Exploring the Phenotypic Stability of Soybean Seed Compositions Using Multi-Trait Stability Index Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Experiments
2.2. Determination of Soybean Seed Protein, Oil, and Fatty Acid Compositions
2.3. Data Analysis
2.3.1. Analysis of Variance
2.3.2. Mean Performance and Stability Indices Based on Multiple Traits
3. Results
3.1. Mean Performance of 135 Soybean Accesions for Seed Composition Traits across Five Environments
3.2. Combined Analysis of Variance
3.3. AMMI Analysis of Variance for Studied Traits
3.4. Mean Performance and Stability of Selected Genotypes
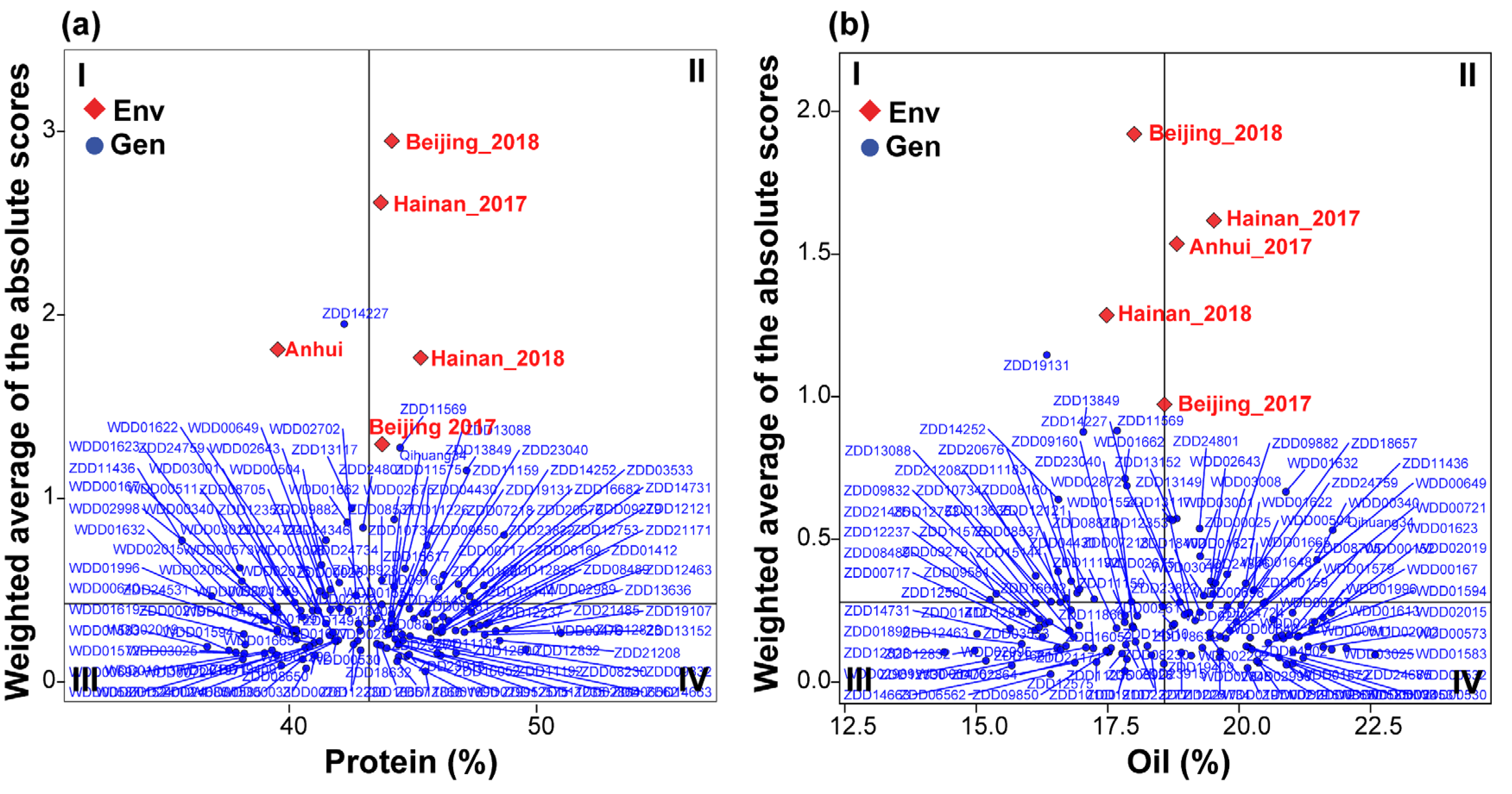
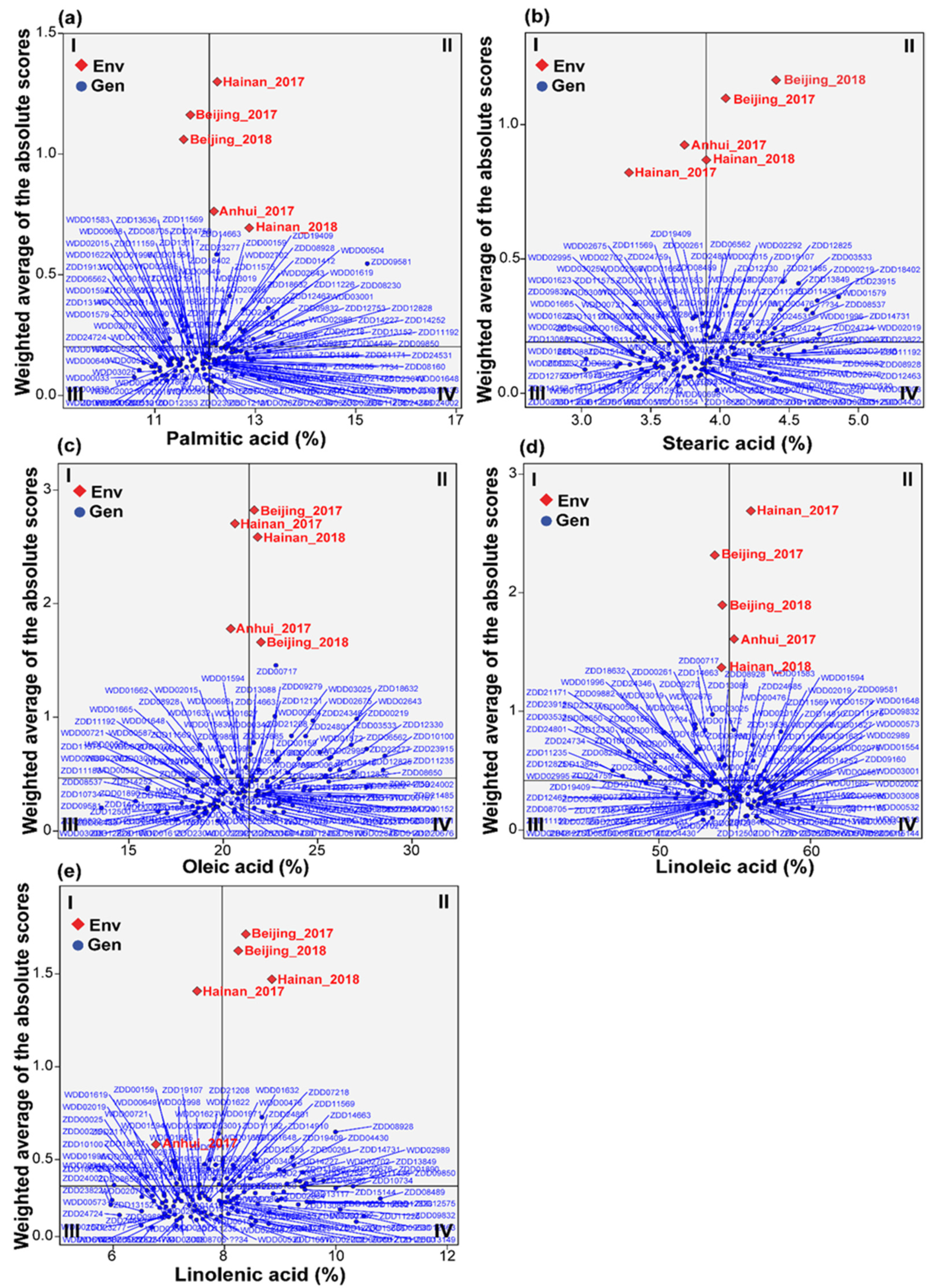
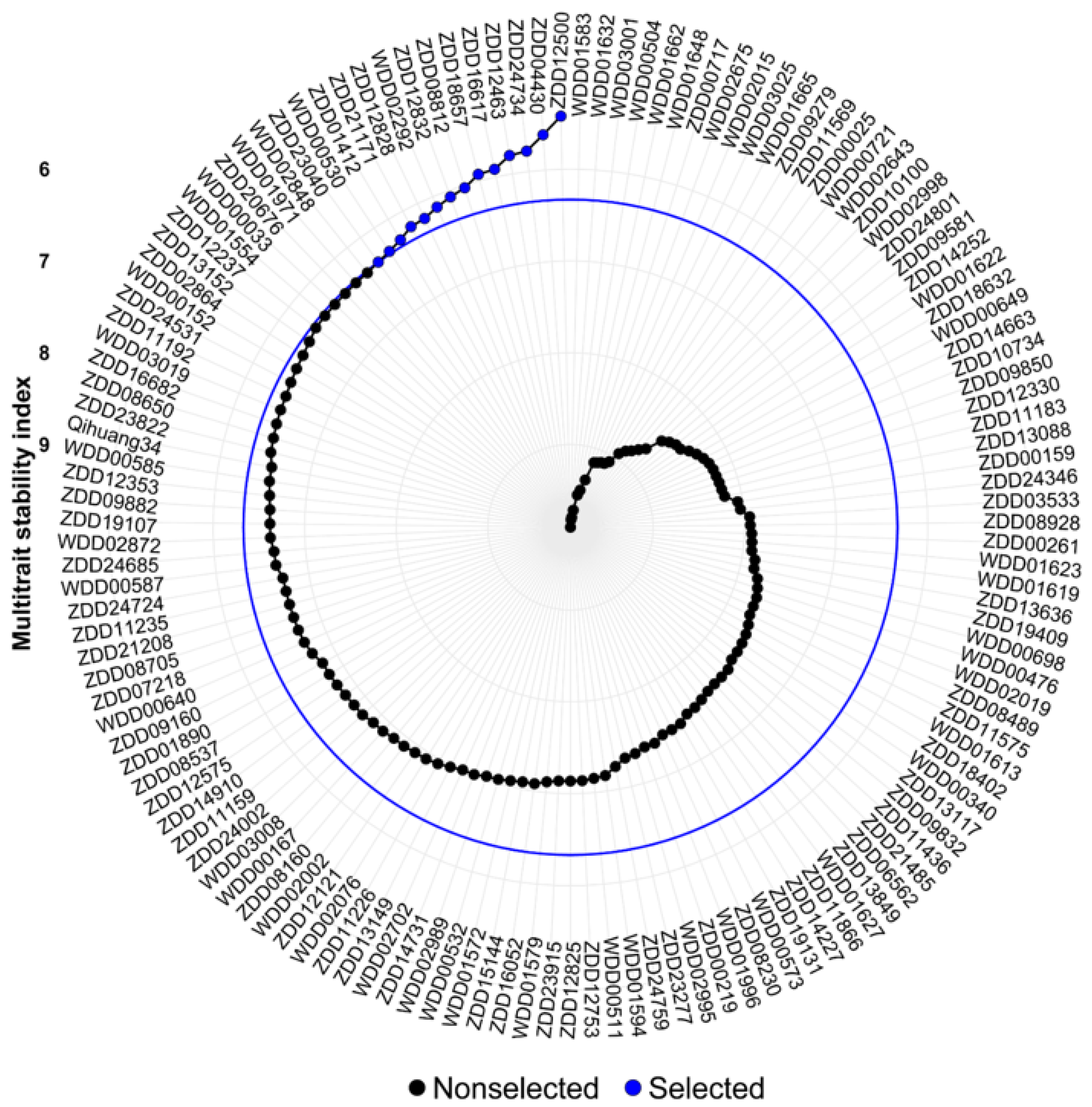
3.5. Multi-Trait Stability Index and Genotype Selection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilcox, J.R.; Shibles, R.M. Interrelationships among seed quality attributes in soybean. Crop Sci. 2001, 41, 11–14. [Google Scholar] [CrossRef]
- Wolf, W.J.; Cowan, J. Soybeans as a Food Source; CRC Press: Boca Raton, FL, USA, 1975; ISBN 0878191127. [Google Scholar]
- Kim, H.-J.; Ha, B.-K.; Ha, K.-S.; Chae, J.-H.; Park, J.-H.; Kim, M.-S.; Asekova, S.; Shannon, J.G.; Son, C.-K.; Lee, J.-D. Comparison of a high oleic acid soybean line to cultivated cultivars for seed yield, protein and oil concentrations. Euphytica 2015, 201, 285–292. [Google Scholar] [CrossRef]
- Boerma, H.R.; Specht, J.E.; Wilson, R.F. Seed Composition. Soybeans Improv. Prod. Uses 2004, 621–677. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Adaawen, S. Understanding climate change and drought perceptions, impact and responses in the rural ravannah, West Africa. Atmosphere 2021, 12, 594. [Google Scholar] [CrossRef]
- Chaudhary, K.R.; Wu, J. Stability analysis for yield and seed quality of soybean [Glycine max (l.) Merril] across different environments in Eastern South Dakota. Conf. Appl. Stat. Agric. 2012. [Google Scholar] [CrossRef] [Green Version]
- Ray, C.L.; Shipe, E.R.; Bridges, W.C. Planting date influence on soybean agronomic traits and seed composition in modified fatty acid breeding lines. Crop Sci. 2008, 48, 181–188. [Google Scholar] [CrossRef]
- Prado, E.E.D.; Hiromoto, D.M.; Godinho, V.D.P.C.; Utumi, M.M.; Ramalho, A.R. Adaptabilidade e estabilidade de cultivares de soja em cinco épocas de plantio no cerrado de Rondônia. Pesqui. Agropecuária Bras. 2001, 36, 625–635. [Google Scholar] [CrossRef] [Green Version]
- Sudarić, A.; Matoša Kočar, M.; Duvnjak, T.; Zdunić, Z.; Markulj Kulundžić, A. Improving Seed Quality of Soybean Suitable for Growing in Europe. In Soybean for Human Consumption and Animal Feed; IntechOpen: London, UK, 2020; pp. 416–680. [Google Scholar]
- Voss-Fels, K.P.; Stahl, A.; Hickey, L.T. Q&A: Modern crop breeding for future food security. BMC Biol. 2019, 17, 1–7. [Google Scholar]
- Gurmu, F.; Mohammed, H.; Alemaw, G. Genotype x environment interactions and stability of soybean for grain yield and nutrition quality. Afr. Crop Sci. J. 2009, 17. [Google Scholar] [CrossRef]
- Tolorunse, K.D.; Gana, A.S.; Bala, A.; Sangodele, E.A. Yield stability studies of soybean (Glycine max (L.) Merrill) under rhizobia inoculation in the savanna region of Nigeria. Plant Breed. 2018, 137, 262–270. [Google Scholar] [CrossRef]
- Becker, H.C.; Leon, J. Stability analysis in plant breeding. Plant Breed. 1988, 101, 1–23. [Google Scholar] [CrossRef]
- Sharma, P.K.; Gupta, P.K.; Govila, O.P. AMMI analysis of a pearl millet yield trial. Indian J. Genet. Plant Breed. 1998, 58, 183–192. [Google Scholar]
- Chandrakar, P.K.; Shrivastava, R.; Agrawal, S.K.; Rao, S.S. Stability analysis of soybean (Glycine max L. merrill) varieties in rice zone of Madhya Pradesh. J. Oilseeds Res. 1998, 15, 247–249. [Google Scholar]
- Poysa, V.; Woodrow, L. Stability of soybean seed composition and its effect on soymilk and tofu yield and quality. Food Res. Int. 2002, 35, 337–345. [Google Scholar] [CrossRef]
- Ramana, M.V.; Satyanarayana, A. Stability of yield and its components in soybean (Glycine max L. Merrill). J. Oilseeds Res. 2005, 22, 18–21. [Google Scholar]
- Arslanoglu, F.; Aytac, S.; Oner, E.K. Effect of genotype and environment interaction on oil and protein content of soybean (Glycine max (L.) Merrill) seed. Afr. J. Biotechnol. 2011, 10, 18409–18417. [Google Scholar] [CrossRef]
- Balešević-Tubić, S.; Đorđević, V.; Miladinović, J.; Đukić, V.; Tatić, M. Stability of soybean seed composition. Genetika 2011, 43, 217–227. [Google Scholar] [CrossRef]
- Akhter, M.; Sneller, C.H. Yield and yield components of early maturing soybean genotypes in the mid-south. Crop Sci. 1996, 36, 877–882. [Google Scholar] [CrossRef]
- Malek, M.A.; Rafii, M.Y.; Afroz, S.S.; Nath, U.K.; Mondal, M. Morphological characterization and assessment of genetic variability, character association, and divergence in soybean mutants. Sci. World J. 2014, 2014, 968796. [Google Scholar] [CrossRef] [Green Version]
- Olivoto, T.; Diel, M.I.; Schmidt, D.; Lúcio, A.D. Multivariate analysis of strawberry experiments: Where are we now and where can we go? bioRxiv 2021, 2012–2020. [Google Scholar] [CrossRef]
- Dalló, S.C.; Zdziarski, A.D.; Woyann, L.G.; Milioli, A.S.; Zanella, R.; Conte, J.; Benin, G. Across year and year-by-year GGE biplot analysis to evaluate soybean performance and stability in multi-environment trials. Euphytica 2019, 215, 1–12. [Google Scholar] [CrossRef]
- Jarquin, D.; Howard, R.; Crossa, J.; Beyene, Y.; Gowda, M.; Martini, J.W.R.; Covarrubias Pazaran, G.; Burgueño, J.; Pacheco, A.; Grondona, M. Genomic prediction enhanced sparse testing for multi-environment trials. G3 Genes Genomes Genet. 2020, 10, 2725–2739. [Google Scholar]
- Woyann, L.G.; Meira, D.; Matei, G.; Zdziarski, A.D.; Dallacorte, L.V.; Madella, L.A.; Benin, G. Selection indexes based on linear-bilinear models applied to soybean breeding. Agron. J. 2020, 112, 175–182. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D.C.; da Silva, J.A.G.; Sari, B.G.; Diel, M.I. Mean performance and stability in multi-environment trials II: Selection based on multiple traits. Agron. J. 2019, 111, 2961–2969. [Google Scholar] [CrossRef]
- Zuffo, A.M.; Steiner, F.; Aguilera, J.G.; Teodoro, P.E.; Teodoro, L.P.R.; Busch, A. Multi-trait stability index: A tool for simultaneous selection of soya bean genotypes in drought and saline stress. J. Agron. Crop Sci. 2020, 206, 815–822. [Google Scholar] [CrossRef]
- Koundinya, A.V.V.; Ajeesh, B.R.; Hegde, V.; Sheela, M.N.; Mohan, C.; Asha, K.I. Genetic parameters, stability and selection of cassava genotypes between rainy and water stress conditions using AMMI, WAAS, BLUP and MTSI. Sci. Hortic. 2021, 281, 109949. [Google Scholar]
- Bornhofen, E.; Todeschini, M.H.; Stoco, M.G.; Madureira, A.; Marchioro, V.S.; Storck, L.; Benin, G. Wheat yield improvements in Brazil: Roles of genetics and environment. Crop Sci. 2018, 58, 1082–1093. [Google Scholar] [CrossRef]
- Mohammadi, R.; Armion, M.; Zadhasan, E.; Ahmadi, M.M.; Amri, A. The use of AMMI model for interpreting genotype× environment interaction in durum wheat. Exp. Agric. 2018, 54, 670–683. [Google Scholar] [CrossRef]
- Nowosad, K.; Liersch, A.; Popławska, W.; Bocianowski, J. Genotype by environment interaction for seed yield in rapeseed (Brassica napus L.) using additive main effects and multiplicative interaction model. Euphytica 2016, 208, 187–194. [Google Scholar] [CrossRef]
- Olivoto, T. Stability Analysis in R Using the WAASB Index. 2019. Available online: https://doi.org/10.13140/RG.2.2.32425.75369/1 (accessed on 21 May 2021).
- Hymowitz, T.; Dudley, J.W.; Collins, F.I.; Brown, C.M. Estimations of protein and oil concentration in corn, soybean, and oat seed by near-infrared light reflectance. Crop Sci. 1974, 14, 713–715. [Google Scholar] [CrossRef]
- Fan, S.; Li, B.; Yu, F.; Han, F.; Yan, S.; Wang, L.; Sun, J. Analysis of additive and epistatic quantitative trait loci underlying fatty acid concentrations in soybean seeds across multiple environments. Euphytica 2015, 206, 689–700. [Google Scholar] [CrossRef] [Green Version]
- Abdelghany, A.M.; Zhang, S.; Azam, M.; Shaibu, A.S.; Feng, Y.; Li, Y.; Tian, Y.; Hong, H.; Li, B.; Sun, J. Profiling of seed fatty acid composition in 1025 Chinese soybean accessions from diverse ecoregions. Crop J. 2020, 8, 635–644. [Google Scholar] [CrossRef]
- Olivoto, T.; Lúcio, A.D. metan: An R package for multi—Environment trial analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Version 4.0.3; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Zhang, G.; Xu, S.; Mao, W.; Hu, Q.; Gong, Y. Determination of the genetic diversity of vegetable soybean [Glycine max (L.) Merr.] using EST-SSR markers. J. Zhejiang Univ. Sci. B 2013, 14, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Perić, V.; Nikolić, A.; Babić, V.; Sudarić, A.; Srebrić, M.; Đorđević, V.; Mladenović-Drinić, S. Genetic relatedness of soybean genotypes based on agromorphological traits and RAPD markers. Genetika-Belgrade 2014, 46, 839–854. [Google Scholar] [CrossRef]
- Adie, M.M.; Krisnawati, A. Soybean yield stability in eight locations and its potential for seed oil source in Indonesia. Energy Procedia 2015, 65, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Sudarić, A.; Šimić, D.; Vratarić, M. Characterization of genotype by environment interactions in soybean breeding programmes of southeast Europe. Plant Breed. 2006, 125, 191–194. [Google Scholar] [CrossRef]
- Bhartiya, A.; Aditya, J.P.; Singh, K.; Purwar, J.P.; Agarwal, A. AMMI & GGE biplot analysis of multi environment yield trial of soybean in North Western Himalayan state Uttarakhand of India. Legum. Res. Int. J. 2017, 40, 306–312. [Google Scholar]
- Bhartiya, A.; Aditya, J.P.; Kumari, V.; Kishore, N.; Purwar, J.P.; Agrawal, A.; Kant, L. GGE biplot & ammi analysis of yield stability in multi-environment trial of soybean [Glycine max (L.) Merrill] genotypes under rainfed condition of north western Himalayan hills. Indian J. Genet. Plant Breed. 2017, 27, 227–238. [Google Scholar]
- Mwiinga, B.; Sibiya, J.; Kondwakwenda, A.; Musvosvi, C.; Chigeza, G. Genotype x environment interaction analysis of soybean (Glycine max (L.) Merrill) grain yield across production environments in Southern Africa. F. Crop. Res. 2020, 256, 107922. [Google Scholar] [CrossRef]
- Amira, J.O.; Ojo, D.K.; Ariyo, O.J.; Oduwaye, O.A.; Ayo-Vaughan, M.A. Relative discriminating powers of GGE and AMMI models in the selection of tropical soybean genotypes. Afr. Crop Sci. J. 2013, 21, 67–73. [Google Scholar]
- Asfaw, A.; Alemayehu, F.; Gurum, F.; Atnaf, M. AMMI and SREG GGE biplot analysis for matching varieties onto soybean production environments in Ethiopia. Sci. Res. Essays 2009, 4, 1322–1330. [Google Scholar]
- Azam, M.; Zhang, S.; Abdelghany, A.M.; Shaibu, A.S.; Feng, Y.; Li, Y.; Tian, Y.; Hong, H.; Li, B.; Sun, J. Seed isoflavone profiling of 1168 soybean accessions from major growing ecoregions in China. Food Res. Int. 2020, 130. [Google Scholar] [CrossRef]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean domestication: The origin, genetic architecture and molecular bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef] [Green Version]
- Primomo, V.S.; Falk, D.E.; Ablett, G.R.; Tanner, J.W.; Rajcan, I. Genotype x environment interactions, stability, and agronomic performance of soybean with altered fatty acid profiles. Crop Sci. 2002, 42, 37–44. [Google Scholar] [CrossRef]
- Assefa, Y.; Bajjalieh, N.; Archontoulis, S.; Casteel, S.; Davidson, D.; Kovács, P.; Naeve, S.; Ciampitti, I.A. Spatial characterization of soybean yield and quality (amino acids, oil, and protein) for United States. Sci. Rep. 2018, 8, 14653. [Google Scholar] [CrossRef] [Green Version]
- Yaklich, R.W.; Vinyard, B.; Camp, M.; Douglass, S. Analysis of seed protein and oil from soybean northern and southern region uniform tests. Crop Sci. 2002, 42, 1504–1515. [Google Scholar] [CrossRef]
- Wu, T.; Yang, X.; Sun, S.; Wang, C.; Wang, Y.; Jia, H.; Man, W.; Fu, L.; Song, W.; Wu, C. Temporal–spatial characterization of seed proteins and oil in widely grown soybean cultivars across a century of breeding in China. Crop Sci. 2017, 57, 748–759. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, X.; Huang, W.; Yang, J.; Zheng, Y.; Wang, S.; Qiu, L. Stability analysis of seven agronomic traits for soybean [(Glycine max (L.) Merr.] Tokachi nagaha and its derived cultivars using the AMMI model. Plant Prod. Sci. 2017, 20, 499–506. [Google Scholar] [CrossRef] [Green Version]
- Nataraj, V.; Pandey, N.; Ramteke, R.; Verghese, P.; Reddy, R.; Onkarappa, T.; Mehtre, S.P.; Gupta, S.; Satpute, G.K.; Mohan, Y. GGE biplot analysis of vegetable type soybean genotypes under multi-environmental conditions in India. J. Environ. Biol. 2021, 42, 247–253. [Google Scholar] [CrossRef]
- Sousa, L.B.; Hamawaki, O.T.; Nogueira, A.P.O.; Batista, R.O.; Oliveira, V.M.; Hamawaki, R.L. Evaluation of soybean lines and environmental stratification using the AMMI, GGE biplot, and factor analysis methods. Genet. Mol. Res. 2015, 14, 12660–16674. [Google Scholar] [CrossRef] [PubMed]
- Cheelo, P.; Lungu, D.; Mwala, M. GGE biplot analysis for identification of ideal soybean [Glycine max L. Merrill] test and production locations in Zambia. J. Exp. Agric. Int. 2017, 1–15. [Google Scholar] [CrossRef]
- Nataraj, V.; Bhartiya, A.; Singh, C.P.; Devi, H.N.; Deshmukh, M.P.; Verghese, P.; Singh, K.; Mehtre, S.P.; Kumari, V.; Maranna, S.; et al. WAASB based stability analysis and simultaneous selection for grain yield and early maturity in soybean. Agron. J. 2021, 113, 3089–3099. [Google Scholar] [CrossRef]
- Hagos, H.G.; Abay, F. AMMI and GGE biplot analysis of bread wheat genotypes in the northern part of Ethiopia. J. Plant Breed. Genet. 2013, 1, 12–18. [Google Scholar]
- Mahmodi, N.; Yaghotipoor, A.; Farshadfar, E. AMMI stability value and simultaneous estimation of yield and yield stability in bread wheat (Triticum aestivum L.). Aust. J. Crop Sci. 2011, 5, 1837–1844. [Google Scholar]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef]
- Sneller, C.H.; Dombek, D. Comparing soybean cultivar ranking and selection for yield with AMMI and full-data performance estimates. Crop Sci. 1995, 35, 1536–1541. [Google Scholar] [CrossRef]
- Ndhlela, T.; Herselman, L.; Magorokosho, C.; Setimela, P.; Mutimaamba, C.; Labuschagne, M. Genotype× environment interaction of maize grain yield using AMMI biplots. Crop Sci. 2014, 54, 1992–1999. [Google Scholar] [CrossRef]
- Shilpashree, N.; Devi, S.N.; Manjunathagowda, D.C.; Muddappa, A.; Abdelmohsen, S.A.M.; Tamam, N.; Elansary, H.O.; El-Abedin, T.K.Z.; Abdelbacki, A.M.M.; Janhavi, V. Morphological characterization, variability and diversity among vegetable soybean (Glycine max L.) genotypes. Plants 2021, 10, 671. [Google Scholar] [CrossRef] [PubMed]
- Whaley, R.; Eskandari, M. Genotypic main effect and genotype-by-environment interaction effect on seed protein concentration and yield in food-grade soybeans (Glycine max (L.) Merrill). Euphytica 2019, 215, 33. [Google Scholar] [CrossRef]
- Rao, P.S.; Reddy, P.S.; Rathore, A.; Reddy, B.V.S.; Panwar, S. Application GGE biplot and AMMI model to evaluate sweet sorghum (Sorghum bicolor) hybrids for genotype× environment interaction and seasonal adaptation. Indian J. Agric. Sci. 2011, 81, 438–444. [Google Scholar]
- Olivoto, T.; Lúcio, A.D.C.; da Silva, J.A.G.; Marchioro, V.S.; de Souza, V.Q.; Jost, E. Mean performance and stability in multi-environment trials I: Combining features of AMMI and BLUP techniques. Agron. J. 2019, 111, 2949–2960. [Google Scholar] [CrossRef]
| Source | Df | Mean Squares | ||||||
|---|---|---|---|---|---|---|---|---|
| Protein Content | Oil Content | Palmitic Acid | Stearic Acid | Oleic Acid | Linoleic Acid | Linolenic Acid | ||
| ENV | 4 | 1970 ** | 288.00 | 74.9 ns | 66 ns | 255 ** | 404 ** | 256 ** |
| REP(ENV) | 10 | 49.4 ** | 49.10 | 44.7 ** | 32.6 ** | 47 ** | 70.4 ** | 60.4 ** |
| GEN | 134 | 152 ** | 54.40 | 7.78 ** | 2.16 ** | 113 ** | 98.8 ** | 16.2 ** |
| GEN × ENV | 536 | 18.3 ** | 4.50 | 1 ** | 0.74 ** | 25.1 ** | 17 ** | 2.78 ** |
| Residuals | 1340 | 0.05 | 0.09 | 0.09 | 0.09 | 0.08 | 0.08 | 0.09 |
| Source | Df | Mean Squares | ||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Oil | Palmitic Acid | Stearic Acid | Oleic Acid | Linoleic Acid | Linolenic Acid | ||
| ENV | 4 | 1970 *** | 282 ** | 72.2 ns | 64.9 ns | 253 ** | 415 ** | 248 * |
| REP(ENV) | 10 | 49.4 *** | 45.9 *** | 47.6 *** | 31.6 *** | 47 *** | 68.8 *** | 61.7 *** |
| GEN | 134 | 152 *** | 53.2 *** | 7.57 *** | 2.16 *** | 112 *** | 97.8 *** | 16.5 *** |
| GEN × ENV | 536 | 18.3 *** | 4.36 *** | 0.886 *** | 0.63 *** | 25.1 *** | 16.9 *** | 2.62 *** |
| IPCA1† | 137 | 33.6 *** | 7.93 *** | 1.53 *** | 1.16 *** | 43.8 *** | 27.9 *** | 5.03 *** |
| IPCA2 | 135 | 18.8 *** | 4.07 *** | 0.825 *** | 0.594 *** | 22 *** | 15.6 *** | 2.7 *** |
| IPCA3 | 133 | 11.6 *** | 3.09 *** | 0.705 *** | 0.461 *** | 19.8 *** | 12.2 *** | 1.54 *** |
| IPCA4 | 131 | 8.39 *** | 2.2 *** | 0.457 *** | 0.283 *** | 14.1 *** | 11.4 *** | 1.13 *** |
| Residuals | 1340 | 0.00321 | 0.001 | 0.00576 | 0.0022 | 0.00238 | 0.0000148 | 0.0092 |
| Total | 2560 | 18.9 | 5.23 | 1.07 | 0.603 | 16.9 | 13.1 | 2.59 |
| Trait | Factor | Xo | Xs | SD | SD (%) | SG | SG (%) | h2 |
|---|---|---|---|---|---|---|---|---|
| Protein | FA 1 | 44.1 | 46 | 1.93 | 4.37 | 1.69 | 3.84 | 0.88 |
| Oil | FA 1 | 19.4 | 18.4 | −0.941 | −4.86 | −0.86 | −4.45 | 0.92 |
| Palmitic acid | FA 1 | 12.9 | 13 | 0.145 | 1.13 | 0.13 | 0.98 | 0.87 |
| Linolenic acid | FA 1 | 8.76 | 9.01 | 0.246 | 2.81 | 0.20 | 2.33 | 0.83 |
| Stearic acid | FA 2 | 4.69 | 4.85 | 0.156 | 3.33 | 0.10 | 2.19 | 0.66 |
| Oleic acid | FA 2 | 22.2 | 21.8 | −0.386 | −1.74 | −0.30 | −1.35 | 0.78 |
| Linoleic acid | FA 3 | 55.4 | 55.3 | −0.12 | −0.216 | −0.099 | −0.18 | 0.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelghany, A.M.; Zhang, S.; Azam, M.; Shaibu, A.S.; Feng, Y.; Qi, J.; Li, J.; Li, Y.; Tian, Y.; Hong, H.; et al. Exploring the Phenotypic Stability of Soybean Seed Compositions Using Multi-Trait Stability Index Approach. Agronomy 2021, 11, 2200. https://doi.org/10.3390/agronomy11112200
Abdelghany AM, Zhang S, Azam M, Shaibu AS, Feng Y, Qi J, Li J, Li Y, Tian Y, Hong H, et al. Exploring the Phenotypic Stability of Soybean Seed Compositions Using Multi-Trait Stability Index Approach. Agronomy. 2021; 11(11):2200. https://doi.org/10.3390/agronomy11112200
Chicago/Turabian StyleAbdelghany, Ahmed M., Shengrui Zhang, Muhammad Azam, Abdulwahab S. Shaibu, Yue Feng, Jie Qi, Jing Li, Yanfei Li, Yu Tian, Huilong Hong, and et al. 2021. "Exploring the Phenotypic Stability of Soybean Seed Compositions Using Multi-Trait Stability Index Approach" Agronomy 11, no. 11: 2200. https://doi.org/10.3390/agronomy11112200
APA StyleAbdelghany, A. M., Zhang, S., Azam, M., Shaibu, A. S., Feng, Y., Qi, J., Li, J., Li, Y., Tian, Y., Hong, H., Lamlom, S. F., Li, B., & Sun, J. (2021). Exploring the Phenotypic Stability of Soybean Seed Compositions Using Multi-Trait Stability Index Approach. Agronomy, 11(11), 2200. https://doi.org/10.3390/agronomy11112200











