Agricultural Utilization of Unused Resources: Liquid Food Waste Material as a New Source of Plant Growth-Promoting Microbes
Abstract
1. Introduction
2. Materials and Methods
2.1. Liquid Food Waste Materials
2.2. Assay of Liquid Food Waste Material (LFM) Utilization
2.3. Isolation of Bacteria from LFM
2.4. DNA Extraction and PCR Amplification for Culturable Bacteria
2.5. High-Throughput DNA Sequencing
2.6. Plant Growth-Promoting Traits of Isolates
2.7. Pot Experiments
2.8. Field Experiment Using LFM
2.9. Statistical Analysis
3. Results
3.1. Isolation of Plant Growth Promoting Bacteria (PGPB)
3.2. Identification of Culturable Bacteria
3.3. PGP Traits
3.4. High-Throughput DNA Sequencing of LFM
3.5. Incubation Study of LFM Utilization
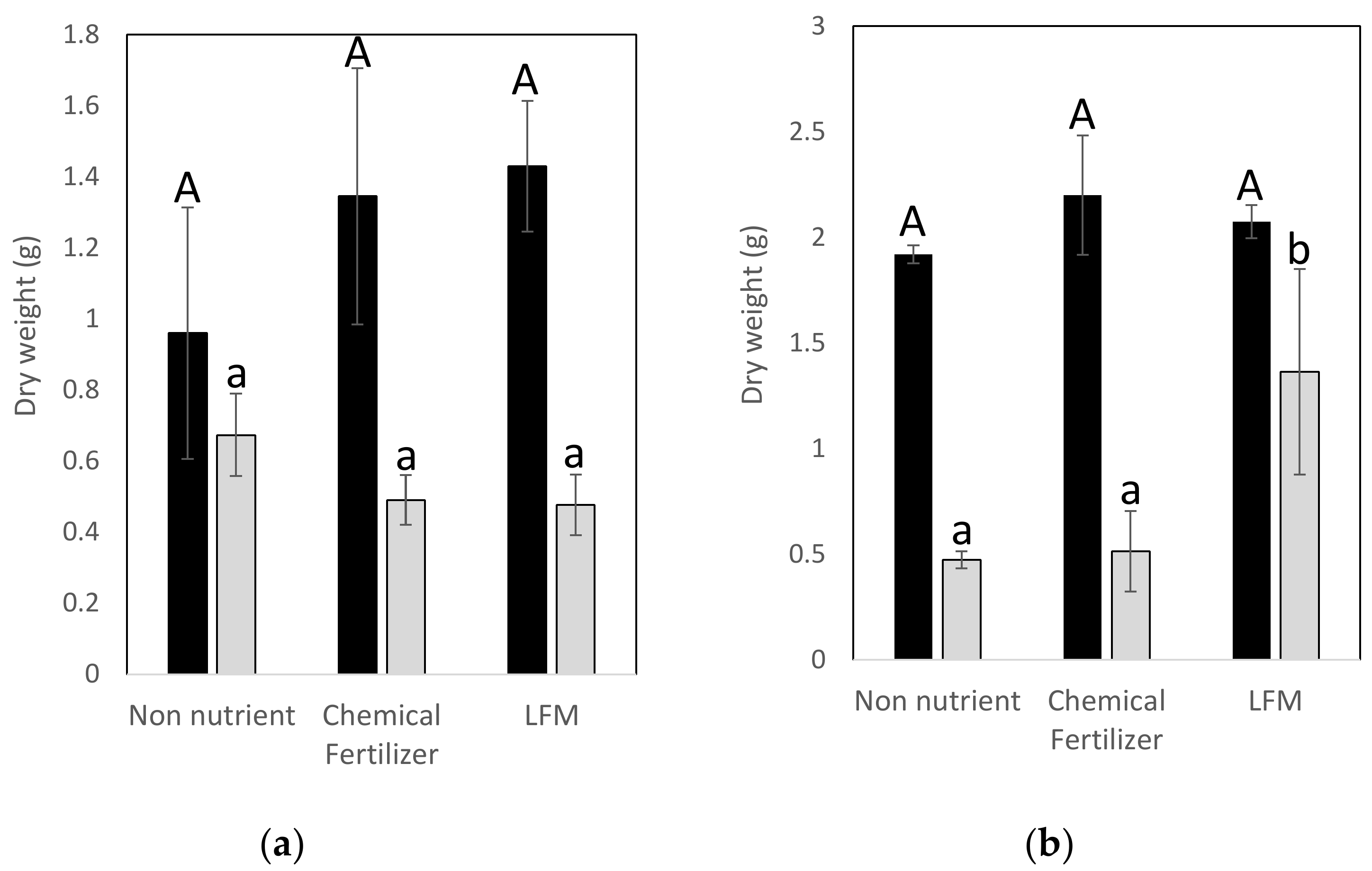
3.6. Pot Experiments
3.7. Field Experiments
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application. BMC Microbiol. 2020, 20, 1–12. [Google Scholar]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 1–19. [Google Scholar]
- Lin, L.; Li, Z.; Hu, C.; Zhang, X.; Chang, S.; Yang, L.; Li, Y.; An, Q. Plant growth-promoting nitrogen-fixing enterobacteria are in association with sugarcane plants growing in Guangxi, China. Microbes Environ. 2012, 27, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Zaheer, A.; Malik, A.; Sher, A.; Qaisrani, M.M.; Mehmood, A.; Khan, S.U.; Ashraf, M.; Mirza, Z.; Karim, S.; Rasool, M. Isolation, characterization, and effect of phosphate-zinc-solubilizing bacterial strains on chickpea (Cicer arietinum L.) growth. Saudi J. Biol. Sci. 2019, 26, 1061–1067. [Google Scholar] [CrossRef]
- Sivasankari, B.; Anandharaj, M.; Daniel, T. Effect of PGR producing bacterial strains isolated from vermisources on germination and growth of Vigna unguiculata (L.) Walp. J. Biochem. Technol. 2014, 5, 808–813. [Google Scholar]
- Grobelak, A.; Kokot, P.; Hutchison, D.; Grosser, A.; Kacprzak, M. Plant growth-promoting rhizobacteria as an alternative to mineral fertilizers in assisted bioremediation-sustainable land and waste management. J. Environ. Manag. 2018, 227, 1–9. [Google Scholar] [CrossRef]
- Sun, G.; Yao, T.; Feng, C.; Chen, L.; Li, J.; Wang, L. Identification and biocontrol potential of antagonistic bacteria strains against Sclerotinia sclerotiorum and their growth-promoting effects on Brassica napus. Biol. Control. 2017, 104, 35–43. [Google Scholar] [CrossRef]
- Etesami, H.; Emami, S.; Alikhani, H.A. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and future prospects A review. J. Soil Sci. Plant. Nutr. 2017, 17, 897–911. [Google Scholar] [CrossRef]
- Padmini, O.S.; Wuryani, S.; Aryani, R. Application of Organic Fertilizer and Plant Growth-Promoting Rhizobacteria (PGPR) to Increase Rice Yield and Quality. In ICoSI: Proceedings of the 2nd International Conference on Sustainable Innovation 2014; Taufik, T., Prabasari, I., Rineksane, I.A., Yaya, R., Widowati, R., Rosyidi, S.A.P., Riyadi, S., Harsanto, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 3–11. [Google Scholar]
- Komatsuzaki, M.; Ohta, H. Soil management practices for sustainable agro-ecosystems. Sustain. Sci. 2007, 2, 103–120. [Google Scholar] [CrossRef]
- Mahmood, A.; Iguchi, R.; Kataoka, R. Multifunctional food waste fertilizer having the capability of Fusarium-growth inhibition and phosphate solubility: A new horizon of food waste recycle using microorganisms. Waste Manag. 2019, 94, 77–84. [Google Scholar] [CrossRef]
- Mahmood, A.; Kataoka, R. Metabolite profiling reveals a complex response of plants to application of plant growth-promoting endophytic bacteria. Microbiol. Res. 2020, 234, 126421. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.K.; Herat, S.; Bharambe, G.; Patil, S.; Bapat, P.; Chauhan, K.; Valani, D. Human waste-as potential resource: Converting trash into treasure by embracing the 5 r’s philosophy for safe and sustainable waste management. Environ. Res. J. 2009, 13, 111–171. [Google Scholar]
- Gustavsson, J.; Cederberg, C.; Sonesson, U.; Van Otterdijk, R.; Meybeck, A. Global Food Losses and Food Waste; FAO: Rome, Italy, 2011. [Google Scholar]
- Liu, C.; Hotta, Y.; Santo, A.; Hengesbaugh, M.; Watabe, A.; Totoki, Y.; Allen, D.; Bengtsson, M. Food waste in Japan: Trends, current practices and key challenges. J. Clean. Prod. 2016, 133, 557–564. [Google Scholar] [CrossRef]
- Lin, C.S.K.; Pfaltzgraff, L.A.; Herrero-Davila, L.; Mubofu, E.B.; Abderrahim, S.; Clark, J.H.; Koutinas, A.A.; Kopsahelis, N.; Stamatelatou, K.; Dickson, F. Food waste as a valuable resource for the production of chemicals, materials and fuels. Current situation and global perspective. Energy Environ. Sci. 2013, 6, 426–464. [Google Scholar] [CrossRef]
- Baroutian, S.; Munir, M.T.; Sun, J.Y.; Eshtiaghi, N.; Young, B.R. Rheological characterisation of biologically treated and non-treated putrescible food waste. Waste Manag. 2018, 71, 494–501. [Google Scholar] [CrossRef]
- Yan, S.; Li, J.; Chen, X.; Wu, J.; Wang, P.; Ye, J.; Yao, J. Enzymatical hydrolysis of food waste and ethanol production from the hydrolysate. Renew. Energy 2011, 36, 1259–1265. [Google Scholar] [CrossRef]
- Leung, C.C.J.; Cheung, A.S.Y.; Zhang, A.Y.-Z.; Lam, K.F.; Lin, C.S.K. Utilisation of waste bread for fermentative succinic acid production. Biochem. Eng. J. 2012, 65, 10–15. [Google Scholar] [CrossRef]
- Marinari, S.; Masciandaro, G.; Ceccanti, B.; Grego, S. Influence of organic and mineral fertilisers on soil biological and physical properties. Bioresour. Technol. 2000, 72, 9–17. [Google Scholar] [CrossRef]
- Ney, L.; Franklin, D.; Mahmud, K.; Cabrera, M.; Hancock, D.; Habteselassie, M.; Newcomer, Q.; Dahal, S. Impact of inoculation with local effective microorganisms on soil nitrogen cycling and legume productivity using composted broiler litter. Appl. Soil Ecol. 2020, 154, 103567. [Google Scholar] [CrossRef]
- Ney, L.; Franklin, D.; Mahmud, K.; Cabrera, M.; Hancock, D.; Habteselassie, M.; Newcomer, Q. Examining trophic-level nematode community structure and nitrogen mineralization to assess local effective microorganisms’ role in nitrogen availability of swine effluent to forage crops. Appl. Soil Ecol. 2018, 130, 209–218. [Google Scholar] [CrossRef]
- Truong, L.; Morash, D.; Liu, Y.; King, A. Food waste in animal feed with a focus on use for broilers. Int. J. Recycl. Org. Waste Agric. 2019, 8, 417–429. [Google Scholar] [CrossRef]
- Torres-León, C.; Ramírez-Guzman, N.; Londoño-Hernandez, L.; Martinez-Medina, G.A.; Díaz-Herrera, R.; Navarro-Macias, V.; Alvarez-Pérez, O.B.; Picazo, B.; Villarreal-Vázquez, M.; Ascacio-Valdes, J.; et al. Food waste and byproducts: An opportunity to minimize malnutrition and hunger in developing countries. Front. Sustain. Food Syst. 2018, 2. [Google Scholar] [CrossRef]
- Girotto, F.; Alibardi, L.; Cossu, R. Food waste generation and industrial uses: A review. Waste Manag. 2015, 45, 32–41. [Google Scholar] [CrossRef]
- Center, K.-C.B. Food waste recycle. Available online: https://www.city.kai.yamanashi.jp/kurashi_tetsuduki/gomi_kankyo_pet/kankyo/3704.html (accessed on 27 May 2020).
- Mahmood, A.; Takagi, K.; Ito, K.; Kataoka, R. Changes in endophytic bacterial communities during different growth stages of cucumber (Cucumis sativus L.). World J. Microbiol. Biotechnol. 2019, 35, 104. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Acuña, J.; Jorquera, M.; Martínez, O.; Menezes-Blackburn, D.; Fernández, M.; Marschner, P.; Greiner, R.; Mora, M. Indole acetic acid and phytase activity produced by rhizosphere bacilli as affected by pH and metals. J. Soil Sci. Plant. Nutr. 2011, 11, 1–12. [Google Scholar]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef]
- Poly, F.; Ranjard, L.; Nazaret, S.; Gourbière, F.; Monrozier, L.J. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 2001, 67, 2255–2262. [Google Scholar] [CrossRef]
- Jha, B.; Gontia, I.; Hartmann, A. The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant. Soil 2012, 356, 265–277. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.; Fernández, F. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef]
- Pikovskaya, R. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 1948, 17, 362–370. [Google Scholar]
- Piggot, P.J.; Hilbert, D.W. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004, 7, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Qin, Y.; Han, Y.; Dong, C.; Li, P.; Shang, Q. Comparative proteomic analysis reveals intracellular targets for bacillomycin L to induce Rhizoctonia solani Kühn hyphal cell death. Biochim. Biophys. Acta 2016, 1864, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.A.; Nobre, B.; da Silva, T.L.; Pinheiro, H.; Reis, A. Dual-mode cultivation of Chlorella protothecoides applying inter-reactors gas transfer improves microalgae biodiesel production. J. Biotechnol. 2014, 184, 74–83. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rahman, A.; Uddin, W.; Wenner, N.G. Induced systemic resistance responses in perennial ryegrass against M agnaporthe oryzae elicited by semi-purified surfactin lipopeptides and live cells of B acillus amyloliquefaciens. Mol. Plant. Pathol. 2015, 16, 546–558. [Google Scholar] [CrossRef]
- Yuan, J.; Ruan, Y.; Wang, B.; Zhang, J.; Waseem, R.; Huang, Q.; Shen, Q. Plant growth-promoting rhizobacteria strain Bacillus amyloliquefaciens NJN-6-enriched bio-organic fertilizer suppressed Fusarium wilt and promoted the growth of banana plants. J. Agric. Food Chem. 2013, 61, 3774–3780. [Google Scholar] [CrossRef] [PubMed]
- Gowtham, H.; Murali, M.; Singh, S.B.; Lakshmeesha, T.; Murthy, K.N.; Amruthesh, K.; Niranjana, S. Plant growth promoting rhizobacteria-Bacillus amyloliquefaciens improves plant growth and induces resistance in chilli against anthracnose disease. Biol. Control. 2018, 126, 209–217. [Google Scholar] [CrossRef]
- Machado, D.L.M.; Lucena, C.C.d.; Santos, D.d.; Siqueira, D.L.d.; Matarazzo, P.H.M.; Struiving, T.B. Slow-release and organic fertilizers on early growth of Rangpur lime. Rev. Ceres 2011, 58, 359–365. [Google Scholar] [CrossRef]
- Cheong, J.C.; Lee, J.T.; Lim, J.W.; Song, S.; Tan, J.K.; Chiam, Z.Y.; Yap, K.Y.; Lim, E.Y.; Zhang, J.; Tan, H.T. Closing the food waste loop: Food waste anaerobic digestate as fertilizer for the cultivation of the leafy vegetable, xiao bai cai (Brassica rapa). Sci. Total Environ. 2020, 715, 136789. [Google Scholar] [CrossRef]
- Sheirdil, R.A.; Hayat, R.; Zhang, X.-X.; Abbasi, N.A.; Ali, S.; Ahmed, M.; Khattak, J.Z.K.; Ahmad, S. Exploring potential soil bacteria for sustainable wheat (Triticum aestivum L.) production. Sustainability 2019, 11, 3361. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, W.; Li, H. The role of GA, IAA and BAP in the regulation of in vitro shoot growth and microtuberization in potato. Acta Physiol. Plant. 2005, 27, 363–369. [Google Scholar] [CrossRef]
- Berrios, L.; Ely, B. Plant growth enhancement is not a conserved feature in the Caulobacter genus. Plant. Soil 2020, 449, 81–95. [Google Scholar] [CrossRef]
- Dey, R.; Pal, K.; Bhatt, D.; Chauhan, S. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 2004, 159, 371–394. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Kumar, U.; Mishra, P.K.; Prakash, V. Efficiency of plant growth promoting rhizobacteria for the enhancement of Cicer arietinum L. growth and germination under salinity. Adv. Biol Res. 2010, 4, 92–96. [Google Scholar]
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of Plant Growth by ACC Deaminase-Producing Soil Bacteria. In New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria, Research; Bakker, P.A.H.M., Raaijmakers, J.M., Bloemberg, G., Höfte, M., Lemanceau, P., Cook, B.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 329–339. [Google Scholar]
- Pandey, S.; Gupta, S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front. Microbiol. 2019, 10, 1506. [Google Scholar]
- Vansuyt, G.; Robin, A.; Briat, J.-F.; Curie, C.; Lemanceau, P. Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol. Plant. Microbe Interact. 2007, 20, 441–447. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Babar, M.A. Metabolic and physiological changes induced by plant growth regulators and plant growth promoting rhizobacteria and their impact on drought tolerance in Cicer arietinum L. PLoS ONE 2019, 14, e0213040. [Google Scholar] [CrossRef] [PubMed]



| Strain No. | Indole-3-Acetic Acid (IAA) | Phosphate Solubilization | Nitrogen Fixation | 1-Aminocyclopropane- 1-Carboxylic Acid Deaminase | Siderophore |
|---|---|---|---|---|---|
| 1 | − | − | − | − | − |
| 2 | + | − | − | + | − |
| 3 | − | − | − | − | + |
| 4 | + | − | − | + | − |
| 5 | - | − | − | + | − |
| 6 | + | − | + | + | − |
| 7 | − | + | − | − | − |
| 8 | − | + | − | − | − |
| 9 | − | − | - | + | + |
| 10 | − | + | − | − | − |
| 11 | + | + | − | + | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asghar, W.; Kondo, S.; Iguchi, R.; Mahmood, A.; Kataoka, R. Agricultural Utilization of Unused Resources: Liquid Food Waste Material as a New Source of Plant Growth-Promoting Microbes. Agronomy 2020, 10, 954. https://doi.org/10.3390/agronomy10070954
Asghar W, Kondo S, Iguchi R, Mahmood A, Kataoka R. Agricultural Utilization of Unused Resources: Liquid Food Waste Material as a New Source of Plant Growth-Promoting Microbes. Agronomy. 2020; 10(7):954. https://doi.org/10.3390/agronomy10070954
Chicago/Turabian StyleAsghar, Waleed, Shiho Kondo, Riho Iguchi, Ahmad Mahmood, and Ryota Kataoka. 2020. "Agricultural Utilization of Unused Resources: Liquid Food Waste Material as a New Source of Plant Growth-Promoting Microbes" Agronomy 10, no. 7: 954. https://doi.org/10.3390/agronomy10070954
APA StyleAsghar, W., Kondo, S., Iguchi, R., Mahmood, A., & Kataoka, R. (2020). Agricultural Utilization of Unused Resources: Liquid Food Waste Material as a New Source of Plant Growth-Promoting Microbes. Agronomy, 10(7), 954. https://doi.org/10.3390/agronomy10070954






