Characterization and Analysis of Anthocyanin-Related Genes in Wild-Type Blueberry and the Pink-Fruited Mutant Cultivar ‘Pink Lemonade’: New Insights into Anthocyanin Biosynthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search for Anthocyanin Pathway Structural Genes
2.2. Search for Anthocyanin Pathway Regulatory Genes
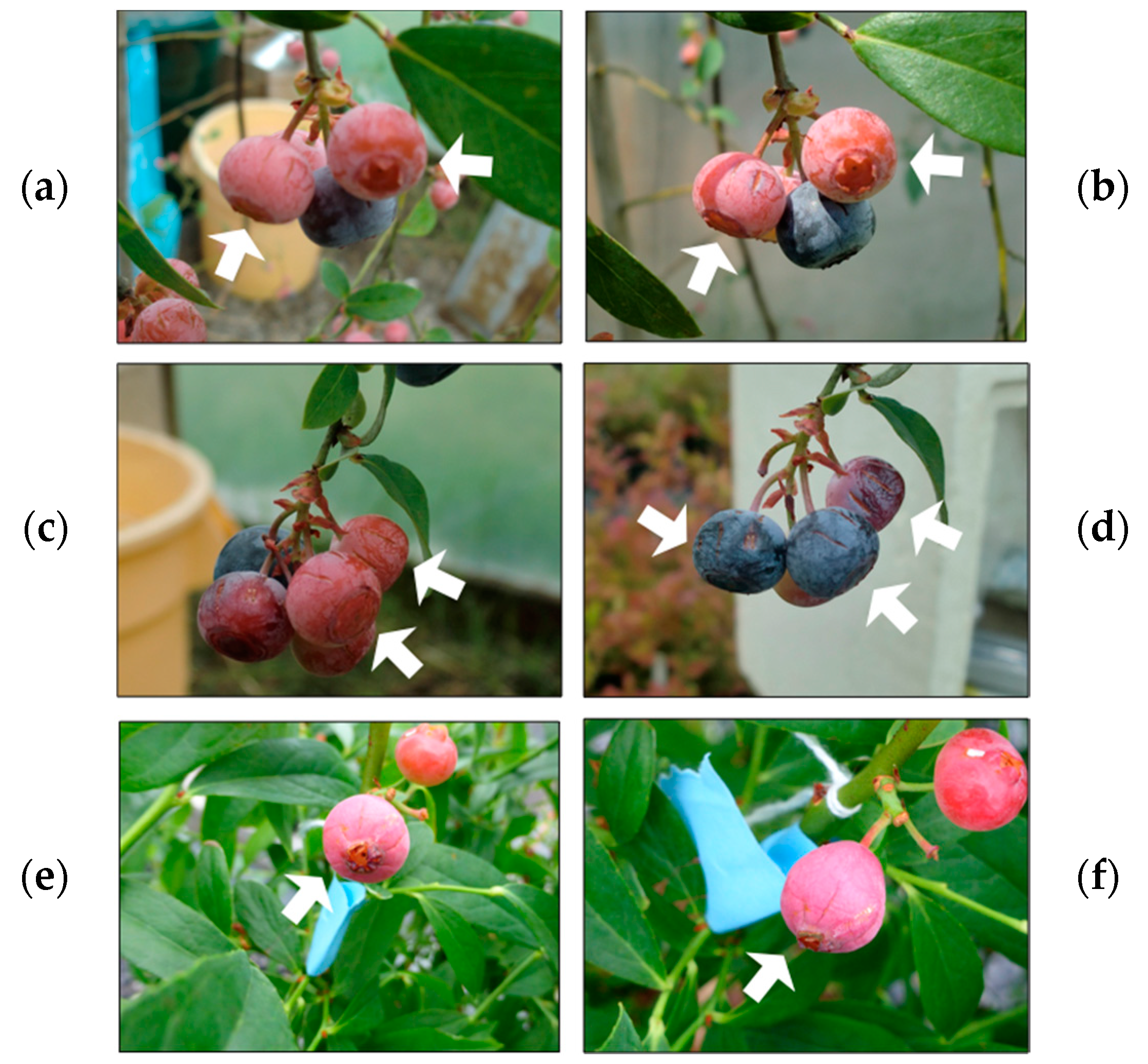
2.3. Plant Material
2.4. RNA Isolation and Quality Controls
2.5. cDNA Synthesis and Quality Controls
2.6. Primer Design and qPCR Assay
2.7. qPCR Data Analysis
2.8. Phylogenetic Analysis
2.9. Transient Expression Assay
2.10. Sequence Analysis
3. Results
3.1. Search for Anthocyanin Genes in the Blueberry Genome and Isolation of Candidate Genes
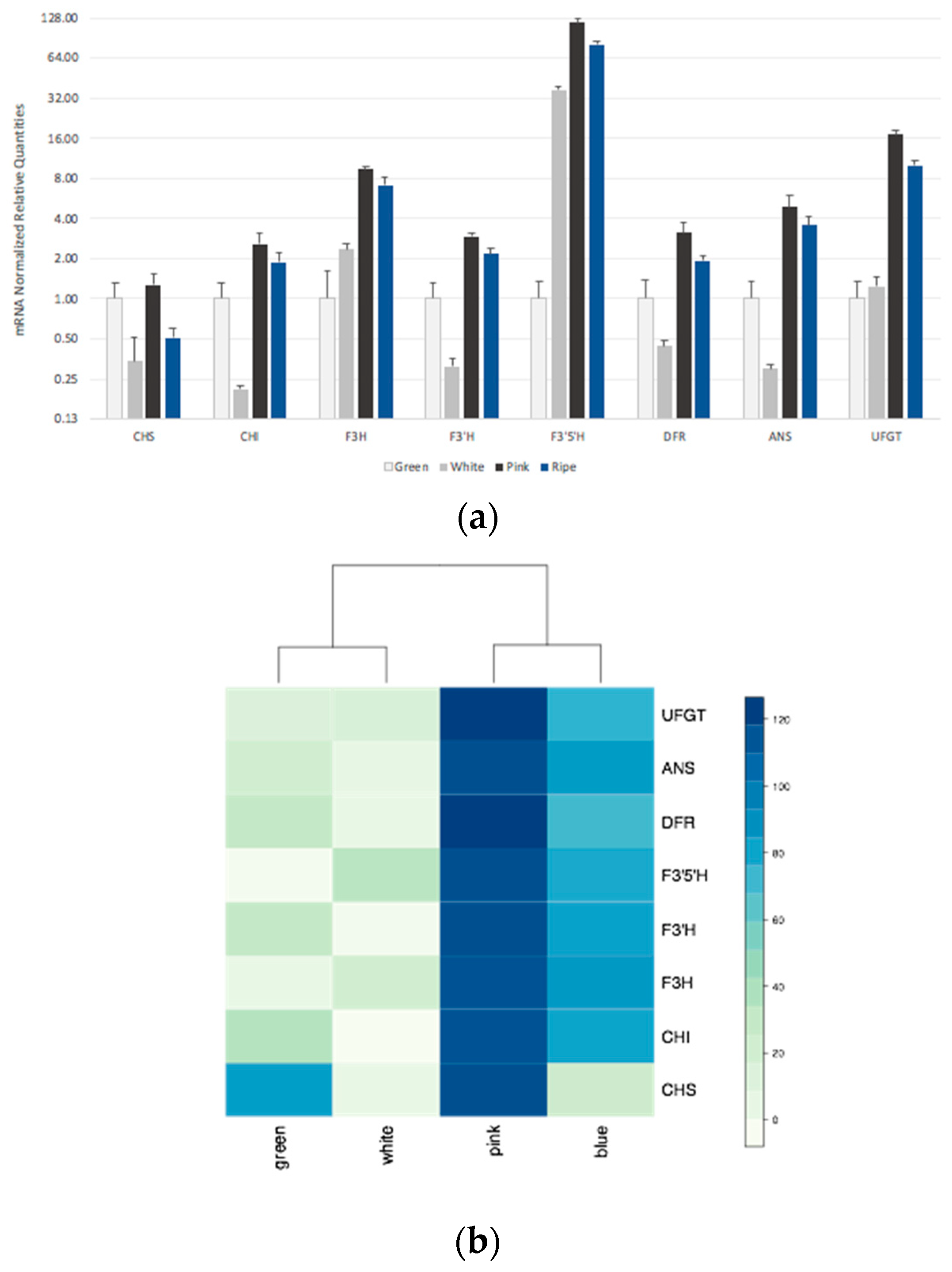
3.2. Expression of Structural Genes in Developing Fruits of Wild-Type Berries
3.3. Transient Expression of F3′5′H in Developing Fruits of Wild-Type and Color Mutants
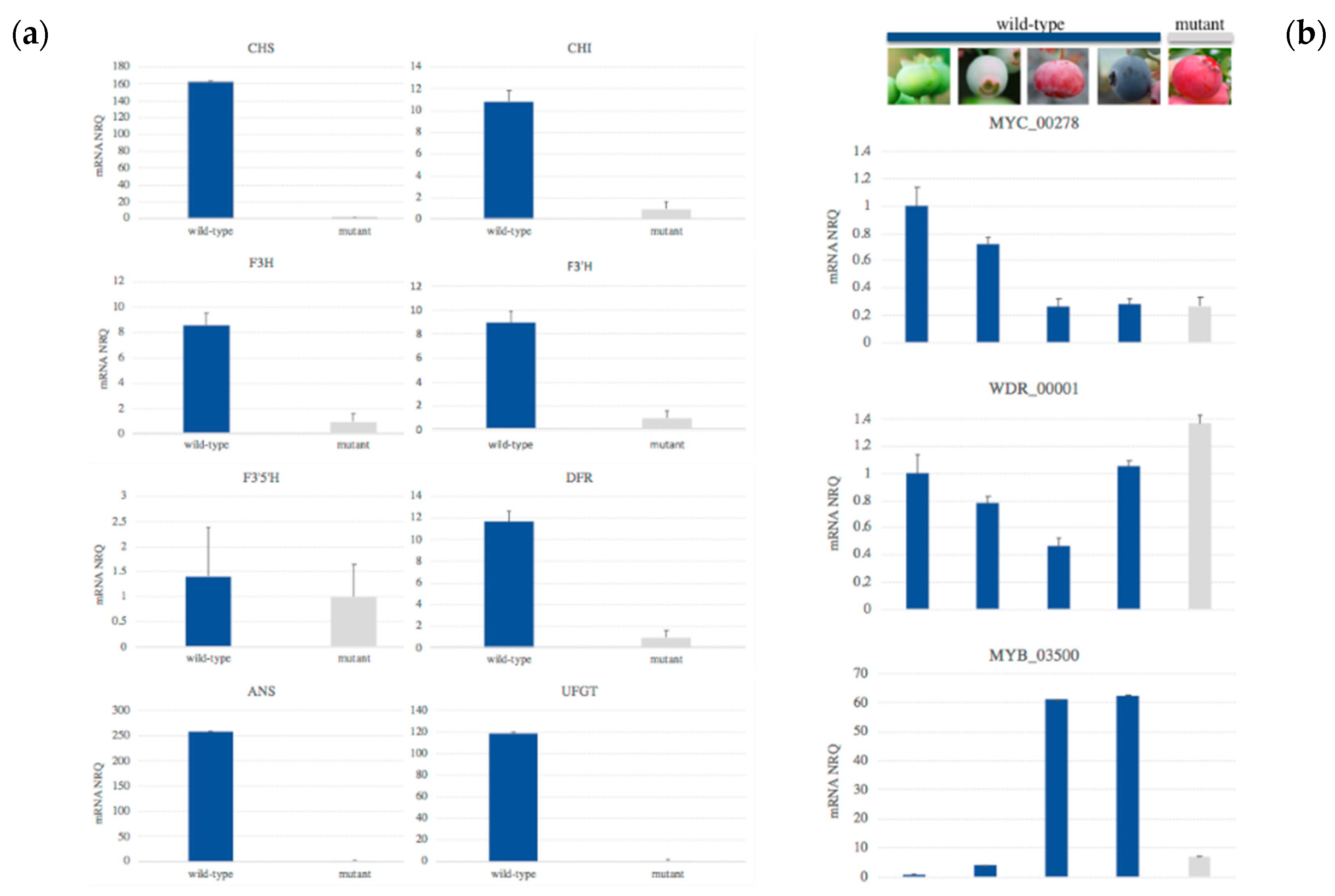
3.4. Expression of Structural Genes in Developed Fruits of Mutant Berries
3.5. Candidate Regulatory Genes
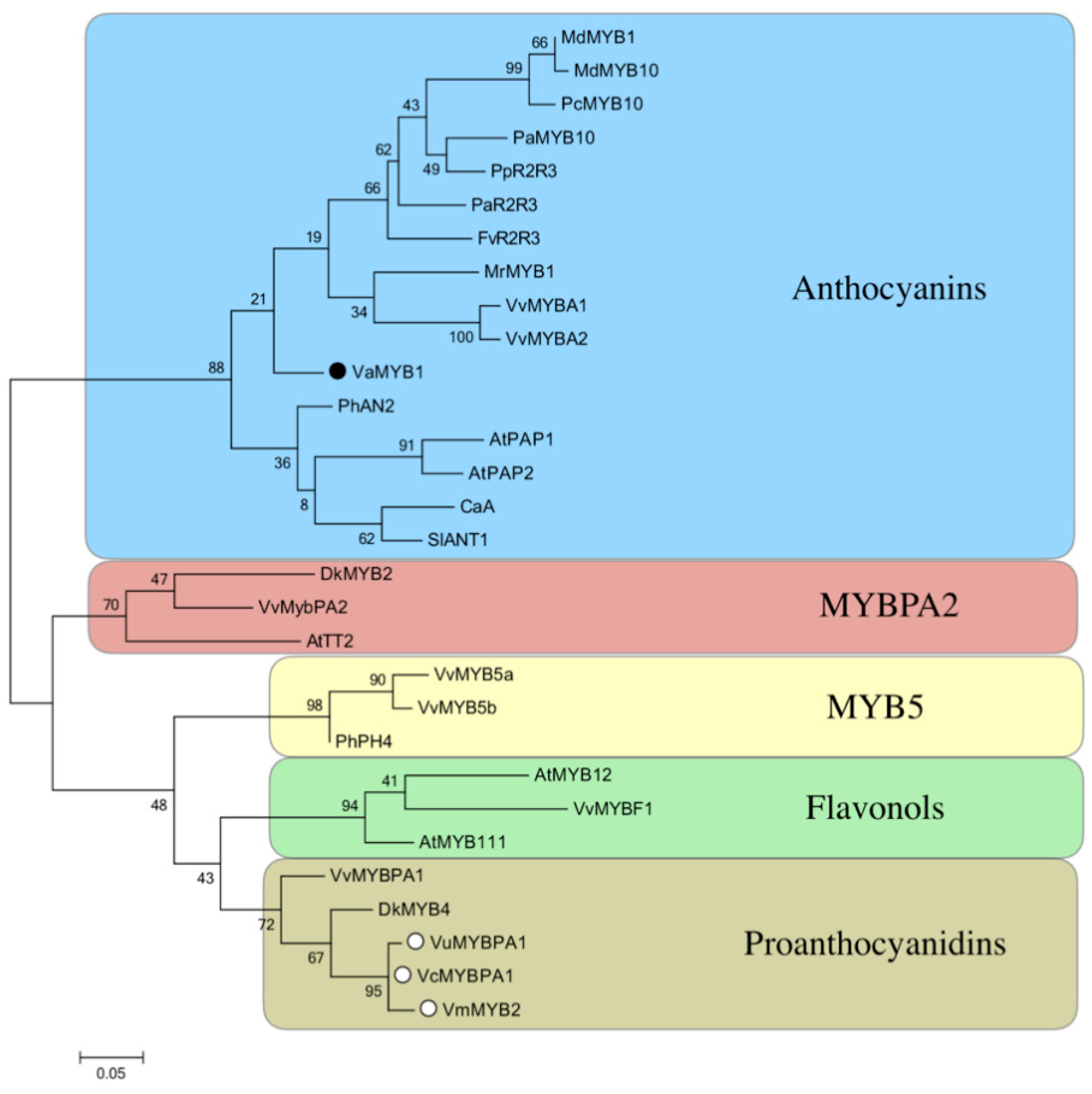
3.6. Phylogenetic Analysis
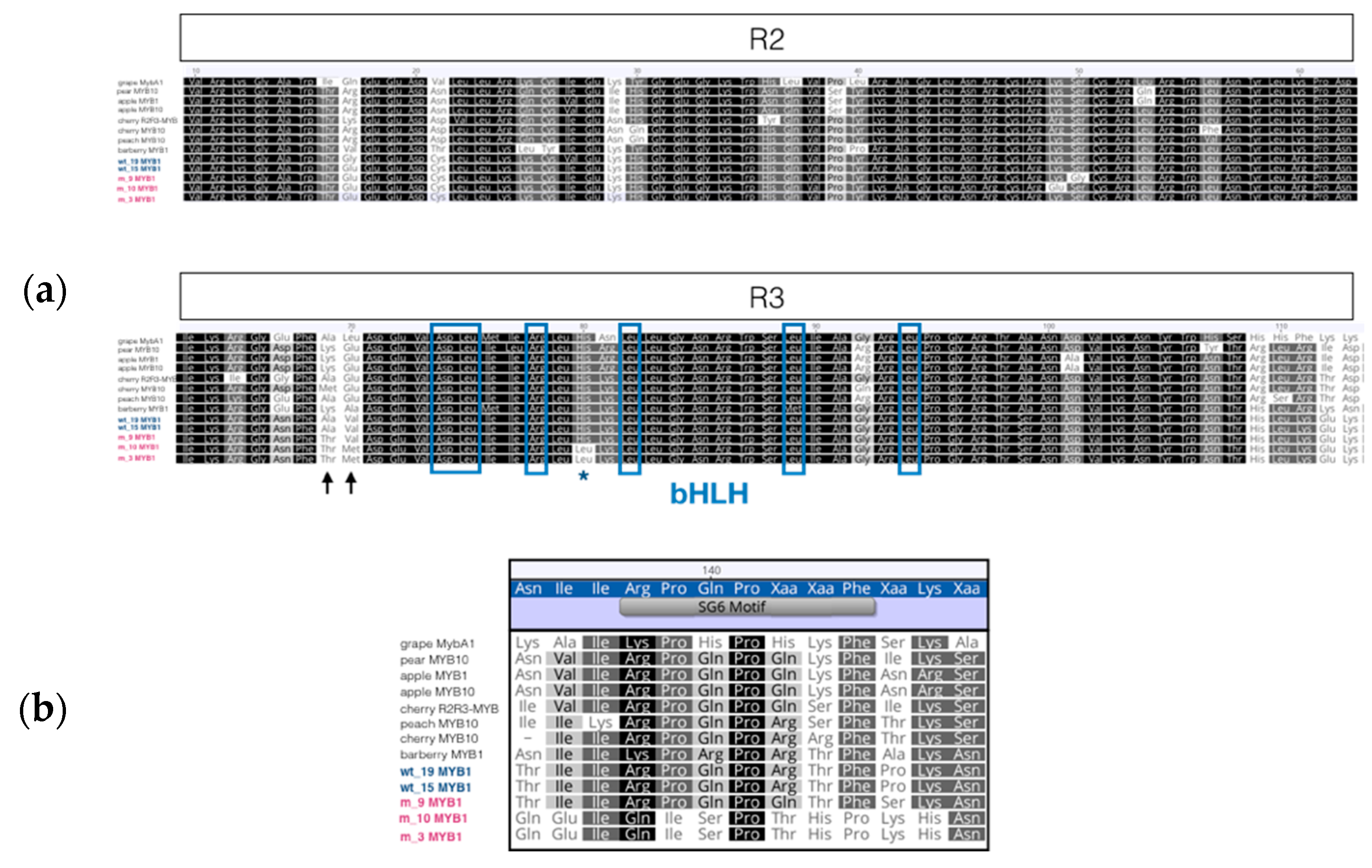
3.7. Sequence Analysis of MYB Transcription Factor
3.8. MYB Activates Anthocyanin Biosynthesis in Planta
4. Discussion
4.1. The Expression Profile of Anthocyanin Biosynthetic Genes Suggests Coordinated Regulation in Blueberry Fruit Development
4.2. Expression of Structural Genes in Developed Fruits of Mutant Berries Suggests the Action of Regulatory Genes
4.3. The MYB Transcription Factor is A Regulator of Anthocyanin Biosynthesis and Might be Mutated in ‘PL’
4.4. Sequence Analysis of MYB-cDNAs from Wild-Type and ‘PL’ Indicates ‘PL’ Expresses Normal and Mutated MYB Alleles
5. Concluding Remarks and Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- NASS; USDA. Noncitrus Fruits and Nuts 2017 Summary. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/ncit0618.pdf#:~:text=104%20Noncitrus%20Fruits%20and%20Nuts%202017%20Summary%20%28June,7%2C200%2C%20up%203%20percent%20from%20the%20previous%20year (accessed on 30 June 2018).
- FAOSTAT; FAO. Crop Statistics. Available online: http://faostat.fao.org (accessed on 20 November 2018).
- Nicoletti, A.M.; Gularte, M.A.; Elias, M.C.; dos Santos, M.S.; Ávila, B.P.; Fernandes, J.L.; Peres, W. Blueberry Bioactive Properties and Their Benefits. IJNTR 2015, 1, 51–57. [Google Scholar]
- Prior, R.L.; Lazarus, S.A.; Cao, G.; Muccitelli, H.; Hammerstone, J.F. Identification of procyanidins and anthocyanins in blueberries and cranberries (Vaccinium spp.) using high-performance liquid chromatography/mass spectrometry. J. Agric. Food Chem. 2001, 49, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Camp, M.J.; Ehlenfeldt, M.K. Antioxidant capacity and α-glucosidase inhibitory activity in peel and flesh of blueberry (Vaccinium spp.) cultivars. Food Chem. 2012, 132, 1759–1768. [Google Scholar] [CrossRef]
- Scalzo, J.; Stevenson, D.; Hedderley, D. Blueberry estimated harvest from seven new cultivars: Fruit and anthocyanins. Food Chem. 2013, 139, 44–50. [Google Scholar] [CrossRef]
- Wang, S.Y.; Chen, H.; Camp, M.J.; Ehlenfeldt, M.K. Genotype and growing season influence blueberry antioxidant capacity and other quality attributes. Int. J. Food Sci. Technol. 2012, 47, 1540–1549. [Google Scholar] [CrossRef]
- Yousef, G.G.; Lila, M.A.; Guzman, I.; Ballington, J.R.; Brown, A.F. Impact of interspecific introgression on anthocyanin profiles of southern highbush blueberry. J. Amer. Soc. Hort. Sci. 2014, 139, 99–112. [Google Scholar] [CrossRef] [Green Version]
- Zoratti, L.; Karppinen, K.; Luengo Escobar, A.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef]
- Jaakola, L.; Määttä, K.; Pirttilä, A.M.; Törrönen, R.; Kärenlampi, S.; Hohtola, A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002, 130, 729–739. [Google Scholar] [CrossRef] [Green Version]
- Zifkin, M.; Jin, A.; Ozga, J.A.; Zaharia, L.I.; Schernthaner, J.P.; Gesell, A.; Abrams, S.R.; Kennedy, J.A.; Constabel, C.P. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 2012, 158, 200–224. [Google Scholar] [CrossRef] [Green Version]
- Sanoner, P.; Guyot, S.; Marnet, N.; Molle, D.; Drilleau, J.P. Polyphenol profiles of French cider apple varieties (Malus domestica spp.). J. Agric. Food Chem. 1999, 47, 4847–4853. [Google Scholar] [CrossRef]
- Willson, M.F.; Whelan, C.J. The evolution of fruit color in fleshy-fruited plants. Am. Nat. 1990, 136, 790–809. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Func. Plant Biol. 2012, 39, 619. [Google Scholar] [CrossRef]
- Wang, S.Y.; Chen, H.; Camp, M.J.; Ehlenfeldt, M.K. Flavonoid constituents and their contribution to antioxidant activity in cultivars and hybrids of rabbiteye blueberry (Vaccinium ashei Reade). Food Chem. 2012, 132, 855–864. [Google Scholar] [CrossRef]
- Timmers, M.A.; Grace, M.H.; Yousef, G.G.; Lila, M.A. Inter- and intra-seasonal changes in anthocyanin accumulation and global metabolite profiling of six blueberry genotypes. J. Food Comp. Anal. 2017, 59, 105–110. [Google Scholar] [CrossRef]
- Rowland, L.J.; Alkharouf, N.; Darwish, O.; Ogden, E.L.; Polashock, J.J.; Bassil, N.V.; Main, D. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012, 12, 46. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Estrada, A.D.; Blakley, I.; Reid, R.; Patel, K.; Meyer, M.D.; Andersen, S.U.; Brown, A.F.; Lila, M.A.; Loraine, A.E. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 2015, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Colle, M.; Leisner, C.P.; Wai, C.M.; Ou, S.; Bird, K.A.; Wang, J.; Wisecaver, J.H.; Yocca, A.E.; Alger, E.I.; Tang, H.; et al. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhang, H.; Liu, Z.; Cui, X.; Zhang, T.; Li, Y.; Zhang, L. Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 2016, 16, 223. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Wang, Y.; Li, B.; Tan, H.; Li, D.; Li, L.; Liu, X.; Han, J.; Meng, X. Comparative transcriptome analysis of genes involved in anthocyanin synthesis in blueberry. Plant Physiol. Biochem. 2018, 127, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Ehlenfeldt, M.K.; Finn, C.E. G-435 and ARS 96-138, Pink-fruited Blueberry Selections. HortScience 2007, 42, 172–173. [Google Scholar] [CrossRef] [Green Version]
- Matus, J.T.; Poupin, M.J.; Cañón, P.; Bordeu, E.; Alcalde, J.A.; Arce-Johnson, P. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol. Biol. 2010, 72, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Cadle-Davidson, M.M.; Owens, C.L. Genomic amplification of the Gret1 retroelement in white-fruited accessions of wild vitis and interspecific hybrids. Theor. Appl. Genet. 2008, 116, 1079–1094. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Die, J.V.; Rowland, L.J. Elucidating cold acclimation pathway in blueberry by transcriptome profiling. Environ. Exp. Bot. 2014, 106, 87–98. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Die, J.V.; Obrero, Á.; González-Verdejo, C.I.; Román, B. Characterization of the 3’:5’ ratio for reliable determination of RNA quality. Anal. Biochem. 2011, 419, 336–338. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3–New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Die, J.V.; Rowland, L.J. Superior cross-species reference genes: A blueberry case study. PLoS ONE 2013, 8, e73354. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Yamagishi, M.; Shimoyamada, Y.; Nakatsuka, T.; Masuda, K. Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 2010, 51, 463–474. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Dhanaraj, A.L.; Slovin, J.P.; Rowland, L.J. Analysis of gene expression associated with cold acclimation in blueberry floral buds using expressed sequence tags. Plant Sci. 2004, 166, 863–872. [Google Scholar] [CrossRef]
- Dhanaraj, A.L.; Alkharouf, N.W.; Beard, H.S.; Chouikha, I.B.; Matthews, B.F.; Wei, H.; Arora, R.; Rowland, L.J. Major differences observed in transcript profiles of blueberry during cold acclimation under field and cold room conditions. Planta 2007, 225, 735–751. [Google Scholar] [CrossRef]
- Li, X.; Sun, H.; Pei, J.; Dong, Y.; Wang, F.; Chen, H.; Sun, Y.; Wang, N.; Li, H.; Li, Y. De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants. Gene 2012, 511, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Ogden, E.L.; Ehlenfeldt, M.K.; Rowland, L.J. Dataset of de novo assembly and functional annotation of the transcriptome of blueberry (Vaccinium spp.). Data Brief 2019, 25, 104390. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Ogden, E.L.; Die, J.V.; Ehlenfeldt, M.K.; Polashock, J.J.; Darwish, O.; Alkharouf, N.; Rowland, L.J. Transcriptome analysis identifies genes related to the waxy coating on blueberry fruit in two northern-adapted rabbiteye breeding populations. BMC Plant Biol. 2019, 19, 460. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karppinen, K.; Zoratti, L.; Nguyenquynh, N.; Häggman, H.; Jaakola, L. On the developmental and environmental regulation of secondary metabolism in Vaccinium spp. Berries. Front. Plant Sci. 2016, 7, 655. [Google Scholar] [CrossRef] [Green Version]
- Primetta, A.K.; Karppinen, K.; Riihinen, K.R.; Jaakola, L. Metabolic and molecular analyses of white mutant Vaccinium berries show down-regulation of MYBPA1-type R2R3 MYB regulatory factor. Planta 2015, 242, 631–643. [Google Scholar] [CrossRef] [Green Version]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verriès, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar] [CrossRef] [Green Version]
- Matus, J.T.; Cavallini, E.; Loyola, R.; Höll, J.; Finezzo, L.; Dal Santo, S.; Vialet, S.; Commisso, M.; Roman, F.; Schubert, A.; et al. A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 2017, 91, 220–236. [Google Scholar] [CrossRef] [Green Version]
- Plunkett, B.J.; Espley, R.V.; Dare, A.P.; Warren, B.A.W.; Grierson, E.R.P.; Cordiner, S.; Turner, J.L.; Allan, A.C.; Albert, N.W.; Davies, K.M.; et al. MYBA from blueberry (Vaccinium section Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Front. Plant Sci. 2018, 9, 1300. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Die, J.V.; Jones, R.W.; Ogden, E.L.; Ehlenfeldt, M.K.; Rowland, L.J. Characterization and Analysis of Anthocyanin-Related Genes in Wild-Type Blueberry and the Pink-Fruited Mutant Cultivar ‘Pink Lemonade’: New Insights into Anthocyanin Biosynthesis. Agronomy 2020, 10, 1296. https://doi.org/10.3390/agronomy10091296
Die JV, Jones RW, Ogden EL, Ehlenfeldt MK, Rowland LJ. Characterization and Analysis of Anthocyanin-Related Genes in Wild-Type Blueberry and the Pink-Fruited Mutant Cultivar ‘Pink Lemonade’: New Insights into Anthocyanin Biosynthesis. Agronomy. 2020; 10(9):1296. https://doi.org/10.3390/agronomy10091296
Chicago/Turabian StyleDie, Jose V., Richard W. Jones, Elizabeth L. Ogden, Mark K. Ehlenfeldt, and Lisa J. Rowland. 2020. "Characterization and Analysis of Anthocyanin-Related Genes in Wild-Type Blueberry and the Pink-Fruited Mutant Cultivar ‘Pink Lemonade’: New Insights into Anthocyanin Biosynthesis" Agronomy 10, no. 9: 1296. https://doi.org/10.3390/agronomy10091296
APA StyleDie, J. V., Jones, R. W., Ogden, E. L., Ehlenfeldt, M. K., & Rowland, L. J. (2020). Characterization and Analysis of Anthocyanin-Related Genes in Wild-Type Blueberry and the Pink-Fruited Mutant Cultivar ‘Pink Lemonade’: New Insights into Anthocyanin Biosynthesis. Agronomy, 10(9), 1296. https://doi.org/10.3390/agronomy10091296






