch. Therefore, researchers began to study the method of combining deep features with support vector machines, that is, using CNN to extract deep features to train support vector machine classifiers to achieve fast, small-sample training of classifiers. In [
13], deep features extracted by 13 CNN models (AlexNet, Vgg16, Vgg19, Xception, Resnet18, Resnet50, Resnet101, Inceptionv3, Inceptionresnetv2, GoogleNet, Densenet201, Mobilenetv2, shufflenet) were adopted to train an SVM classifier. The results show that the classifier trained by features extracted by Resnet50 is superior to other networks for rice disease recognition, and a F1 score of 0.9838 was obtained on the test dataset. In the same year, the method was adopted by the team to detect nitrogen deficiency of rice. The SVM classifier trained with ResNet50 deep features achieved the best performance with an accuracy of 99.84% [
14]. In addition, an SVM classifier with the same idea as [
13,
14] was established by Jiang et al. [
15] for rice leaf diseases diagnosis, and a mean correct identification accuracy of 96.8% was achieved.
The above research shows that using the deep features extracted using a CNN model to train the SVM classifier can obtain classification performance that is not inferior to those applying a deep network directly. In addition, compared with training a deep network from scratch, the computing resources and training time of training a support vector machine classifier are significantly reduced. The problem of the proposed deep features plus support vector machine classification method is that the current research is mainly focused on finding the best deep feature for classification, i.e., evaluating the performance of an SVM model trained using deep features of a specific layer of a single CNN model. There is no research on further processing the deep features extracted from different CNN networks to further improve the identification performance. To solve the above-mentioned problem in the current research, a method to train an SVM classifier with fused deep features is proposed to further improve the diagnosis performance of grape leaf disease. Compared with the single type of deep features, the fused deep features from different networks can make the SVM classifier learn more features and improve the classification performance. The main contributions of this study are as follows:
- (1)
The deep features extracted by CNN models were adopted to train a support vector machine (SVM) classifier for the classification of grape leaf disease.
- (2)
Three deep feature fusion methods were adopted to fuse deep features extracted from different CNN models to improve the classification performance of the classifier.
- (3)
A comprehensive analysis of the deep feature plus SVM, fused deep features plus SVM, and conventional deep learning methods was carried out.
The rest of the paper is organized in the following manner. In
Section 2, the studied dataset and the proposed method is given. Then, in
Section 3, a comprehensive discussion based on the experiment results is presented. Finally, a conclusion of the research is given in
Section 4.
2. Materials and Methods
2.1. Dataset
The dataset adopted to evaluate the performance in this study is a publicly available grape leaf disease dataset, which can be downloaded at
http://www.kaggle.com (1 August 2021). The database of Kaggle, which is the largest database in the world, contains a large number of plant disease images. The dataset contains 4062 images (resolution: 256 × 256) with a total of 4 kinds of grape leaves (black rot, esca measles, leaf spot, and healthy). A detailed distribution of the dataset is shown in
Table 1, and the images of grape leaves in 4 categories are shown in
Figure 1.
2.2. Network Architecture and Deep Features Layers
In this study, the deep features extracted from three state-of-the-art CNN models, i.e., AlexNet [
16], GoogLeNet [
17], and ResNet [
18], were adopted to evaluate the performance of the proposed method. All the deep features are extracted from a fully connected layer of a CNN model. Generally, a CNN network may contain several different fully connected layers (deep feature layers), e.g., the AlexNet has three fully connected layers of fc6, fc7, and fc8. Then, in this research, only some typical deep feature layers were examined, and detailed information of the selected layers is listed in
Table 2.
2.2.1. AlexNet
AlexNet was proposed by Alex Krizhevsky et al. [
16]. and won first place in the ImageNet competition in 2012. The proposal of AlexNet is regarded as the beginning of deep learning. AlexNet, as shown in
Figure 2, is a basic, simple, and effective CNN architecture, which is mainly composed of a convolutional layer, pooling layer, rectified linear unit (ReLU) layer, and fully connected layer. The success of AlexNet can be attributed to some practical strategies: (1) using ReLU nonlinear layers instead of a sigmoid function as activation functions, which can significantly accelerate the training phase and prevent overfitting; (2) a dropout strategy, which can be considered as a regularization to reduce the co-adaptation of neurons by setting the number of input neurons or hidden neurons to zero at random, was adopted to suppress overfitting; (3) the network was trained using multi-GPU to speed up the training phase. In this research, the deep features of fc6, fc7, and fc8 of AlexNet were examined.
2.2.2. GoogLeNet
GoogLeNetwas proposed by Christian Szegedy in 2014; before that, the deep learning networks obtained better performance by increasing the depth of the network (layers). However, with the increase of layers many problems, such as overfitting, gradient disappearance, and gradient explosion, may occur. In addition, when designing a network, only one operation such as convolution or pooling was used in a layer. Moreover, the size of the convolution kernel for the convolution operation is fixed. However, in practical situations, for different sizes of images, different sizes of convolution kernels are needed to produce the best performance, or for the same image, different sizes of convolution kernels behave differently because they have a different perceptual field. To address the above problems, GoogLeNet, constructed by Inception, was proposed. Inception puts multiple convolutions parallel together as a unit to form a network. Then, the model can choose the optimal convolutional kernels by adjusting the parameters during training. Networks constructed through inception modules can use computing resources more efficiently and can extract more features with the same amount of calculation. In this research, the deep feature layer of the loss3-classifier was examined.
2.2.3. ResNet
The residual network (ResNet) was proposed by He et al. [
18], which could solve the degradation problem via the introduction of a residual module. The problem of network degradation refers to the decline of network accuracy with the deepening of network layers. It is certain that the performance degradation is not caused by overfitting, in which situation the accuracy should be high enough. Theoretically, for the problem of “accuracy decreases as the network deepens”, the residual block provides two options, i.e., identity mapping and residual mapping, where identity mapping (usually called “shortcut connection”) and residual mapping correspond to the
x and
F(
x), respectively. The output of a residual block is
y =
F(
x) +
x (do not consider nonlinear activation). In the training phase, when the network has reached the optimum state, even if the network deepens, the residual mapping will be pushed to 0, leaving only identity mapping; then, the network is kept in optimum state and the performance will not decrease.
As shown in Equation (1), when the dimensions of
and
are different, a linear projection
should be applied on
such that the dimensions of
x could match the dimensions of
.
In this article, three widely used ResNet architectures, i.e., ResNet18, ResNet50, and ResNet101, were chosen as the deep feature extraction network. In addition, the deep features extraction layers shown in
Table 2 were examined.
2.3. Fusion of Deep Features by Canonical Correlation Analysis
In this research, the canonical correlation analysis (CCA) [
19] algorithm was adopted to fuse two kinds of deep features extracted by different networks of different deep features layers into a single feature vector. The fused feature is more discriminative than any of the input feature vectors. Canonical correlation analysis (CCA) has been widely adopted to analyze associations between two sets of variables.
Suppose that two ways are adopted to extract the and dimensional deep features of each sample, and two matrices, and , are obtained respectively, where is the number of samples. Then, a total of dimensional features of each sample are extracted.
Let
and
denote the within-sets covariance matrices of
and
and
denote the between-set covariance matrix between
and
. The matrix
shown below is the overall
covariance matrix, which contains all the information on associations between the pairs of deep features.
However, the correlation between these two sets of deep feature vectors may not follow a consistent pattern, and therefore, it is difficult to understand the relationship between these two sets of deep features from this matrix [
20]. The aim of CCA is to find a linear transformation,
and
, and to maximize the pair-wise correlation between the two datasets:
where
,
and
. The covariance between
and
(
are known as canonical variables) is maximized using the Lagrange multiplier method, and the constraint condition is
. Further, the linear transformation matrix
and
can be obtained by solving the eigenvalue equation as below [
20]:
where
and
are the eigenvectors, and
is a diagonal matrix of eigenvalues or squares of the canonical correlations.
The number of non-zero eigenvalues of each equation is
, further arranged in descending order,
. The transformation matrix
and
is composed of eigenvectors corresponding to sorted non-zero eigenvalues. For the transformed data, the form of the sample covariance matrix defined in Equation (2) is as follows:
As shown in the above matrix, the upper left and lower right identity matrices indicate that the canonical variates are uncorrelated within each data set, and canonical variates have none zero correlation only on their corresponding indices.
As defined in [
19], the deep features extracted by different CNN models could be fused via concatenation or summation of the transformed features (canonical variates
and
), and the fusion equations are shown in Equations (6) and (7).
where
and
are named canonical correlation discriminant features (CCDFs). In this research, both the fusion methods shown in Equations (6) and (7) were adopted to achieve the fusion of deep features extracted from different CNN networks. In addition, the fusion method of a direct concatenation of two kinds of deep features extracted from different CNN networks was also evaluated in the experiment.
2.4. Proposed Methodology
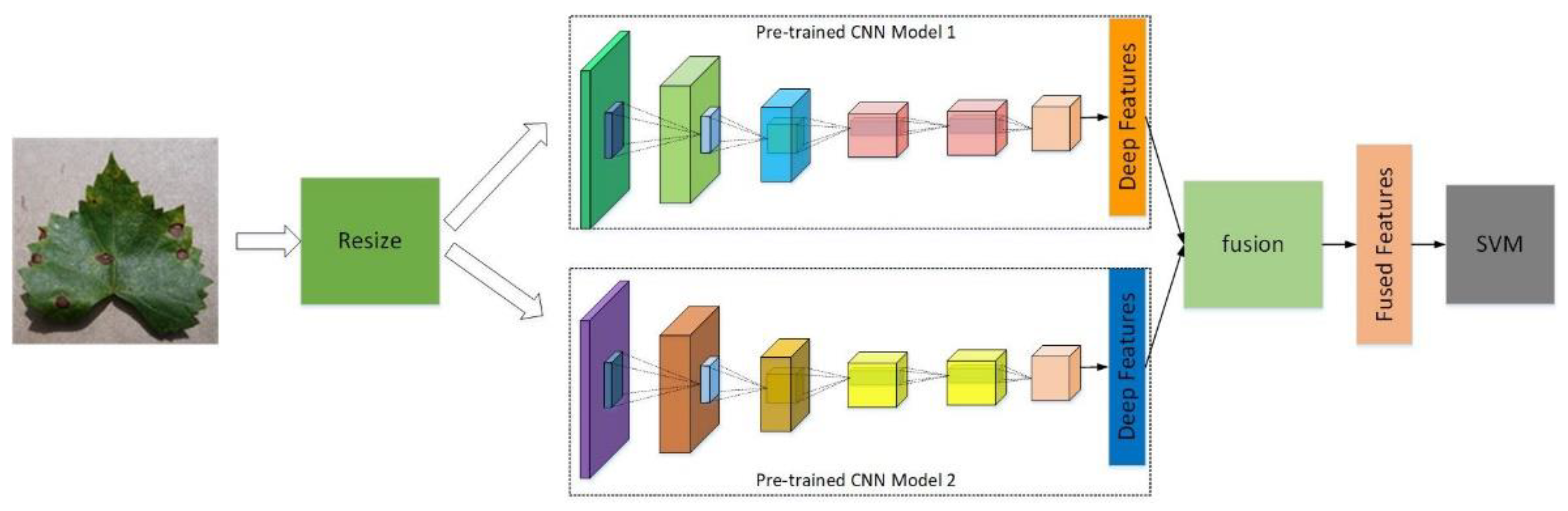
The processing flow of the proposed method is demonstrated in
Figure 3:
First, adjust the image size to make it fit the input requirement of the CNN models. The input requirements of the selected CNN models (AlexNet, ResNet, and GoogLeNet) are 227 × 227 × 3, 224 × 224 × 3, and 224 × 224 × 3, respectively.
Second, extract the deep features at specific layers of the CNN model. By inputting the image to the pre-trained CNN model and getting the parameter values on specified layers of the network, the specified deep feature can be obtained. The selected CNNs are pre-trained using ImageNet, which is a famous dataset for different applications. ImageNet contains more than 14 million images, covering more than 20,000 categories. As a result, more effective and meaningful deep features could be extracted by the pre-trained CNN models.
Third, make a fusion of the extracted deep features using one of the following methods, i.e., direct concatenation, canonical correlation analysis (CCA) concatenation, and canonical correlation analysis (CCA) sum.
Finally, feed the fused deep features into a fine-trained SVM classifier; then, the classifier can output the disease types of the input grape leaves. In the training stage, the “fit class error correcting output codes” (fitcecoc) function (MATLAB 2020b) with its default parameters was used, which can train a multi-class SVM classifier. The function of “fitcecoc” uses a K(K-1)/2 binary SVM model with a one-vs-one coding design, which enhances the classification performance of the classifier. Part of the default parameters adopted in the training stage are shown in
Table 3.
4. Conclusions and Future Works
Considering the need for diagnosis of various diseases during grape growth, an SVM plus fused deep feature method was proposed to identify three common grape leaves diseases and healthy leaves. In this paper, aiming at the identification of grape leaf diseases, a fast and accurate detection method based on fused deep, which extracted from a convolutional neural network (CNN), plus a support vector machine (SVM) is proposed. In the research, based on an open dataset, three types of state-of-the-art CNN networks, seven species of deep feature layers, three features fusion methods, and a multi-class SVM classifier were studied.
When using one type of deep feature to train the SVM classifier, the Fc1000 deep features can achieve the best classification performance, and its accuracy, precision, recall, and F1 scores are 99.08%, 99.26%, 99.24%, and 99.25%, respectively. When the feature fusion method is adopted, the performance is usually better, and the classification performance of direct concatenation is better than that of the CCA correlation fusion method. The best classification performance is obtained from the direct fusion of Fc1000 features of Resnet50 and Resnet101. Its accuracy, precision, recall and F1 scores are 99.77%, 99.81%, 99.81%, and 99.81%, respectively. The performance improvement verified that the proposed method does make sense. Furthermore, compared with using the CNN network directly, the proposed algorithm can also achieve a better classification performance. Especially, from the perspective of training time, in the experimental environment of this study, it usually takes tens of minutes to train a CNN network, while training the SVM with fused deep features only takes less than one second, which shows an obvious advantage.
In the future, work will be focused on model deployment. Many studies have implemented their algorithm on the smartphone [
28,
29,
30,
31], which is more convenient for end-users to diagnose diseases in situ. Generally, there are two candidate solutions to implement the proposed method on smartphones: (1) make the algorithm proposed in this article into a library file and develop an APP base on it; (2) deploy the algorithm in a cloud server, and the smartphone is responsible for sending image to the cloud server and receiving diagnosis results. The first scheme can enable the application to be used in the non-network environment, but depends on the computational ability, while the latter method needs good network bandwidth. In addition, although the proposed method could be applied to the diagnosis of other plant diseases theoretically, its versatility and effectiveness need to be further verified on other datasets in the future.