Semi-Quantification of Lectins in Rice (Oryza sativa L.) Genotypes via Hemagglutination
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Plant Material
2.2. Extraction of Lectins from Rice
2.3. Preparation of RBCs
2.4. HA Assay
2.5. Total Protein Estimation
2.6. Hemagglutination Inhibition Assay (HIA)
2.7. Screening Blood Affinity for Rice Lectins
3. Results
3.1. Rice Lectins Extraction
3.2. HA Assay
3.3. Total Protein Content
3.4. HAI
3.5. Affinity Comparison of Various Types of Blood for Rice Lectins
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peumans, W.J.; van Damme, E.J.M. Plant Lectins: Versatile Proteins with Important Perspectives in Biotechnology. Biotechnol. Genet. Eng. Rev. 1998, 15, 199–228. [Google Scholar] [CrossRef]
- Sharon, N.; Lis, H. Lectins as cell recognition molecules. Science 1989, 246, 227–234. [Google Scholar] [CrossRef]
- Zhang, N.; Ping, Q.; Huang, G.; Xu, W.; Cheng, Y.; Han, X. Lectin-modified solid lipid nanoparticles as carriers for oral administration of insulin. Int. J. Pharm. 2006, 327, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Dan, X.; Liu, W.; Ng, T.B. Development and Applications of Lectins as Biological Tools in Biomedical Research. Med. Res. Rev. 2016, 36, 221–247. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.; Dan, X.; Ng, C.C.W.; Ng, T.B. Lectins with Potential for Anti-Cancer Therapy. Molecules 2015, 20, 3791–3810. [Google Scholar] [CrossRef]
- Fu, L.-L.; Zhou, C.-C.; Yao, S.; Yu, J.-Y.; Liu, B.; Bao, J.-K. Plant lectins: Targeting programmed cell death pathways as antitumor agents. Int. J. Biochem. Cell Biol. 2011, 43, 1442–1449. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Bian, H.-J.; Bao, J.-K. Plant lectins: Potential antineoplastic drugs from bench to clinic. Cancer Lett. 2010, 287, 1–12. [Google Scholar] [CrossRef]
- Peumans, W.J.; Cammue, B.P.A. Gramineae Lectins: A Special Class of Plant Lectins. In Proceedings of the IUB Symposium No. 144, The Seventh International Lectin Meeting, Bruxelles, Belgium, 18–23 August 1985; pp. 31–38. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, J.; Ilic, S.; Xue, S.J.; Kakuda, Y. Biological Properties and Characterization of Lectin from Red Kidney Bean (Phaseolus Vulgaris). Food Rev. Int. 2008, 25, 12–27. [Google Scholar] [CrossRef]
- Gautam, A.K.; Shrivastava, N.; Sharma, B.; Bhagyawant, S.S. Current Scenario of Legume Lectins and Their Practical Applications. J. Crop. Sci. Biotechnol. 2018, 21, 217–227. [Google Scholar] [CrossRef]
- Gulzar, H.; Pervez, S.; Jan, A.; Nawaz, M.A. Phylogenetics of rice (Oryza sativa L.) genotypes from Pakistan based on diver-sity of biochemical markers. Biosci. Res. 2021, 18, 1681–1693. [Google Scholar] [CrossRef]
- Souza, M.A.; Carvalho, F.C.; Ruas, L.; Ricci-Azevedo, R.; Roque-Barreira, M.C. The immunomodulatory effect of plant lectins: A review with emphasis on ArtinM properties. Glycoconj. J. 2013, 30, 641–657. [Google Scholar] [CrossRef]
- Chrispeels, M.J.; Raikhel, N.V. Lectins, lectin genes, and their role in plant defense. Plant Cell 1991, 3, 1–9. [Google Scholar] [CrossRef]
- Powell, K.; Gatehouse, A.; Hilder, V.A.; Gatehouse, J. Antimetabolic effects of plant lectins and plant and fungal enzymes on the nymphal stages of two important rice pests, Nilaparvata lugens and Nephotettix cinciteps. Entomol. Exp. Appl. 1993, 66, 119–126. [Google Scholar] [CrossRef]
- Passricha, N.; Saifi, S.K.; Kharb, P.; Tuteja, N. Rice lectin receptor-like kinase provides salinity tolerance by ion homeostasis. Biotechnol. Bioeng. 2020, 117, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Teraoka, T.; Yamanaka, H.; Harashima, A.; Kunisaki, A.; Takahashi, H.; Hosokawa, D. Novel mannose-binding rice lectin composed of some isolectins and its relation to a stress-inducible salT gene. Plant Cell Physiol. 2000, 41, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Reddy, I.N.B.L.; Kim, B.-K.; Yoon, I.-S.; Kim, K.-H.; Kwon, T.-R. Salt Tolerance in Rice: Focus on Mechanisms and Approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Claes, B.; DeKeyser, R.; Villarroel, R.; Bulcke, M.V.D.; Bauw, G.; Van Montagu, M.; Caplan, A. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 1990, 2, 19–27. [Google Scholar] [CrossRef]
- Garcia, A.B.; Engler, J.A.; Claes, B.; Villarroel, R.; van Montagu, M.V.; Gerats, T.; Caplan, A. The expression of the salt-responsive gene sal T from rice is regulated by hormonal and developmental cues. Planta 1998, 207, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Barre, A.; Astoul, C.H.; Rovira, P.; Proost, P.; Truffa-Bachi, P.; Jalali, A.A.H.; van Damme, E.; Zhang, W. Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa L.) plants. Planta 2000, 210, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Branco, A.T.; Bernabé, R.B.; Ferreira, B.D.S.; de Oliveira, M.V.V.; Garcia, A.B.; Filho, G.A.D.S. Expression and purification of the recombinant SALT lectin from rice (Oryza sativa L.). Protein Expr. Purif. 2004, 33, 34–38. [Google Scholar] [CrossRef]
- Raikhel, N.V.; Lerner, D.R. Expression and regulation of lectin genes in cereals and rice. Dev. Genet. 1991, 12, 255–260. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Ma, Z.; Ramachandran, S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bellande, K.; Bono, J.-J.; Savelli, B.; Jamet, E.; Canut, H. Plant Lectins and Lectin Receptor-Like Kinases: How Do They Sense the Outside? Int. J. Mol. Sci. 2017, 18, 1164. [Google Scholar] [CrossRef]
- Munday, J.; Chua, N.-H. Abscisic acid and water stress induce the expression of a novel rice gene. EMBO J. 1988, 7, 2279–2286. [Google Scholar] [CrossRef]
- Fadzilla, N.A.M.; Finch, R.P.; Burdon, R.H. Salinity, oxidative stress and antioxidant responses in shoot cultures of rice. J. Exp. Bot. 1997, 48, 325–331. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Arai, R. Lectin engineering: The possible and the actual. Interface Focus 2019, 9, 20180068. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, S.; Chaudhary, V.K. Recombinant fusion proteins for haemagglutination-based rapid detection of antibodies to HIV in whole blood. J. Immunol. Methods 2001, 256, 121–140. [Google Scholar] [CrossRef]
- Aubert, D.; Foudrinier, F.; Kaltenbach, M.L.; Guyot-Walser, D.; Marx-Chemla, C.; Geers, R.; Lepan, H.; Pinon, J.M. Automated reading and processing of quantitative IgG, IgM, IgA, and IgE isotypic agglutination results in microplates Development and application in parasitology-mycology. J. Immunol. Methods 1995, 186, 323–328. [Google Scholar] [CrossRef]
- Lorbach, J.N.; Wang, L.; Nolting, J.M.; Benjamin, M.G.; Killian, M.L.; Zhang, Y.; Bowman, A.S. Porcine Hemagglutinating Encephalomyelitis Virus and Respiratory Disease in Exhibition Swine, Michigan, USA, 2015. Emerg. Infect. Dis. 2017, 23, 1168–1171. [Google Scholar] [CrossRef]
- Wood, J.M.; Major, D.; Heath, A.; Newman, R.W.; Höschler, K.; Stephenson, I.; Clark, T.; Katz, J.M.; Zambon, M.C. Reproducibility of serology assays for pandemic influenza H1N1: Collaborative study to evaluate a candidate WHO International Standard. Vaccine 2012, 30, 210–217. [Google Scholar] [CrossRef]
- Gulzar, H.; Gul, J.; Hassan, G. Protein profiles of phytoagglutinins from indigenous species of Euphorbiaceae, Leguminosae and Moraceae from Pakistan. Pak. J. Bot. 2015, 47, 119–126. [Google Scholar]
- Bhagyawant, S.S.; Gautam, A.; Chaturvedi, S.K.; Shrivastava, N. Hemagglutinating activity of Chickpea extracts for lectin. Int. J. Pharm. Phytopharm. Res. 2017, 5, 15–21. [Google Scholar]
- Bing, D.H.; Weyand, J.G.M.; Stavitsky, A.B. Hemagglutination with Aldehyde-Fixed Erythrocytes for Assay of Antigens and Antibodies. Exp. Biol. Med. 1967, 124, 1166–1170. [Google Scholar] [CrossRef]
- Heard, D.H. Factors influencing the agglutinability of red cells. Variation of the red cells of the rabbit in susceptibility to ag-glutination by homologous iso-antisera. Epidemiol. Infect. 1955, 53, 408–419. [Google Scholar] [CrossRef][Green Version]
- Levine, P.; Landsteiner, K. On immune isoagglutinins in rabbits. J. Immunol. 1929, 17, 559–564. [Google Scholar]
- Kellner, A.; Hedal, E.F. Experimental Erythroblastosis Fetalis in Rabbits. J. Exp. Med. 1953, 97, 33–49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nakata, H.; Lin, C.Y.; Abolhassani, M.; Ogawa, T.; Tateno, H.; Hirabayashi, J.; Muramoto, K. Isolation of Rice Bran Lectins and Characterization of Their Unique Behavior in Caco-2 Cells: Characterization of a pair of allelic blood group fac-tors and their specific immune isoantibodies. Int. J. Mol. Sci. 2017, 18, 1052. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ng, T.; Liu, Q.B. Isolation of a new heterodimeric lectin with mitogenic activity from fruiting bodies of the mushroom Agrocybe cylindracea. Life Sci. 2002, 70, 877–885. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sun, J.-P.; Hou, C.-Y.; Feng, J.; Wang, X. Determination of the protein content in rice by the digital chromatic method. J. Food Qual. 2008, 31, 250–263. [Google Scholar] [CrossRef]
- Adenike, K.; Eretan, O.B. Purification and partial characterization of a lectin from the fresh leaves of Kalanchoe crenata (Andr.) Haw. J. Biochem. Mol. Biol. 2004, 37, 229–233. [Google Scholar] [CrossRef] [PubMed]
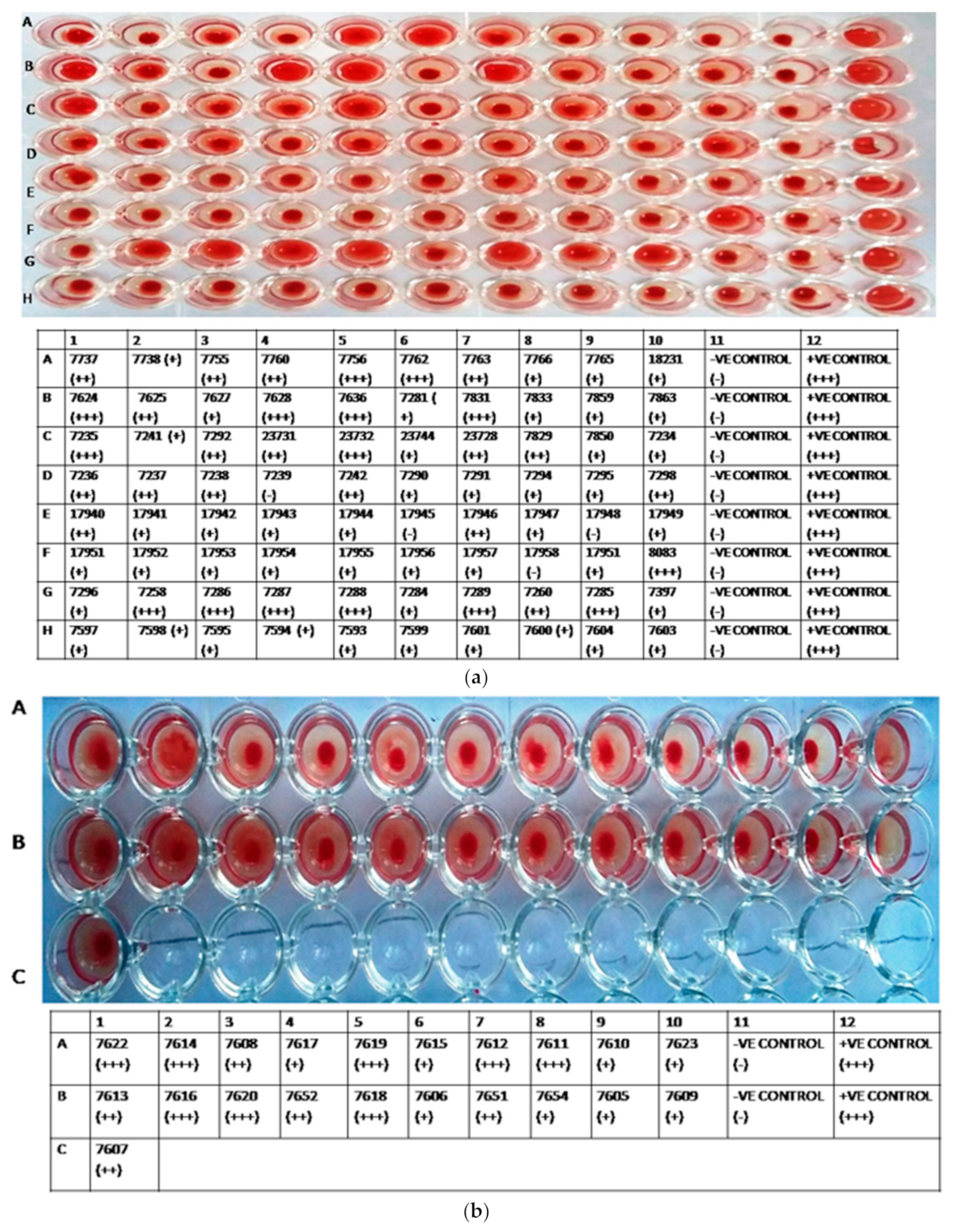
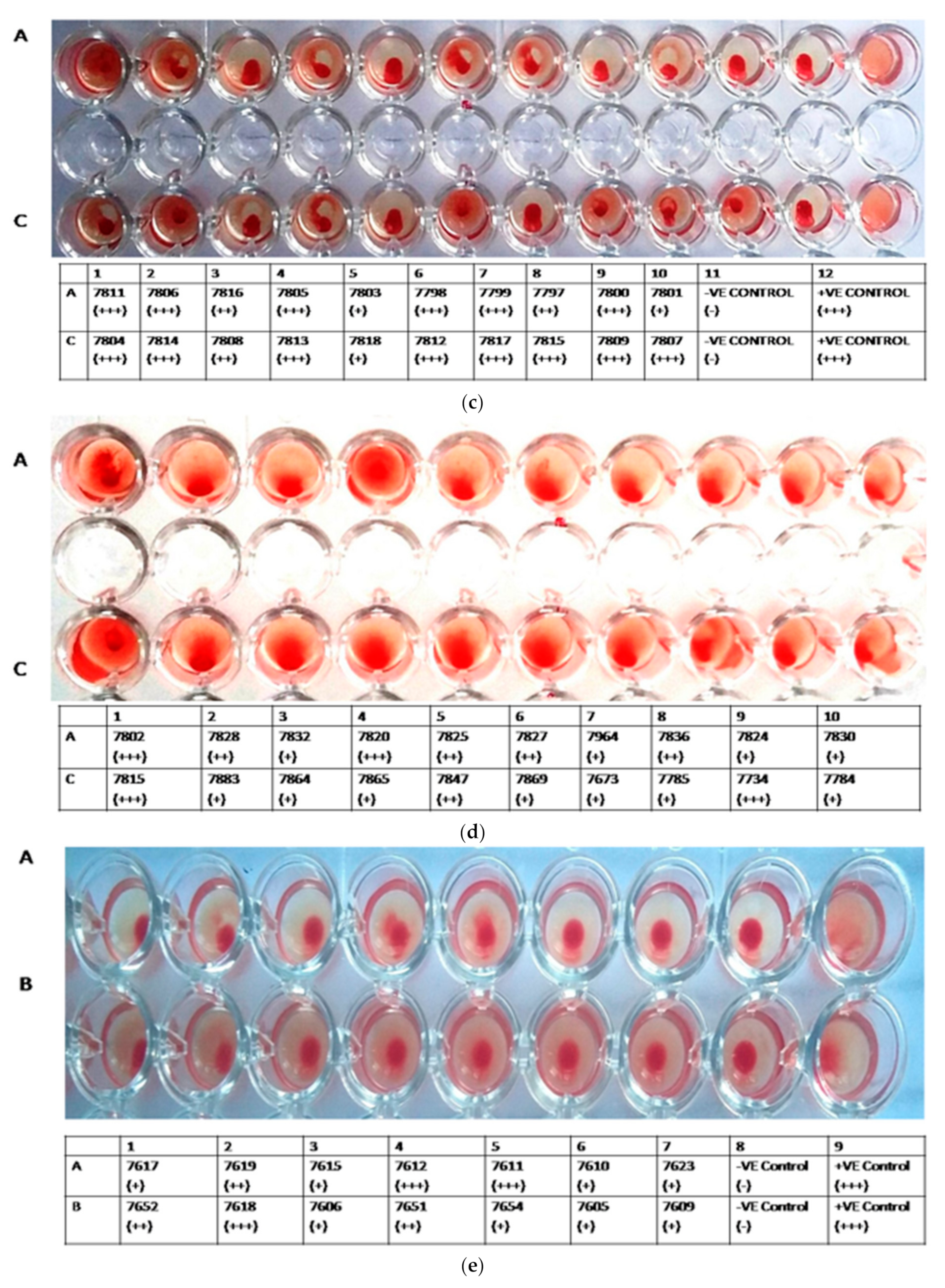
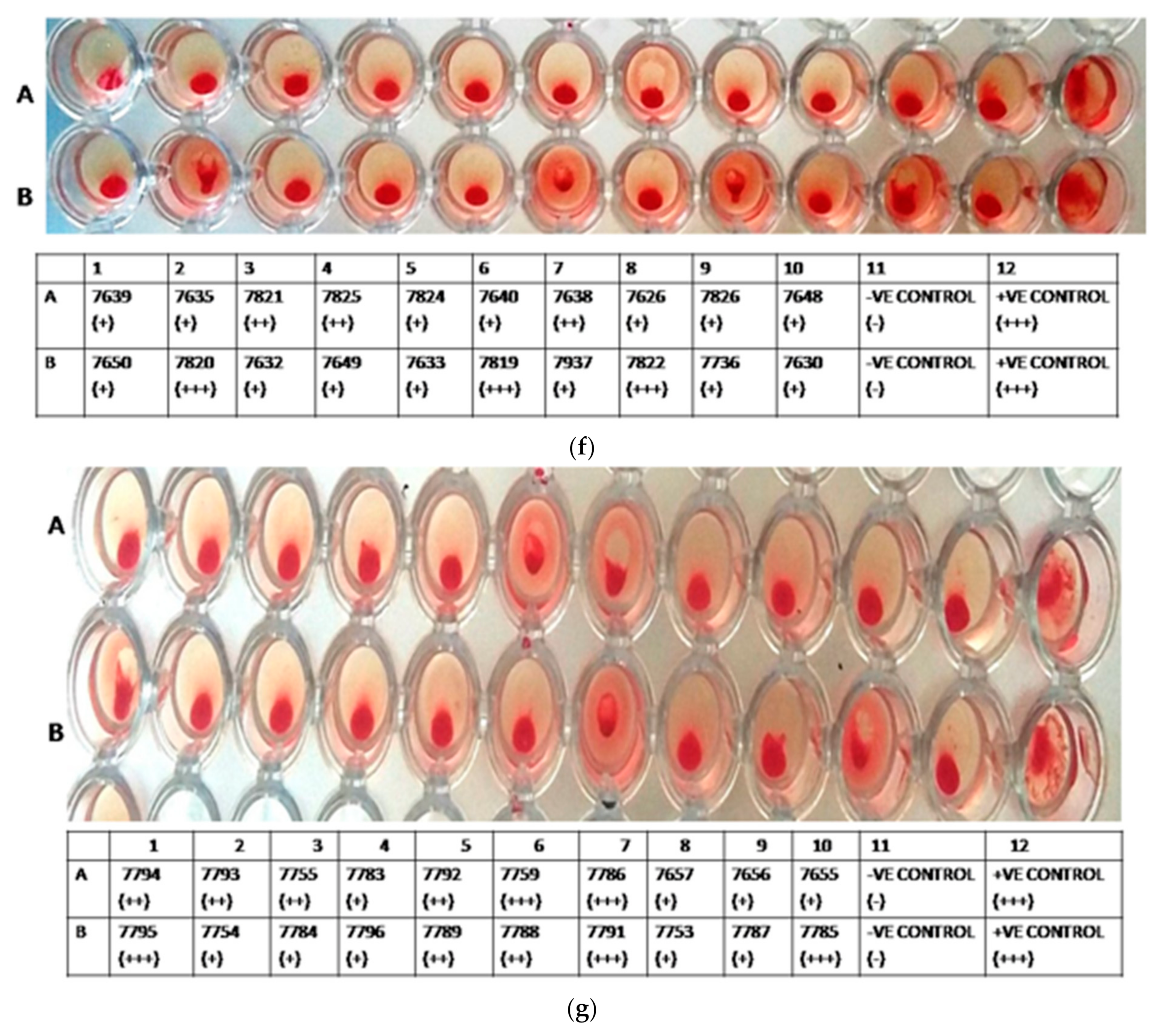
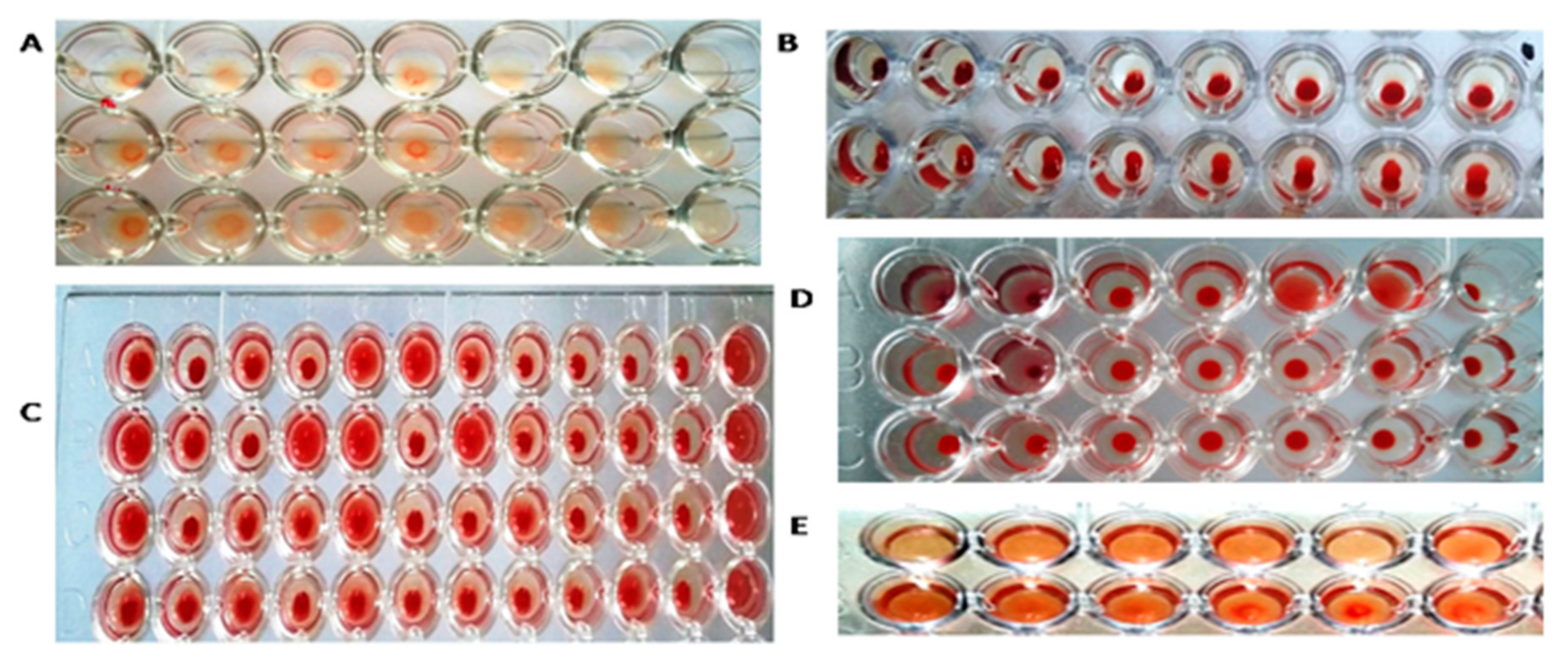
| S. No | Accession No. | Country, Year of Release | Province/ Donor No/Area | District |
|---|---|---|---|---|
| 1–4 | 7216–7219 | China, 1983 | C-12, C-13, C-14, C-15 respectively | NA |
| 5 | 7225 | Pakistan, 1983 | Punjab | Sheikhupura |
| 6 | 7234 | // | Khyber Pakhtunkhwa | Unknown |
| 7 | 7235 | // | // | Malakand |
| 8–11 | 7236–7239 | // | // | Unknown |
| 12–13 | 7241–7242 | // | // | Unknown |
| 14 | 7258 | Pakistan, 1984 | // | Parachinar |
| 15 | 7260 | // | // | Kurrum Agency |
| 16–18 | 7261–7263 | // | // | Pachinar |
| 19 | 7268 | // | // | Parachinar |
| 20 | 7269 | // | // | Unknown |
| 21 | 7271 | // | // | Unknown |
| 22 | 7280 | // | // | Unknown |
| 23 | 7281 | Pakistan, 1987 | // | Mansehra |
| 24–29 | 7284–89 | Pakistan, 1984 | // | Chitral |
| 30–35 | 7290–7295 | // | // | Swat |
| 36–39 | 7296–99 | // | // | Mansehra |
| 40 | 7396 | Pakistan, 1987 | // | // |
| 41 | 7397 | // | // | Swat |
| 42 | 7411 | // | Unknown | Unknown |
| 43 | 7413 | // | // | // |
| 44 | 7414 | // | // | // |
| 45 | 7416 | // | // | // |
| 46–47 | 7418–7419 | // | // | // |
| 48 | 7420 | // | Gilgit, Baltistan | NA |
| 49–51 | 7593–7595 | // | Khyber Pakhtunkhwa | Malakand |
| 52–55 | 7597–7600 | // | // | // |
| 56 | 7601 | Pakistan, 1989 | // | Dir |
| 57–64 | 7603–7610 | // | // | // |
| 63–84 | 7611–7630 | // | // | Chitral |
| 85–93 | 7632–7640 | // | // | Swat |
| 94 | 7641 | // | // | Unknown |
| 95–104 | 7648–7657 | // | // | Mansehra |
| 105–106 | 7672–7673 | // | Punjab | Gujrat |
| 107–108 | 7734–7735 | // | AJK | NA |
| 109–110 | 7737–7738 | // | Khyber Pakhtunkhwa | Unknown |
| 111–113 | 7753–7755 | // | // | Besham |
| 114 | 7756 | // | // | Unknown |
| 115 | 7759 | // | // | Mansehra |
| 116 | 7760 | // | // | Unknown |
| 117 | 7762 | // | // | // |
| 118 | 7763 | // | Punjab | Gujranwala |
| 119 | 7765 | // | // | Bahawalnagar |
| 120 | 7766 | // | // | Bahawalpur |
| 121–147 | 7783–7809 | Pakistan, 1991 | Khyber Pakhtunkhwa | Chitral |
| 148–155 | 7811–7818 | // | // | // |
| 156–159 | 7819–7822 | // | // | // |
| 160–163 | 7824–7827 | // | // | // |
| 164–166 | 7828–7830 | // | // | Unknown |
| 167–169 | 7831–7833 | // | // | // |
| 170 | 7836 | // | // | // |
| 171 | 7839 | // | // | Unknown |
| 172 | 7847 | Pakistan, 1973 | RGP-ARC | NA |
| 173 | 7850 | // | // | // |
| 174 | 7859 | // | // | // |
| 175 | 7863 | // | // | // |
| 176–177 | 7864–7865 | Pakistan, 1972 | // | // |
| 178 | 7869 | Pakistan, 1973 | // | // |
| 179 | 7883 | // | // | // |
| 180 | 7886 | // | // | // |
| 181 | 7937 | Pakistan,1991 | // | Chitral |
| 182 | 8083 | // | // | // |
| 183 | 8085 | // | // | // |
| 184 | 17940 | // | // | Swat |
| 185–186 | 17,941–17,942 | Pakistan, 2000 | // | Nowshehra |
| 187–189 | 17,943–17,945 | Pakistan, 1996 | // | Chitral |
| 190 | 17,946 | Pakistan, 1997 | // | Mansehra |
| 191–202 | 17,947–17,958 | IRRI, 1999 | NA | NA |
| 203 | 18,231 | Source not mentioned in the catalog // // // | ||
| 204 | 23,718 | |||
| 205–206 | 23,727–23,728 | |||
| 207–208 | 23,731–23,732 | |||
| 209 | 23,736 | // | ||
| 210 | 23,744 | |||
| 211 | Boti | Pakistan | Khyber, Pakhtunkhwa | Swat |
| 212 | Fakhre Malakand | // | // | Malakand |
| 213–217 | Five Transgenic seeds | NA | NA | NA |
| Dilution Number | Ratio and %Age Concentration of the Diluted Sample | Dilution Factor | HA Titer Value (HU/mL) | Specific Activity = Titre/mg of Protein | |
|---|---|---|---|---|---|
| Original sample | 1:1 (100%) | 1/20 | 1 | 1 | e.g., Protein Concentration = 6.2 mg/mL and titre = 16 Therefore, Specific activity of the plant/animal lectins showing visible HA will be 16/6.2 = 2.58 |
| 1 | 1:2 (50%) | 1/21 | 1/2 | 2 | |
| 2 | 1:4 (25%) | 1/22 | 1/4 | 4 | |
| 3 | 1:8 (12.5%) | 1/23 | 1/8 | 8 | |
| 4 | 1:16 (0.063%) | 1/24 | 1/16 | 16 | |
| 5 | 1:32 (0.0313%) | 1/25 | 1/32 | 32 | |
| 6 | 1:64 (0.01563%) | 1/26 | 1/64 | 64 | |
| 7 | 1:128 (0.007813%) | 1/27 | 1/128 | 128 | |
| 8 | 1:256 (0.0039063%) | 1/28 | 1/256 | 256 | |
| 9 | 1:512 (0.00195313%) | 1/29 | 1/512 | 512 | |
| 10 | 1:1024 (0.0009766%) | 1/210 | 1/1024 | 1024 | |
| 11 | 1:2048 (0.0004883%) | 1/211 | 1/2048 | 2048 | |
| 12 | 1:4096 (0.000244141%) | 1/212 | 1/4096 | 4096 | |
| S. No | Accession No. | HA | Protein Conc. mg/mL | S. No | Accession No. | HA | Protein Conc. mg/mL | S. No | Accession No. | HA | Protein Conc. mg/mL | S. No | Accession No. | HA | Protein Conc. mg/mL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7216 | + | 5.815 | 54 | 7599 | + | 5.38 | 107 | 7734 | +3 | 4.269 | 160 | 7822 | +3 | 6.425 |
| 2 | 7217 | + | 5.18 | 55 | 7600 | + | 8.03 | 108 | 7735 | +2 | 4.808 | 161 | 7824 | + | 4.608 |
| 3 | 7218 | + | 5.865 | 56 | 7601 | + | 6.33 | 109 | 7736 | + | 6.245 | 162 | 7825 | +2 | 6.545 |
| 4 | 7219 | + | 5.77 | 57 | 7603 | + | 5.67 | 110 | 7737 | +2 | 4.931 | 163 | 7826 | + | 6.785 |
| 5 | 7225 | - | 7.3 | 58 | 7604 | + | 4.994 | 111 | 7738 | + | 4.998 | 164 | 7827 | +2 | 4.956 |
| 6 | 7234 | + | 4.621 | 59 | 7605 | + | 6.65 | 112 | 7753 | + | 3.056 | 165 | 7828 | +2 | 4.897 |
| 7 | 7235 | +3 | 4.996 | 60 | 7606 | + | 5.965 | 113 | 7754 | + | 4.703 | 166 | 7829 | +2 | 4.672 |
| 8 | 7236 | + | 3.87 | 61 | 7607 | +2 | 4.878 | 114 | 7755 | +2 | 5.42 | 167 | 7830 | + | 5.125 |
| 9 | 7237 | +2 | 4.731 | 62 | 7608 | +2 | 5.2 | 115 | 7756 | +3 | 5.285 | 168 | 7831 | +3 | 5.585 |
| 10 | 7238 | +2 | 4.344 | 63 | 7609 | + | 6.25 | 116 | 7759 | +3 | 7.84 | 169 | 7832 | + | 4.28 |
| 11 | 7239 | - | 4.293 | 64 | 7610 | + | 6.94 | 117 | 7760 | +3 | 4.770 | 170 | 7833 | + | 6.335 |
| 12 | 7241 | + | 4.624 | 65 | 7611 | +3 | 6.115 | 118 | 7762 | +3 | 5.375 | 171 | 7836 | +3 | 5.685 |
| 13 | 7242 | +2 | 4.871 | 66 | 7612 | +3 | 5.965 | 119 | 7763 | +2 | 4.167 | 172 | 7839 | +2 | 4.354 |
| 14 | 7258 | +3 | 5.735 | 67 | 7613 | +2 | 6.575 | 120 | 7765 | + | 4.463 | 173 | 7847 | +2 | 4.912 |
| 15 | 7260 | +3 | 6.26 | 68 | 7614 | +3 | 6.995 | 121 | 7766 | + | 5.155 | 174 | 7850 | + | 4.504 |
| 16 | 7261 | +2 | 4.98 | 69 | 7615 | + | 6.665 | 122 | 7783 | + | 6.04 | 175 | 7859 | + | 5.79 |
| 17 | 7262 | +3 | 4.326 | 70 | 7616 | +3 | 6.82 | 123 | 7784 | + | 4.884 | 176 | 7863 | + | 5.045 |
| 18 | 7263 | + | 3.372 | 71 | 7617 | + | 5.16 | 124 | 7785 | +3 | 6.845 | 177 | 7864 | + | 4.197 |
| 19 | 7268 | +3 | 4.365 | 72 | 7618 | +3 | 6.235 | 125 | 7786 | +3 | 6.675 | 178 | 7865 | + | 4.398 |
| 20 | 7269 | +2 | 4.706 | 73 | 7619 | +2 | 6.465 | 126 | 7787 | + | 6.825 | 179 | 7869 | + | 4.515 |
| 21 | 7271 | +3 | 4.326 | 74 | 7620 | +3 | 5.3 | 127 | 7788 | +2 | 7.13 | 180 | 7883 | + | 4.327 |
| 22 | 7280 | +3 | 5.105 | 75 | 7621 | +3 | 5.815 | 128 | 7789 | +2 | 6.14 | 181 | 7886 | +3 | 5.665 |
| 23 | 7281 | + | 5 | 76 | 7622 | +3 | 6.305 | 129 | 7790 | +3 | 6.815 | 182 | 7937 | + | 5.94 |
| 24 | 7284 | + | 6.165 | 77 | 7623 | + | 4.915 | 130 | 7791 | +3 | 7.585 | 183 | 8083 | +3 | 6.215 |
| 25 | 7285 | +3 | 5.445 | 78 | 7624 | +3 | 4.421 | 131 | 7792 | +2 | 6.62 | 184 | 8085 | +3 | 6.78 |
| 26 | 7286 | +3 | 6.95 | 79 | 7625 | +2 | 5.755 | 132 | 7793 | +2 | 6.8 | 185 | 17940 | +2 | 6.765 |
| 27 | 7287 | +3 | 6.565 | 80 | 7626 | + | 6.785 | 133 | 7794 | +2 | 5.76 | 186 | 17941 | + | 5.51 |
| 28 | 7288 | +3 | 6.13 | 81 | 7627 | + | 6.98 | 134 | 7795 | +3 | 6.185 | 187 | 17942 | + | 5.54 |
| 29 | 7289 | +3 | 6.995 | 82 | 7628 | +3 | 5.375 | 135 | 7796 | + | 7.28 | 188 | 17943 | + | 6.055 |
| 30 | 7290 | + | 5.07 | 83 | 7629 | + | 5.6 | 136 | 7797 | +2 | 7.56 | 189 | 17944 | + | 4.716 |
| 31 | 7291 | + | 4.418 | 84 | 7630 | + | 6.855 | 137 | 7798 | +3 | 7.525 | 190 | 17945 | - | 5.395 |
| 32 | 7292 | +2 | 4.037 | 85 | 7632 | + | 6.43 | 138 | 7799 | +3 | 7.51 | 191 | 17946 | +2 | 6.915 |
| 33 | 7293 | +2 | 3.761 | 86 | 7633 | + | 5.1 | 139 | 7800 | +3 | 5.765 | 192 | 17947 | + | 5.375 |
| 34 | 7294 | + | 3.929 | 87 | 7634 | +2 | 5.315 | 140 | 7801 | + | 3.545 | 193 | 17948 | - | 5.795 |
| 35 | 7295 | + | 4.376 | 88 | 7635 | + | 5.25 | 141 | 7802 | +3 | 3.352 | 194 | 17949 | + | 5.515 |
| 36 | 7296 | + | 7.245 | 89 | 7636 | +3 | 5.22 | 142 | 7803 | + | 4.465 | 195 | 17951 | + | 5.27 |
| 37 | 7297 | + | 4.712 | 90 | 7637 | +3 | 4.695 | 143 | 7804 | +3 | 7.29 | 196 | 17952 | + | 4.99 |
| 38 | 7298 | +2 | 4.560 | 91 | 7638 | +2 | 4.382 | 144 | 7805 | +3 | 5.31 | 197 | 17953 | + | 5.755 |
| 39 | 7299 | + | 3.718 | 92 | 7639 | + | 7.16 | 145 | 7806 | +3 | 5.505 | 198 | 17954 | + | 4.9185 |
| 40 | 7396 | +3 | 7.35 | 93 | 7640 | + | 6.81 | 146 | 7807 | +3 | 3.845 | 199 | 17955 | + | 6.26 |
| 41 | 7397 | + | 5.265 | 94 | 7641 | +2 | 5.11 | 147 | 7808 | +2 | 4.815 | 200 | 17956 | + | 5.875 |
| 42 | 7411 | +3 | 6.26 | 95 | 7648 | + | 4.913 | 148 | 7809 | +3 | 6.081 | 201 | 17957 | + | 5.38 |
| 43 | 7413 | +3 | 5.1 | 96 | 7649 | + | 5.175 | 149 | 7811 | +3 | 4.56 | 202 | 17958 | - | 5.59 |
| 44 | 7414 | +3 | 5.65 | 97 | 7650 | + | 4.617 | 150 | 7812 | +3 | 5.67 | 203 | 18231 | + | 4.0485 |
| 45 | 7416 | +3 | 4.985 | 98 | 7651 | +2 | 4.105 | 151 | 7813 | +3 | 6.12 | 204 | 23718 | + | 5.68 |
| 46 | 7418 | +3 | 5.4 | 99 | 7652 | +2 | 4.565 | 152 | 7814 | +3 | 4.84 | 205 | 23727 | + | 5.575 |
| 47 | 7419 | +3 | 6 | 100 | 7653 | +2 | 5.78 | 153 | 7815 | +3 | 4.495 | 206 | 23728 | +2 | 5.66 |
| 48 | 7420 | +3 | 5.63 | 101 | 7654 | + | 6.785 | 154 | 7816 | +2 | 4.315 | 207 | 23731 | +2 | 3.577 |
| 49 | 7593 | + | 4.168 | 102 | 7655 | + | 5.63 | 155 | 7817 | +3 | 6.31 | 208 | 23732 | +3 | 3.8585 |
| 50 | 7594 | + | 7.5 | 103 | 7656 | + | 7.335 | 156 | 7818 | + | 4.39 | 209 | 23736 | +3 | 6.22 |
| 51 | 7595 | + | 4.522 | 104 | 7657 | + | 6.23 | 157 | 7819 | +3 | 7.07 | 210 | 23744 | + | 3.6395 |
| 52 | 7597 | + | 4.579 | 105 | 7672 | +2 | 4.114 | 158 | 7820 | +3 | 5.64 | 211 | Boti | +2 | …… |
| 53 | 7598 | + | 5.29 | 106 | 7673 | + | 4.958 | 159 | 7821 | +2 | 6.56 | 212 | F.M | +2 | 2.79 |
| 213–217 = Five transgenic seeds of rice, where only two showed visible HA (+2) with 3.54 and 3.79 protein content. | |||||||||||||||
| S. No | Accession No. | HA | Protein Conc. mg/mL | Titre Value | Specific activity HU/mg | S. No | Accession No. | HA | Protein Conc. mg/mL | Titer Value | Specific activity HU/mg | S. No | Accession No. | HA | Protein Conc. mg/mL | Titre Value | Specific activity HU/mg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7235 | +3 | 4.996 | 27 = 128 | 25.621 | 37 | 7619 | +2 | 6.465 | 23 = 8 | 1.237 | 73 | 7800 | +3 | 5.765 | 22 = 4 | 0.693 |
| 2 | 7236 | +2 | 3.87 | 25 = 32 | 8.268 | 38 | 7620 | +3 | 5.3 | 25 = 32 | 6.037 | 74 | 7802 | +3 | 3.352 | 23 = 8 | 2.386 |
| 3 | 7237 | +2 | 4.731 | 27 = 128 | 27.055 | 39 | 7622 | +3 | 6.305 | 23 = 8 | 1.268 | 75 | 7804 | +3 | 7.29 | 23 = 8 | 1.097 |
| 4 | 7238 | +2 | 4.34 | 27 = 128 | 29.49 | 40 | 7624 | +3 | 4.4205 | 24 = 16 | 3.6195 | 76 | 7805 | +3 | 5.31 | 22 = 4 | 0.753 |
| 5 | 7242 | +2 | 4.871 | 27 = 128 | 26.28 | 41 | 7625 | +2 | 5.755 | 22 = 4 | 0.695 | 77 | 7806 | +3 | 5.505 | 23 = 8 | 1.453 |
| 6 | 7258 | +3 | 5.735 | 28 = 256 | 44.63 | 42 | 7627 | + | 6.98 | 21 = 2 | 0.286 | 78 | 7807 | +3 | 3.845 | 23 = 8 | 2.081 |
| 7 | 7260 | +3 | 6.26 | 22 = 4 | 0.638 | 43 | 7628 | +3 | 5.375 | 29 = 512 | 95.25 | 79 | 7808 | +2 | 4.815 | 22 = 4 | 0.8307 |
| 8 | 7261 | +2 | 4.98 | 22 = 4 | 0.803 | 44 | 7634 | +2 | 5.315 | 22 = 4 | 0.752 | 80 | 7809 | +3 | 6.08 | 21 = 2 | 0.3289 |
| 9 | 7262 | +3 | 2.163 | 22 = 4 | 1.849 | 45 | 7636 | +3 | 5.22 | 24=16 | 3.065 | 81 | 7811 | +3 | 4.56 | 23 = 8 | 1.754 |
| 10 | 7268 | +3 | 4.365 | 23 =8 | 1.8328 | 46 | 7637 | +3 | 4.695 | 24 = 16 | 3.407 | 82 | 7812 | +3 | 5.67 | 23 = 8 | 1.411 |
| 11 | 7269 | +2 | 4.706 | 22 = 4 | 0.8499 | 47 | 7641 | +2 | 5.11 | 24 = 16 | 3.131 | 83 | 7813 | +3 | 6.12 | 22 = 4 | 0.654 |
| 12 | 7271 | +3 | 4.326 | 210 =1024 | 236.70 | 48 | 7651 | +2 | 4.105 | 22 = 4 | 0.974 | 84 | 7814 | +3 | 4.84 | 23 = 8 | 1.653 |
| 13 | 7280 | +3 | 5.105 | 23 = 8 | 1.5670 | 49 | 7652 | +2 | 4.565 | 22 = 4 | 0.876 | 85 | 7815 | +3 | 4.495 | 25 = 32 | 7.119 |
| 14 | 7285 | +3 | 5.445 | 23 = 8 | 1.469 | 50 | 7655 | + | 5.63 | 28 = 256 | 45.47 | 86 | 7816 | +2 | 4.315 | 20 = 1 | 0.231 |
| 15 | 7286 | +3 | 6.95 | 23 = 8 | 1.151 | 51 | 7672 | +2 | 4.114 | 22 = 4 | 0.972 | 87 | 7819 | +3 | 7.07 | 23 = 8 | 1.1315 |
| 16 | 7287 | +3 | 6.565 | 23 = 8 | 1.2185 | 52 | 7735 | +2 | 4.8085 | 26 = 64 | 13.309 | 88 | 7820 | +3 | 5.64 | 24 = 16 | 2.8368 |
| 17 | 7288 | +3 | 6.13 | 23 = 8 | 1.3050 | 53 | 7737 | +2 | 4.9315 | 27 = 128 | 25.955 | 89 | 7821 | +2 | 6.56 | 22 = 4 | 0.609 |
| 18 | 7289 | +3 | 6.995 | 23 = 8 | 1.1436 | 54 | 7755 | +2 | 5.42 | 25 = 32 | 5.9040 | 90 | 7822 | +3 | 6.425 | 23 = 8 | 1.245 |
| 19 | 7292 | +2 | 4.0375 | 27 = 128 | 31.702 | 55 | 7756 | +3 | 5.285 | 22 = 4 | 0.7568 | 91 | 7825 | +2 | 6.545 | 24 = 16 | 2.444 |
| 20 | 7293 | +2 | 3.761 | 22 = 4 | 1.0635 | 56 | 7759 | +3 | 5.2266 | 23 = 8 | 1.2374 | 92 | 7827 | +2 | 4.956 | 25 = 32 | 6.456 |
| 21 | 7298 | +2 | 4.5605 | 27 = 128 | 28.067 | 57 | 7760 | +3 | 3.1803 | 25 = 32 | 10.07 | 93 | 7828 | +2 | 4.897 | 25 = 32 | 6.5346 |
| 22 | 7396 | +3 | 7.35 | 22 = 4 | 0.5442 | 58 | 7762 | +3 | 3.5833 | 21 = 2 | 0.558 | 94 | 7829 | +2 | 4.672 | 26 = 64 | 13.698 |
| 23 | 7411 | +3 | 6.26 | 210 = 1024 | 163.57 | 59 | 7763 | +3 | 2.7783 | 22 = 4 | 1.4397 | 95 | 7831 | +3 | 5.585 | 210 = 1024 | 183.34 |
| 24 | 7413 | +3 | 5.1 | 23 = 8 | 1.5686 | 60 | 7785 | +3 | 4.5633 | 24 = 16 | 3.506 | 96 | 7836 | +3 | 5.685 | 22 = 4 | 0.7036 |
| 25 | 7414 | +3 | 5.65 | 23 = 8 | 1.4159 | 61 | 7786 | +3 | 4.45 | 23 = 8 | 1.797 | 97 | 7839 | +2 | 4.354 | 22 = 4 | 0.9188 |
| 26 | 7416 | +3 | 4.985 | 23 = 8 | 1.6048 | 62 | 7788 | +2 | 4.7533 | 28 = 256 | 53.857 | 98 | 7886 | +3 | 5.665 | 23 = 8 | 1.412 |
| 27 | 7418 | +3 | 5.4 | 24 = 16 | 2.9629 | 63 | 7789 | +2 | 4.0933 | 25 = 32 | 7.8176 | 99 | 8083 | +3 | 6.215 | 23 = 8 | 1.287 |
| 28 | 7419 | +3 | 6 | 210 = 1024 | 170.66 | 64 | 7790 | +3 | 4.5433 | 26 = 64 | 14.086 | 100 | 8085 | +3 | 6.78 | 29 = 512 | 75.516 |
| 29 | 7607 | +2 | 4.8785 | 22 = 4 | 0.8199 | 65 | 7791 | +3 | 5.0566 | 23 = 8 | 1.582 | 101 | 17940 | +2 | 6.765 | 22 = 4 | 0.591 |
| 30 | 7608 | +2 | 5.2 | 22 = 4 | 0.7692 | 66 | 7792 | +2 | 4.4133 | 22 = 4 | 0.906 | 102 | 17946 | +2 | 6.915 | 21 = 2 | 0.289 |
| 31 | 7611 | +3 | 6.115 | 23 = 8 | 1.3082 | 67 | 7793 | +2 | 4.5333 | 22 = 4 | 0.882 | 103 | 23728 | +2 | 5.66 | 29 = 512 | 90.45 |
| 32 | 7612 | +3 | 5.965 | 23 = 8 | 1.3411 | 68 | 7794 | +2 | 3.84 | 22 = 4 | 1.0416 | 104 | 23731 | +2 | 3.577 | 26 = 64 | 17.89 |
| 33 | 7613 | +2 | 6.575 | 23 = 8 | 1.2167 | 69 | 7795 | +3 | 4.1233 | 22 = 4 | 0.9700 | 105 | 23732 | +3 | 3.859 | 26 = 64 | 16.586 |
| 34 | 7614 | +3 | 6.9995 | 24 = 16 | 2.2858 | 70 | 7797 | +2 | 5.04 | 20 = 1 | 0.198 | 106 | 23736 | +3 | 6.22 | 23 = 8 | 1.286 |
| 35 | 7616 | +3 | 6.82 | 23 = 8 | 1.1730 | 71 | 7798 | +3 | 5.0166 | 23 = 8 | 1.594 | 107 | F.M | +2 | 2.79 | 21 = 2 | 0.71 |
| 36 | 7618 | +3 | 6.235 | 23 = 8 | 1.2830 | 72 | 7799 | +3 | 5.0066 | 23 = 8 | 1.597 | 108 | 109T.S | +2 | 3.54,3.79 | 21 = 2 | 0.28, 0.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gulzar, H.; Nawaz, M.A.; Jan, A.; Khan, F.A.; Naz, S.; Zahoor, M.; Naz, D.; Ullah, R.; Ali, E.A.; Hussain, H. Semi-Quantification of Lectins in Rice (Oryza sativa L.) Genotypes via Hemagglutination. Agronomy 2021, 11, 1899. https://doi.org/10.3390/agronomy11101899
Gulzar H, Nawaz MA, Jan A, Khan FA, Naz S, Zahoor M, Naz D, Ullah R, Ali EA, Hussain H. Semi-Quantification of Lectins in Rice (Oryza sativa L.) Genotypes via Hemagglutination. Agronomy. 2021; 11(10):1899. https://doi.org/10.3390/agronomy11101899
Chicago/Turabian StyleGulzar, Haseena, Muhammad Asif Nawaz, Asad Jan, Farhat Ali Khan, Sumaira Naz, Muhammad Zahoor, Dil Naz, Riaz Ullah, Essam A. Ali, and Hidayat Hussain. 2021. "Semi-Quantification of Lectins in Rice (Oryza sativa L.) Genotypes via Hemagglutination" Agronomy 11, no. 10: 1899. https://doi.org/10.3390/agronomy11101899
APA StyleGulzar, H., Nawaz, M. A., Jan, A., Khan, F. A., Naz, S., Zahoor, M., Naz, D., Ullah, R., Ali, E. A., & Hussain, H. (2021). Semi-Quantification of Lectins in Rice (Oryza sativa L.) Genotypes via Hemagglutination. Agronomy, 11(10), 1899. https://doi.org/10.3390/agronomy11101899








