Trait-Based Root Phenotyping as a Necessary Tool for Crop Selection and Improvement
Abstract
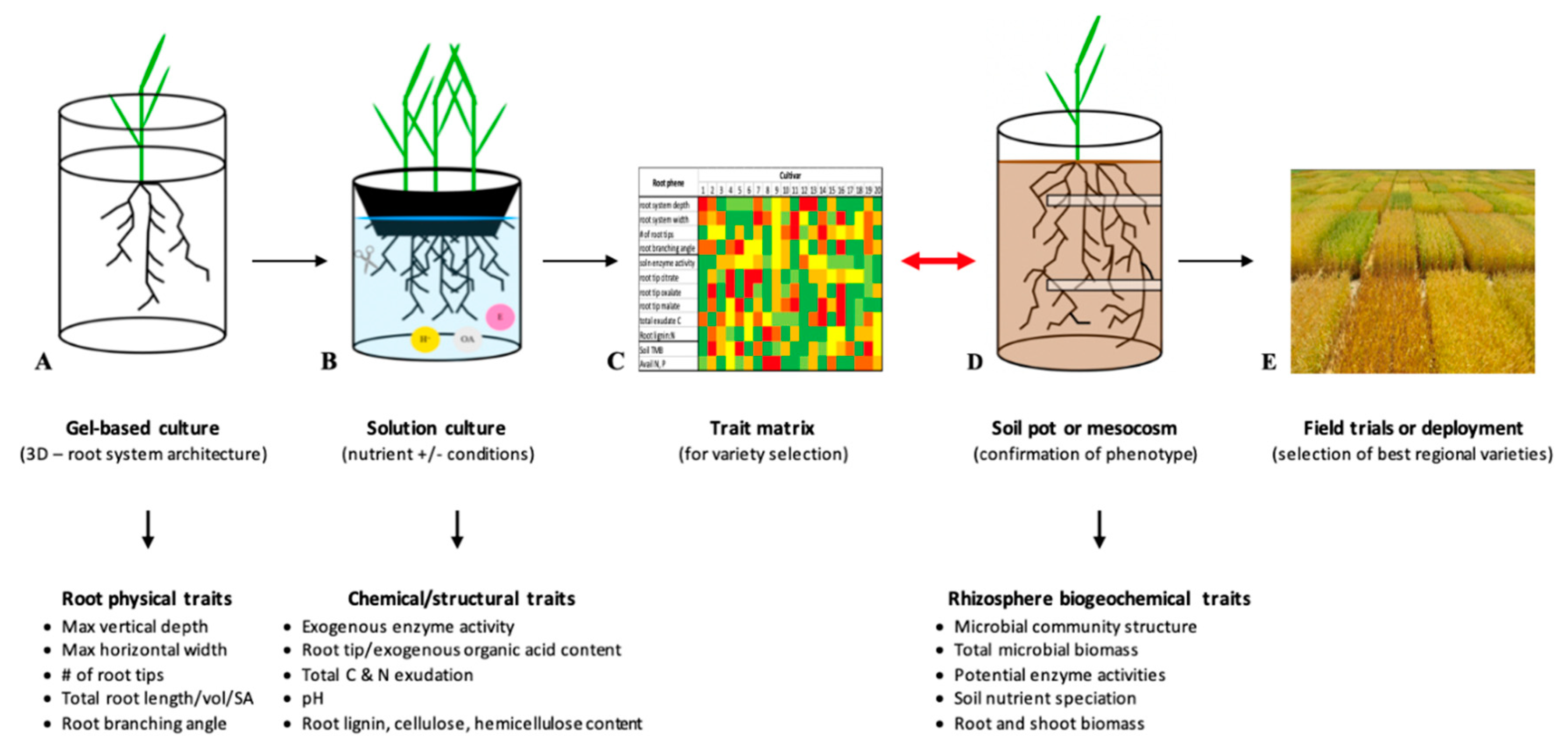
:1. The Need for Root Breeding
2. Current Efforts for Root Breeding
2.1. Field-Based Phenotyping for Root System Architecture
2.2. Lab and Greenhouse-Based Phenotyping for Root System Architecture
2.3. Marker-Based Profiling for Root System Architectural Traits
3. Importance of Looking Beyond Root System Architecture
3.1. Structural Profiling
3.2. Chemical Profiling
4. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects: The 2017 Revision, Key Findings and Advance Tables; Working Paper No. ESA/P/WP/248; United Nations, Department of Economic and Social Affairs, Population Division: New York, NY, USA, 2017. [Google Scholar]
- Lynch, J.P. Roots of the Second Green Revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Pingali, P.L. Green Revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302. [Google Scholar] [CrossRef] [Green Version]
- Isherwood, K.F. Mineral Fertilizer Use and the Environment; International Fertilizer Industry Association/United Nations Environment Programme: Paris, France, 2000. [Google Scholar]
- Baligar, V.C.; Bennett, O.L. Outlook on fertilizer use efficienty in the tropics. Fertil. Res. 1986, 10, 83–96. [Google Scholar] [CrossRef]
- Topp, C.N.; Bray, A.L.; Ellis, N.A.; Liu, Z. How can we harness quantitative genetic variation in crop root systems for agricultural improvement? J. Integr. Plant Biol. 2016, 58, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Baishya, L.K. Nutrient Use Efficiency. In Essential Plant Nutrients: Uptake, Use Efficiency, and Management; Naeem, M., Ansari, A.A., Gill, S.S., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 119–146. [Google Scholar] [CrossRef]
- Ryser, P.; Eek, L. Consequences of phenotypic plasticity vs. interspecific differences in leaf and root traits for acquisition of aboveground and belowground resources. Am. J. Bot. 2000, 87, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Bardgett, R.D. Plant trait-based approaches for interrogating belowground function. Biol. Environ. Proc. R. Ir. Acad. 2017, 117B, 1–13. [Google Scholar] [CrossRef]
- Craine, J.M.; Lee, W.G. Covariation in leaf and root traits for native and non-native grasses along an altitudinal gradient in New Zealand. Oecologia 2003, 134, 471–478. [Google Scholar] [CrossRef]
- Craine, J.M.; Lee, W.G.; Bond, W.J.; Williams, R.J.; Johnson, L.C. Environmental constraints on a global relationship among leaf and root traits of grasses. Ecology 2005, 86, 12–19. [Google Scholar] [CrossRef]
- Kerkhoff, A.J.; Fagan, W.F.; Elser, J.J.; Enquist, B.J. Phylogenetic and Growth Form Variation in the Scaling of Nitrogen and Phosphorus in the Seed Plants. Am. Nat. 2006, 168, E103–E122. [Google Scholar] [CrossRef] [Green Version]
- Ryser, P. Intra- and interspecific variation in root length, root turnover and the underlying parameters. In Variation in Plant Growth; Lambers, H., Poorter, H., Van Vuuren, M.M.I., Eds.; Backhys Publishers: Leiden, The Netherlands, 1998; pp. 441–465. [Google Scholar]
- Ryser, P.; Lambers, H. Root and leaf attributes accounting for the performance of fast- and slow-growing grasses at different nutrient supply. Plant Soil 1995, 170, 251–265. [Google Scholar] [CrossRef]
- Lupton, F.G.H.; Oliver, R.H.; Ellis, F.B.; Barnes, B.T.; Welbank, P.J.; Taylor, P.J. Root and shoot growth of semi-dwarf and taller winter wheats. Ann. Appl. Biol. 1974, 77, 129–144. [Google Scholar] [CrossRef]
- Holbrook, F.S. Rooting Depths of Selected Wheat Cultivars; Colorado State University: Fort Collins, CO, USA, 1973. [Google Scholar]
- Cholick, F.A.; Welsh, J.R.; Cole, V.C. Rooting Patterns of Semi-dwarf and Tall Winter Wheat Cultivars under Dryland Field Conditions. Crop Sci. 1977, 17, 637. [Google Scholar] [CrossRef]
- Wojciechowski, T.; Gooding, M.J.; Ramsay, L.; Gregory, P.J. The effects of dwarfing genes on seedling root growth of wheat. J. Exp. Bot. 2009, 60, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, B.V.; Katyal, J.C.; Narasimham, R.L.; Dakshinamurti, C. Preliminary investigations on root distribution of high yielding wheat varieties. Int. J. Appl. Radiat. Isot. 1968, 19, 385–390. [Google Scholar] [CrossRef]
- Qi, Y.; Wei, W.; Chen, C.; Chen, L. Plant root-shoot biomass allocation over diverse biomes: A global synthesis. Glob. Ecol. Conserv. 2019, 18, e00606. [Google Scholar] [CrossRef]
- Ubeda-Tomás, S.; Swarup, R.; Coates, J.; Swarup, K.; Laplaze, L.; Beemster, G.T.S.; Hedden, P.; Bhalerao, R.; Bennett, M.J. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 2008, 10, 625. [Google Scholar] [CrossRef]
- Richard, C.; Christopher, J.; Chenu, K.; Borrell, A.; Christopher, M.; Hickey, L. Selection in early generations to shift allele frequency for seminal root angle in wheat. Plant Genome 2018. [Google Scholar] [CrossRef] [Green Version]
- Sanguineti, M.C.; Li, S.; Maccaferri, M.; Corneti, S.; Rotondo, F.; Chiari, T.; Tuberosa, R. Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann. Appl. Biol. 2007, 151, 291–305. [Google Scholar] [CrossRef]
- Xie, Q.; Fernando, K.M.C.; Mayes, S.; Sparkes, D.L. Identifying seedling root architectural traits associated with yield and yield components in wheat. Ann. Bot. 2017, 119, 1115–1129. [Google Scholar] [CrossRef]
- Mac Key, J.; Jordan, W.R.; Zobel, R.W. Lecture 3: Shoot:Root interactions in cereals and grasses. In Plant Roots: A compilation of Ten Seminars Given at Iowa State University in Feburary and March, 1980; Iowa State University, Agronomy Department: Ames, Iowa, 1980. [Google Scholar]
- Bektas, H.; Hohn, C.E.; Waines, J.G. Root and shoot traits of bread wheat (Triticum aestivum L.) landraces and cultivars. Euphytica 2016, 212, 297–311. [Google Scholar] [CrossRef]
- Siddique, K.H.M.; Belford, R.K.; Tennant, D. Root:Shoot ratios of old and modern, tall and semi-dwarf wheats in a mediterranean environment. Plant Soil 1990, 121, 89–98. [Google Scholar] [CrossRef]
- Waines, J.G.; Ehdaie, B. Domestication and Crop Physiology: Roots of Green-Revolution Wheat. Ann. Bot. 2007, 100, 991–998. [Google Scholar] [CrossRef] [Green Version]
- Roucou, A.; Violle, C.; Fort, F.; Roumet, P.; Ecarnot, M.; Vile, D. Shifts in plant functional strategies over the course of wheat domestication. J. Appl. Ecol. 2018, 55, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, P.; Robertson, M.; Cooper, M.; Hammer, G. The role of physiological understanding in plant breeding; from a breeding perspective. Field Crop. Res. 1996, 49, 11–37. [Google Scholar] [CrossRef]
- Richards, R.A. Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 2000, 51, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Oosterom, E.J.; Jordan, D.R.; Hunt, C.H.; Hammer, G.L. Genetic variability and control of nodal root angle in sorghum. Crop Sci. 2011, 51, 2011–2020. [Google Scholar] [CrossRef]
- Uga, Y.; Okuno, K.; Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 2011, 62, 2485–2494. [Google Scholar] [CrossRef] [Green Version]
- Kumar, B.; Abdel-Ghani, A.H.; Reyes-Matamoros, J.; Hochholdinger, F.; Lübberstedt, T. Genotypic variation for root architecture traits in seedlings of maize (Zea mays L.) inbred lines. Plant Breed. 2012, 131, 465–478. [Google Scholar] [CrossRef]
- Mace, E.; Singh, V.; Oosterom, E.; Hammer, G.; Hunt, C.; Jordan, D. QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theor. Appl. Genet. 2012, 124, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Christopher, J.; Christopher, M.; Jennings, R.; Jones, S.; Fletcher, S.; Borrell, A.; Manschadi, A.; Jordan, D.; Mace, E.; Hammer, G. QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theor. Appl. Genet. 2013, 126, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Nyquist, W.E.; Baker, R.J. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar] [CrossRef]
- Omori, F.; Mano, Y. QTL Mapping of root angle in F2 populations from maize ‘B73’ × teosinte ‘Zea luxurians’. Plant Root 2007, 1, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Richard, C.A.; Hickey, L.T.; Fletcher, S.; Jennings, R.; Chenu, K.; Christopher, J.T. High-throughput phenotyping of seminal root traits in wheat. Plant Methods 2015, 11, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, I.; Shimelis, H.; Mwadzingeni, L.; Zengeni, R.; Mutema, M.; Chaplot, V. Variance components and heritability of traits related to root: Shoot biomass allocation and drought tolerance in wheat. Euphytica 2018, 214, 225. [Google Scholar] [CrossRef]
- Ekanayake, I.J.; O’Toole, J.C.; Garrity, D.P.; Masajo, T.M. Inheritance of root characters and their relations to drought resistance in rice. Crop Sci. 1985, 25, 927–933. [Google Scholar] [CrossRef]
- Singh Gahoonia, T.; Nielsen, N.E. Root traits as tools for creating phosphorus efficient crop varieties. Plant Soil 2004, 260, 47–57. [Google Scholar] [CrossRef]
- Fakrudin, B.; Kavil, S.P.; Girma, Y.; Arun, S.S.; Dadakhalandar, D.; Gurusiddesh, B.H.; Patil, A.M.; Thudi, M.; Bhairappanavar, S.B.; Narayana, Y.D.; et al. Molecular mapping of genomic regions harbouring QTLs for root and yield traits in sorghum (Sorghum bicolor L. Moench). Physiol. Mol. Biol. Plants 2013, 19, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Shi, T.; Broadley, M.R.; White, P.J.; Long, Y.; Meng, J.; Xu, F.; Hammond, J.P. High-throughput root phenotyping screens identify genetic loci associated with root architectural traits in Brassica napus under contrasting phosphate availabilities. Ann. Bot. 2013, 112, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Pandey, S.; Pandey, A. Plant roots and carbon sequestration. Curr. Sci. 2006, 91, 885–890. [Google Scholar]
- Slota, M.; Maluszynski, M.; Szarejko, I. An automated, cost-effective and scalable, flood-and-drain based root phenotyping system for cereals. Plant Methods 2016, 12, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, J.E. The Ecological Relations of Roots; Carnegie Institution of Washington: Washington, DC, USA, 1919; p. 172. [Google Scholar]
- Trachsel, S.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 2011, 341, 75–87. [Google Scholar] [CrossRef]
- Das, A.; Schneider, H.; Burridge, J.; Ascanio, A.K.M.; Wojciechowski, T.; Topp, C.N.; Lynch, J.P.; Weitz, J.S.; Bucksch, A. Digital imaging of root traits (DIRT): A high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 2015, 11, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, R.T.; MacCurdy, R.B.; Jung, J.K.; Shaff, J.E.; McCouch, S.R.; Aneshansley, D.J.; Kochian, L.V. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 2011, 156, 455–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauhus, J.; Messier, C. Evaluation of fine root length and diameter measurements obtained using RHIZO image analysis. Agron. J. 1999, 91, 142–147. [Google Scholar] [CrossRef] [Green Version]
- Galkovsky, T.; Mileyko, Y.; Bucksch, A.; Moore, B.; Symonova, O.; Prince, C.A.; Topp, C.N.; Iyer-Pascuzzi, A.S.; Zurek, P.R.; Fang, S.; et al. GiA Roots: Software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 2012, 12, 1–12. [Google Scholar] [CrossRef] [Green Version]
- York, L.M.; Slack, S.; Bennett, M.J.; Foulkes, M.J. Wheat shovelomics I: A field phenotyping approach for characterising the structure and function of root systems in tillering species. bioRxiv 2018, 280875. [Google Scholar] [CrossRef]
- Burridge, J.; Jochua, C.N.; Bucksch, A.; Lynch, J.P. Legume shovelomics: High—throughput phenotyping of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata subsp, unguiculata) root architecture in the field. Field Crop. Res. 2016, 192, 21–32. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Picon-Cochard, C.; Pilon, R.; Tarroux, E.; Pagès, L.; Robertson, J.; Dawson, L. Effect of species, root branching order and season on the root traits of 13 perennial grass species. Plant Soil 2012, 353, 47–57. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Q.; Blaszkiewicz, Z.; Sun, J.; Sun, Q.; He, R.; Li, Y. Phenotyping for the dynamics of field wheat root system architecture. Sci. Rep. 2017, 7, 37649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.G.; Tingey, D.T.; Phillips, D.L.; Storm, M.J. Advancing fine root research with minirhizotrons. Environ. Exp. Bot. 2001, 45, 263–289. [Google Scholar] [CrossRef]
- Bragg, P.L.; Govi, G.; Cannell, R.Q. A comparison of methods, including angled and vertical minirhizotrons, for studying root growth and distribution in a spring oat crop. Plant Soil 1983, 73, 435–440. [Google Scholar] [CrossRef]
- Aulen, M.; Shipley, B. Non-destructive estimation of root mass using electrical capacitance on ten herbaceous species. Plant Soil 2012, 355, 41–49. [Google Scholar] [CrossRef]
- Ellis, T.W.; Williams, C.; Murray, W.; Paul, K.; Kavalieris, L.; Brophy, J.; Maass, M. Electrical capacitance as a rapid and non-invasive indicator of root length. Tree Physiol. 2012, 33, 3–17. [Google Scholar] [CrossRef] [Green Version]
- Amato, M.; Basso, B.; Celano, G.; Bitella, G.; Morelli, G.; Rossi, R. In situ detection of tree root distribution and biomass by multi-electrode resistivity imaging. Tree Physiol. 2008, 28, 1441–1448. [Google Scholar] [CrossRef]
- Srayeddin, I.; Doussan, C. Estimation of the spatial variability of root water uptake of maize and sorghum at the field scale by electrical resistivity tomography. Plant Soil 2009, 319, 185–207. [Google Scholar] [CrossRef]
- Van Dusschoten, D.; Metzner, R.; Kochs, J.; Postma, J.A.; Pflugfelder, D.; Bühler, J.; Schurr, U.; Jahnke, S. Quantitative 3D analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol. 2016, 170, 1176–1188. [Google Scholar] [CrossRef] [Green Version]
- Pflugfelder, D.; Metzner, R.; van Dusschoten, D.; Reichel, R.; Jahnke, S.; Koller, R. Non-invasive imaging of plant roots in different soils using magnetic resonance imaging (MRI). Plant Methods 2017, 13, 102. [Google Scholar] [CrossRef]
- Jahnke, S.; Menzel, M.I.; Van Dusschoten, D.; Roeb, G.W.; Bühler, J.; Minwuyelet, S.; Blümler, P.; Temperton, V.M.; Hombach, T.; Streun, M.; et al. Combined MRI–PET dissects dynamic changes in plant structures and functions. Plant J. 2009, 59, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Garbout, A.; Munkholm, L.J.; Hansen, S.B.; Petersen, B.M.; Munk, O.L.; Pajor, R. The use of PET/CT scanning technique for 3D visualization and quantification of real-time soil/plant interactions. Plant Soil 2012, 352, 113–127. [Google Scholar] [CrossRef]
- Mooney, S.J.; Pridmore, T.P.; Helliwell, J.; Bennett, M.J. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 2012, 352, 1–22. [Google Scholar] [CrossRef]
- Paya, A.M.; Silverberg, J.L.; Padgett, J.; Bauerle, T.L. X-ray computed tomography uncovers root–root interactions: Quantifying spatial relationships between interacting root systems in three dimensions. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Dalton, F.N. In-situ root extent measurements by electrical capacitance methods. Plant Soil 1995, 173, 157–165. [Google Scholar] [CrossRef]
- Atkinson, J.A.; Pound, M.P.; Bennett, M.J.; Wells, D.M. Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotechnol. 2019, 55, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, P.A.T.; Loh, W.W.; Dickin, F.J. Three-dimensional reconstruction algorithm for electrical resistance tomography. IEE Proc.-Sci. Meas. Technol. 1998, 145, 85–93. [Google Scholar] [CrossRef]
- Rossi, R.; Amato, M.; Bitella, G.; Bochicchio, R.; Ferreira Gomes, J.; Lovelli, S.; Martorella, E.; Favale, P. Electrical resistivity tomography as a nonadestructive method for mapping root biomass in an orchard. Eur. J. Soil Sci. 2011, 62, 206–215. [Google Scholar] [CrossRef]
- Paglis, C.M. Application of electrical eesistivity tomography for detecting root biomass in coffee trees. Int. J. Geophys. 2013, 2013. [Google Scholar] [CrossRef]
- Metzner, R.; Eggert, A.; van Dusschoten, D.; Pflugfelder, D.; Gerth, S.; Schurr, U.; Uhlmann, N.; Jahnke, S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: Potential and challenges for root trait quantification. Plant Methods 2015, 11, 17. [Google Scholar] [CrossRef]
- Zappala, S.; Mairhofer, S.; Tracy, S.; Sturrock, C.J.; Bennett, M.; Pridmore, T.; Mooney, S.J. Quantifying the effect of soil moisture content on segmenting root system architecture in X-ray computed tomography images. Plant Soil 2013, 370, 35–45. [Google Scholar] [CrossRef]
- Metzner, R.; van Dusschoten, D.; Bühler, J.; Schurr, U.; Jahnke, S. Belowground plant development measured with magnetic resonance imaging (MRI): Exploiting the potential for non-invasive trait quantification using sugar beet as a proxy. Front. Plant Sci. 2014, 5, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hainsworth, J.; Aylmore, L. The use of computer assisted tomography to determine spatial distribution of soil water content. Soil Res. 1983, 21, 435–443. [Google Scholar] [CrossRef]
- Mairhofer, S.; Sturrock, C.J.; Bennett, M.J.; Mooney, S.J.; Pridmore, T.P. Extracting multiple interacting root systems using X-ray microcomputed tomography. Plant J. 2015, 84, 1034–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, D.M.; Atkinson, J.A.; Bennett, M.J.; Griffiths, M.; Pound, M.P.; Wingen, L.U.; Griffiths, S.; King, J.; Foulkes, M.J.; Gaju, O.; et al. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot. 2015, 66, 2283–2292. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Clark, R.T.; Zheng, Y.; Iyer-Pascuzzi, A.S.; Weitz, J.S.; Kochian, L.V.; Edelsbrunner, H.; Liao, H.; Benfey, P.N. Genotypic recognition and spatial responses by rice roots. Proc. Natl. Acad. Sci. USA 2013, 110, 2670–2675. [Google Scholar] [CrossRef] [Green Version]
- Iyer-Pascuzzi, A.S.; Zurek, P.R.; Benfey, P.N. High-throughput, noninvasive imaging of root systems. Methods Mol. Biol. 2013, 959, 177–187. [Google Scholar] [CrossRef]
- Yan, J.; Wang, B.; Zhou, Y. A root penetration model of Arabidopsis thaliana in phytagel medium with different strength. J. Plant Res. 2017, 130, 941–950. [Google Scholar] [CrossRef]
- Finch, J.A.; Guillaume, G.; French, S.A.; Colaço, R.D.D.R.; Davies, J.M.; Swarbreck, S.M. Wheat root length and not branching is altered in the presence of neighbours, including blackgrass. PLoS ONE 2017, 12, e0178176. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Pace, J.; Gardner, C.; Romay, C.; Ganapathysubramanian, B.; Lübberstedt, T. Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genom. 2015, 16, 47. [Google Scholar] [CrossRef] [Green Version]
- Bentsink, L.; Yuan, K.; Koornneef, M.; Vreugdenhil, D. The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor. Appl. Genet. 2003, 106, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Vreugdenhil, D.; Aarts, M.G.M.; Koornneef, M.; Nelissen, H.; Ernst, W.H.O. Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ. 2004, 27, 828–839. [Google Scholar] [CrossRef]
- Reymond, M.; Svistoonoff, S.; Loudet, O.; Nussaume, L.; Desnos, T. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 2006, 29, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Domanski, G. Carbon input by plants into the soil. Review. J. Plant Nutr. Soil Sci. 2000, 163, 421–431. [Google Scholar] [CrossRef]
- Redin, M.; Guénon, R.; Recous, S.; Schmatz, R.; de Freitas, L.L.; Aita, C.; Giacomini, S.J. Carbon mineralization in soil of roots from twenty crop species, as affected by their chemical composition and botanical family. Plant Soil 2014, 378, 205–214. [Google Scholar] [CrossRef]
- Lavallee, J.M.; Conant, R.T.; Paul, E.A.; Cotrufo, M.F. Incorporation of shoot versus root-derived 13C and 15N into mineral-associated organic matter fractions: Results of a soil slurry incubation with dual-labelled plant material. Biogeochemistry 2018, 137, 379–393. [Google Scholar] [CrossRef]
- Gan, Y.T.; Liang, B.C.; Liu, L.P.; Wang, X.Y.; McDonald, C.L. C:N ratios and carbon distribution profile across rooting zones in oilseed and pulse crops. Crop Pasture Sci. 2011, 62, 496–503. [Google Scholar] [CrossRef]
- Silver, W.L.; Miya, R.K. Global patterns in root decomposition: Comparisons of climate and litter quality effects. Oecologia 2001, 129, 407–419. [Google Scholar] [CrossRef]
- McLaughlin, S.B.; Wimmer, R. Tansley Review No. 104, Calcium Physiology and Terrestrial Ecosystem Processes. New Phytol. 1999, 142, 373–417. [Google Scholar] [CrossRef]
- Cromack, K.; Sollins, P.; Graustein, W.C.; Speidel, K.; Todd, A.W.; Spycher, G.; Li, C.Y.; Todd, R.L. Calcium oxalate accumulation and soil weathering in mats of the hypogeous fungus Hysterangium crassum. Soil Biol. Biochem. 1979, 11, 463–468. [Google Scholar] [CrossRef] [Green Version]
- Grabovich, M.Y.; Dubinina, G.A.; Churikova, V.V.; Churikov, S.N.; Korovina, T.I. Mechnisms of synthesis and utilization of oxalate inclusion in the colorless sulfur bacterium Macromonas bipunctata. Microbiology 1995, 64, 630–636. [Google Scholar]
- Jones, D.L. Organic acids in the rhizosphere–A critical review. Plant Soil 1998, 205, 25–44. [Google Scholar] [CrossRef]
- Frei, M. Lignin: Characterization of a multifaceted crop component. Sci. World J. 2013, 2013, 436517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.; Whipps, J.M. Substrate flow in the rhizosphere. Plant Soil 1990, 129, 1–10. [Google Scholar] [CrossRef]
- Whipps, J.M.; Lynch, J.M. Substrate flow and utilization in the rhizosphere of cereals. New Phytol. 1983, 95, 605–623. [Google Scholar] [CrossRef]
- Whipps, J.M. Environmental factors affecting the loss of carbon from the roots of wheat and barley seedlings. J. Exp. Bot. 1984, 35, 767–773. [Google Scholar] [CrossRef]
- Whipps, J.M. Effect of CO2concentration on growth, carbon distribution and loss of carbon from the roots of maize. J. Exp. Bot. 1985, 36, 644–651. [Google Scholar] [CrossRef]
- Whipps, J. Carbon loss from the roots of tomato and pea seedlings grown in soil. Plant Soil 1987, 103, 95–100. [Google Scholar] [CrossRef]
- Oburger, E.; Jones, D.L. Sampling root exudates-Mission impossible? Rhizosphere 2018, 6. [Google Scholar] [CrossRef]
- Oburger, E.; Dell‘mour, M.; Hann, S.; Wieshammer, G.; Puschenreiter, M.; Wenzel, W.W. Evaluation of a novel tool for sampling root exudates from soil-grown plants compared to conventional techniques. Environ. Exp. Bot. 2013, 87, 235–247. [Google Scholar] [CrossRef]
- Oburger, E.; Gruber, B.; Schindlegger, Y.; Schenkeveld, W.D.C.; Hann, S.; Kraemer, S.M.; Wenzel, W.W.; Puschenreiter, M. Root exudation of phytosiderophores from soil-grown wheat. New Phytol. 2014, 203, 1161–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Dakora, F.D.; Phillips, D.A. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 2002, 245, 35–47. [Google Scholar] [CrossRef]
- Neumann, G.; Römheld, V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 1999, 211, 121–130. [Google Scholar] [CrossRef]
- Lipton, D.S.; Blanchar, R.W.; Blevins, D.G. Citrate, Malate, and Succinate Concentration in Exudates from P-Sufficient and P-Stressed Medicago sativa L. Seedlings. Plant Physiol. 1987, 85, 315–317. [Google Scholar] [CrossRef] [Green Version]
- Römheld, V. Different strategies for iron acquisition in higher plants. Physiol. Plant. 1987, 70, 231–234. [Google Scholar] [CrossRef]
- Römheld, V. The role of phytosiderophores in acquisition of iron and other micronutrients in graminaceous species: An ecological approach. Plant Soil 1991, 130, 127–134. [Google Scholar] [CrossRef]
- Treeby, M.; Marschner, H.; Römheld, V. Mobilization of iron and other micronutrient cations from a calcareous soil by plant-borne, microbial, and synthetic metal chelators. Plant Soil 1989, 114, 217–226. [Google Scholar] [CrossRef]
- Rengel, Z. Physiological mechanisms underlying different nutirent efficiency of crop genotypes. In Mineral Nutrition of Crops: Fundamental Mechanisms and Implications; Rengel, Z., Ed.; Haworth Press, Inc.: New York, NY, USA, 1999; pp. 227–265. [Google Scholar]
- Asmar, F. Variation in activity of root extracellular phytase between genotypes of barley. Plant Soil 1997, 195, 61–64. [Google Scholar] [CrossRef]
- Asmar, F.; Singh Gahoonia, T.; Nielsen, N.E. Barley genotypes differ in activity of soluble extracellular phosphatase and depletion of organic phosphorus in the rhizosphere soil. Plant Soil 1995, 172, 117–122. [Google Scholar] [CrossRef]
- Richardson, A.E.; Hadobas, P.A.; Hayes, J.E. Acid phosphomonoesterase and phytase activities of wheat (Triticum aestivum L.) roots and utilization of organic phosphorus substrates by seedlings grown in sterile culture. Plant Cell Environ. 2000, 23, 397–405. [Google Scholar] [CrossRef]
- McLachlan, K.D.; Marco, D. Acid phosphatase activity of intact roots and phosphorus nutrition in plants. III. Its relation to phosphorus garnering by wheat and a comparison with leaf activity as a measure of phosphorus status. Aust. J. Agric. Res. 1982, 33, 1–11. [Google Scholar] [CrossRef]
- De Vos, C.R.; Lubberding, H.J.; Bienfait, H.F. Rhizosphere Acidification as a Response to Iron Deficiency in Bean Plants. Plant Physiol. 1986, 81, 842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, J.; Rui, Y.; Zhang, F.; Rengel, Z.; Shen, J. Localized application of phosphorus and ammonium improves growth of maize seedlings by stimulating root proliferation and rhizosphere acidification. Field Crop. Res. 2010, 119, 355–364. [Google Scholar] [CrossRef]
- Ding, X.; Fu, L.; Liu, C.; Chen, F.; Hoffland, E.; Shen, J.; Zhang, F.; Feng, G. Positive feedback between acidification and organic phosphate mineralization in the rhizosphere of maize (Zea mays L.). Plant Soil 2011, 349, 13–24. [Google Scholar] [CrossRef]
- Zhou, L.L.; Cao, J.; Zhang, F.S.; Li, L. Rhizosphere acidification of faba bean, soybean and maize. Sci. Total Environ. 2009, 407, 4356–4362. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, H.; Wang, Y.; Jia, W.; Xu, X.; Zhang, X.; Han, Z.; Wu, T.; Zhang, H.; Wang, Y.; et al. Induction of root Fe(III) reductase activity and proton extrusion by iron deficiency is mediated by auxin-based systemic signaling in Malys xiaojinensis. J. Expt. Bot. 2012, 63, 859–870. [Google Scholar] [CrossRef] [Green Version]
- Nunez, G.H.; Olmstead, J.W.; Darnell, R.L. Rhizosphere acidification is not part of the strategy I iron deficiency response of vaccinium arboreum and the southern highbush blueberry. HortScience 2015, 50, 1064–1069. [Google Scholar] [CrossRef] [Green Version]
- Palta, J.A.; Turner, N.C. Crop root system traits cannot be seen as a silver bullet delivering drought resistance. Plant Soil 2018. [Google Scholar] [CrossRef]
- Hodge, A. The plastic plant: Root responses to heterogeneous supplies of nutrients. New Phytol. 2004, 162, 9–24. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McGrail, R.K.; Van Sanford, D.A.; McNear, D.H. Trait-Based Root Phenotyping as a Necessary Tool for Crop Selection and Improvement. Agronomy 2020, 10, 1328. https://doi.org/10.3390/agronomy10091328
McGrail RK, Van Sanford DA, McNear DH. Trait-Based Root Phenotyping as a Necessary Tool for Crop Selection and Improvement. Agronomy. 2020; 10(9):1328. https://doi.org/10.3390/agronomy10091328
Chicago/Turabian StyleMcGrail, Rebecca K., David A. Van Sanford, and David H. McNear. 2020. "Trait-Based Root Phenotyping as a Necessary Tool for Crop Selection and Improvement" Agronomy 10, no. 9: 1328. https://doi.org/10.3390/agronomy10091328
APA StyleMcGrail, R. K., Van Sanford, D. A., & McNear, D. H. (2020). Trait-Based Root Phenotyping as a Necessary Tool for Crop Selection and Improvement. Agronomy, 10(9), 1328. https://doi.org/10.3390/agronomy10091328





