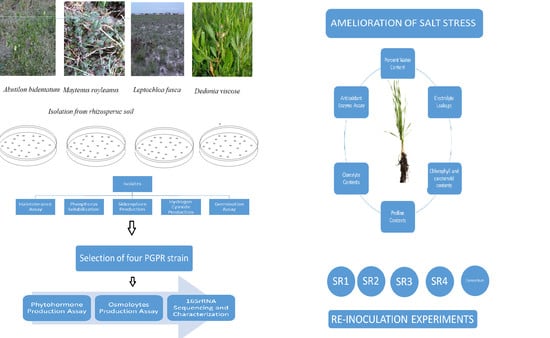
Rhizobacteria Isolated from Saline Soil Induce Systemic Tolerance in Wheat (Triticum aestivum L.) against Salinity Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling and Physicochemical Analysis
2.2. Strain Isolation and HaloTolerance Assay
2.3. Plant Growth-Promoting (PGP) Traits
2.4. Germination Experiment
2.5. Production of Osmolytes
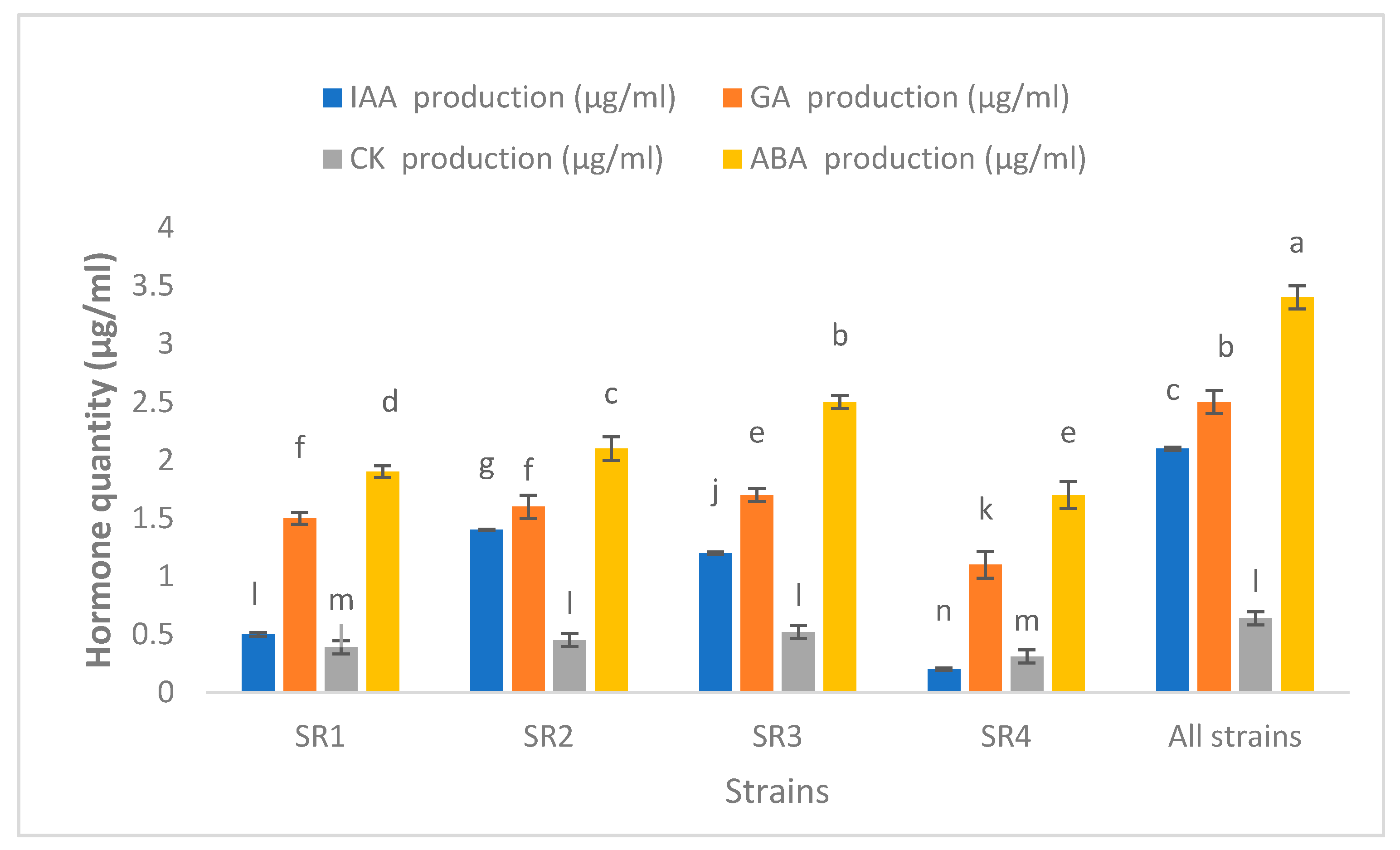
2.6. Phytohormone Production
2.7. 16S rRNA Gene Sequence and Phylogenetic Analysis
2.8. Plant Inoculation
2.9. Electrolyte Leakage (%)
2.10. Chlorophyll and Carotenoid Content
2.11. Proline Content
2.12. Total Soluble Sugar and Amino Acid
2.13. Total Protein Content
2.14. Antioxidant Enzyme Assay
2.15. Statistical Analysis
3. Results
3.1. Soil Analysis
3.2. Isolation and Screening of Salt-Tolerant PGPR Strains
3.3. Effect of Bacterial Isolates on Germination of Wheat
3.4. Identification of Isolates
3.5. Production of Phytohormones
3.6. Production of Compatible Solutes
3.7. Effect of PGPR Inoculation on the Biomass of Wheat (Triticum aestivum L.) Plants Grown under Salinity Stress
3.8. Effect on the Membrane Stability Index and Water Content
3.9. Chlorophyll Contents
3.10. Proline Contents
3.11. Amino Acid Content
3.12. Total Soluble Sugar
3.13. Antioxidants Enzyme Assay
3.14. Heatmap Responses of Pearson’s Correlation Coefficient (r)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Ethical Statement
References
- GAP Report. Global Agricultural Productivity Report (GAP Report) Global Harvest Initiative, Washington. 2018. Available online: https://globalagriculturalproductivity.org/wp-content/uploads/2019/01/GHI_2018-GAP-Report_FINAL-10.03.pdf (accessed on 5 May 2020).
- Abbas, R.; Rasul, S.; Aslam, K.; Baber, M.; Shahid, M.; Mobeen, F.; Naqqash, T. Halo-tolerant PGPR: A hope for cultivation of saline soils. J. King Saud Univ. Sci. 2019, 31, 1195–1201. [Google Scholar] [CrossRef]
- Mishra, J.; Fatima, T.; Arora, N.K. Role of secondary metabolites from plant growth-promoting rhizobacteria in combating salinity stress. In Plant Microbiome: Stress Response; Ahmad, P., Egamberdieva, D., Eds.; Springer: Singapore, 2018; pp. 127–163. [Google Scholar]
- Yasin, N.A.; Khan, W.; Ahmad, S.R.; Ali, A.; Ahmad, A.; Akram, W. Imperative roles of halo-tolerant plant growth-promoting rhizobacteria and kinetin in improving salt tolerance and growth of black gram (Phaseolusmungo). Environ. Sci. Pollut. Res. 2017, 25, 4491–4505. [Google Scholar] [CrossRef]
- Abdel-Latef, A.A.H.; Chaoxing, H. Does Inoculation with Glomus mosseae Improve Salt Tolerance in Pepper Plants? J. Plant Growth Regul. 2014, 33, 644–653. [Google Scholar] [CrossRef]
- Liu, K.; McInroy, J.A.; Hu, C.-H.; Kloepper, J.W. Mixtures of Plant-Growth-Promoting Rhizobacteria Enhance Biological Control of Multiple Plant Diseases and Plant-Growth Promotion in the Presence of Pathogens. Plant Dis. 2018, 102, 67–72. [Google Scholar] [CrossRef]
- Khan, A.; Zhao, X.Q.; Javed, M.T.; Khan, K.S.; Bano, A.; Shen, R.F. Bacillus pumilus enhances tolerance in rice (Oryza sativa L.) to combined stresses of NaCl and high boron due to limited uptake of Na+. Environ. Exp. Bot. 2016, 124, 120–129. [Google Scholar] [CrossRef]
- Yasmin, H.; Nosheen, A.; Naz, R.; Keyani, R.; Anjum, S. Regulatory role of rhizobacteria to induce drought and salt stress tolerance in plants. In Field Crops: Sustainable Management by PGPR. Sustainable Development and Biodiversity; Maheshwari, D., Dheeman, S., Eds.; Springer: Cham, Switzerland, 2019; Volume 23, p. 23. [Google Scholar]
- Niu, X.; Song, L.; Xiao, Y.; Ge, W. Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 2018, 8, 2580. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.N.; Tran, B.T.H.; Van-Bui, L.; Hoang, M.T.T. Plant growth-promoting rhizobacterium Pseudomonas PS01 induces salt tolerance in Arabidopsis thaliana. BMC Res. Notes 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Radojevic, M.; Bashkin, V.N. Practical Environmental Analysis; The Royal Society of Chemistry: Cambridge, UK, 1999; p. 466. [Google Scholar]
- Soltanpour, P.N.; Schwab, A.P. A new soil test for simultaneous extraction of macro- and micro-nutrients in alkaline soils. Commun. Soil Sci. Plant Anal. 1977, 8, 195–207. [Google Scholar] [CrossRef]
- Kingston, H.M. Microwave Assisted Acid Digestion of Sediments, Sludges, Soils and Oils; Duquesne University: Pittsburgh, PA, USA, 1994; p. 4. [Google Scholar]
- Somasegaran, P.; Hoben, H.J. Counting rhizobia by a plant infection method. In Handbook for Rhizobia; Springer: New York, NY, USA, 1994; pp. 58–64. [Google Scholar]
- James, G.C. Native Sherman Rockland Community College, State University of New York; The Benjamin/Coming Publishing Company Inc.: San Francisco, CA, USA, 1987; pp. 75–80. [Google Scholar]
- Robinson, R.J.; Fraaije, B.A.; Clark, I.M.; Jackson, R.W.; Hirsch, P.R.; Mauchline, T.H. Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil 2016, 405, 381–396. [Google Scholar] [CrossRef]
- Pikovskaya, R. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 1948, 17, 362–370. [Google Scholar]
- Murphy, J.A.M.E.S.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Lorck, H. Production of hydrocyanic acid by bacteria. Physiol. Plant. 1948, 1, 142–146. [Google Scholar] [CrossRef]
- Noreen, Z.; Ashraf, M.; Hassan, M.U. Inter-accessional variation for salt tolerance in pea (Pisum sativum L.) at germination and screening stage. Pak. J. Bot. 2007, 39, 2075–2085. [Google Scholar]
- Upadhyay, S.K.; Singh, J.S.; Singh, D.P. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Tien, T.M.; Gaskins, M.H.; Hubbell, D.H. Plant growth substances produced by Azospirillum brasilense and their effect on the growth of pearl millet (Pennisetum americanum L.). Appl. Environ. Microbiol. 1979, 37, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.P.; Kuo, T.T. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993, 21, 2260. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Eck, R.V.; Dayhoff, M.O. Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Silverspring, MD, USA, 1966; p. 12. [Google Scholar]
- Unyayer, S.; Keles, Y.; Cekic, F.O. The antioxidative response of two tomato species with different tolerances as a result of drought and cadmium stress combination. Plant Soil Environ. 2005, 51, 57–64. [Google Scholar] [CrossRef]
- Sriram, S.; Raguchander, T.; Babu, S.; Nandakumar, R.; Shanmugam, V.; Vidhyasekaran, P. Inactivation of phytotoxin produced by the rice sheath blight pathogen Rhizoctonia solani. Can. J. Microbiol. 2000, 46, 520–524. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.B.; VanSlyke, D.D. Amino acid determination with ninhydrin. J. Biol. Chem. 1943, 150, 231–233. [Google Scholar]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram, quantitation of proteins utilizing the principle of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in Vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [CrossRef]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P. Ultraviolet-B- and ozone induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 1996, 110, 125–136. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutase in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Steel, R.G.D.; Torrie, G.H. Principles and Procedures of Statistics Singapore, 2nd ed.; McGraw Hill Book Co Inc.: New York, NY, USA, 1980. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Boil. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Ramadoss, D.; Lakkineni, V.K.; Bose, P.; Ali, S.; Annapurna, K. Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. SpringerPlus 2013, 2, 6. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Ghosh, P.K.; Pramanik, K.; Mitra, S.; Soren, T.; Pandey, S. A halo-tolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Microbiol. Res. 2018, 169, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Saraf, M. Influence of soil ameliorants and microflora on induction of antioxidant enzymes and growth promotion of (Jatropha curcas L.) under saline condition. Eur. J. Soil Biol. 2013, 55, 47–54. [Google Scholar] [CrossRef]
- Abd-Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.A. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 2017, 3, 37–44. [Google Scholar] [CrossRef]
- TrParray, A.P.; Jan, S.; Kamili, A.N.; Qadri, R.A.; Egamberdieva, D.; Ahmad, P. Current perspectives on plant growth promoting rhizobacteria. J. Plant Growth Regul. 2016, 35, 877–902. [Google Scholar] [CrossRef]
- Salomon, M.; Bottini, R.; de Souza Filho, G.A.; Cohen, A.C.; Moreno, D.; Gil, M. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 2014, 151, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Bottini, R.; Cassán, F.; Piccoli, P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 2004, 65, 497–503. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effect on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, J.P. Does plant-microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 2018, 207, 41–52. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Walker, V.; Couillerot, O.; Felten, A.V.; Bellvert, F.; Jansa, J.; Maurhofer, M.; Bally, R.; Moënne-Loccoz, Y.; Comte, G. Variation of secondary metabolite levels in maize seedling roots induced by inoculation with Azospirillum, Pseudomonas and Glomus consortium under field conditions. Plant Soil 2012, 356, 151–163. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Zahir, Z.A.; Naveed, M.; Arshad, M. Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can. J. Microbiol. 2007, 53, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Rakshapal, S.; Sumit, K.S.; Rajendra, P.P.; Alok, K. Technology for improving essential oil yield of Ocimum basilicum L. (sweet basil) by application of bioinoculant colonized seeds under organic field conditions. Ind. Crop. Prod. 2013, 45, 335–342. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Arif, M.S.; Ali, S.; Ali, B.; Hameed, S. Plant growth promoting bacteria confer salt tolerance in Vigna radiata by up-regulating antioxidant defense and biological soil fertility. Plant Growth Regul. 2016, 80, 23–36. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Alamri, S.A.; Ali, H.M.; Alayafi, A.A. Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression. Plant Physiol. Biochem. 2018, 132, 375–384. [Google Scholar] [CrossRef]
- Garg, N.; Manchanda, G. Role of arbuscular mycorrhizae in the alleviation of ionic, osmotic and oxidative stresses induced by salinity in Cajanus cajan (L.) Millsp. (pigeonpea). J. Agron. Crop Sci. 2009, 195, 110–123. [Google Scholar] [CrossRef]
- Bojórquez-Quintal, E.; Velarde-Buendía, A.; Ku-González, Á.; Carillo-Pech, M.; Ortega-Camacho, D.; Echevarría-Machado, I.; Pottosin, I.; Martínez-Estévez, M. Mechanisms of salt tolerance in habanero pepper plants (Capsicum chinense Jacq.): Proline accumulation, ions dynamics and sodium root-shoot partition and compartmentation. Front. Plant Sci. 2014, 5, 605. [Google Scholar] [CrossRef]
- Kaya, C.; Tuna, A.L.; Okant, A.M. Effect of foliar applied kinetin and indole acetic acid on maize plants grown under saline conditions. Turk. J. Agric. For. 2010, 34, 529–538. [Google Scholar]
- Habib, S.H.; Kausar, H.; Saud, H.M. Plant Growth-Promoting Rhizobacteria Enhance Salinity Stress Tolerance in Okra through ROS-Scavenging Enzymes. BioMed Res. Int. 2016, 2016, 6284547. [Google Scholar] [CrossRef]
- Bremer, E.; Kramer, R. Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 2019, 73, 313–334. [Google Scholar] [CrossRef]
- Nautiyal, C.S.; Srivastava, S.; Chauhan, P.S.; Seem, K.; Mishra, A.; Sopory, S.K. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Biochem. 2013, 66, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dodd, I.C.; Belimov, A.A.; Jiang, F. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct. Plant Biol. 2016, 43, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Prado, F.E.; Boero, C.; Gallardo, M.; González, J.A. Effect of NaCl on germination, growth, and soluble sugar content in Chenopodium quinoa wild seeds. Bot. Bull. Acad. Sin. 2000, 41, 27–343. [Google Scholar]
- Upadhyay, S.K.; Singh, D.P. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. (Stuttg.) 2015, 17, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.; Farhadi, N.; Panjtandoust, M.; Ghanati, F. Seed germination, antioxidant enzymes activity and proline content in medicinal plant Tagetes minuta under salinity stress. Plant Biosyst. 2019. [Google Scholar] [CrossRef]
- Wang, C.J.; Yang, W.; Wang, C.; Gu, C.; Niu, D.D.; Liu, H.X.; Wang, Y.P.; Guo, J. Induction of drought tolerance in cucumber plants by a consortium of three plant growth-promoting rhizobacterium strains. PLoS ONE 2012, 7, e52565. [Google Scholar] [CrossRef]
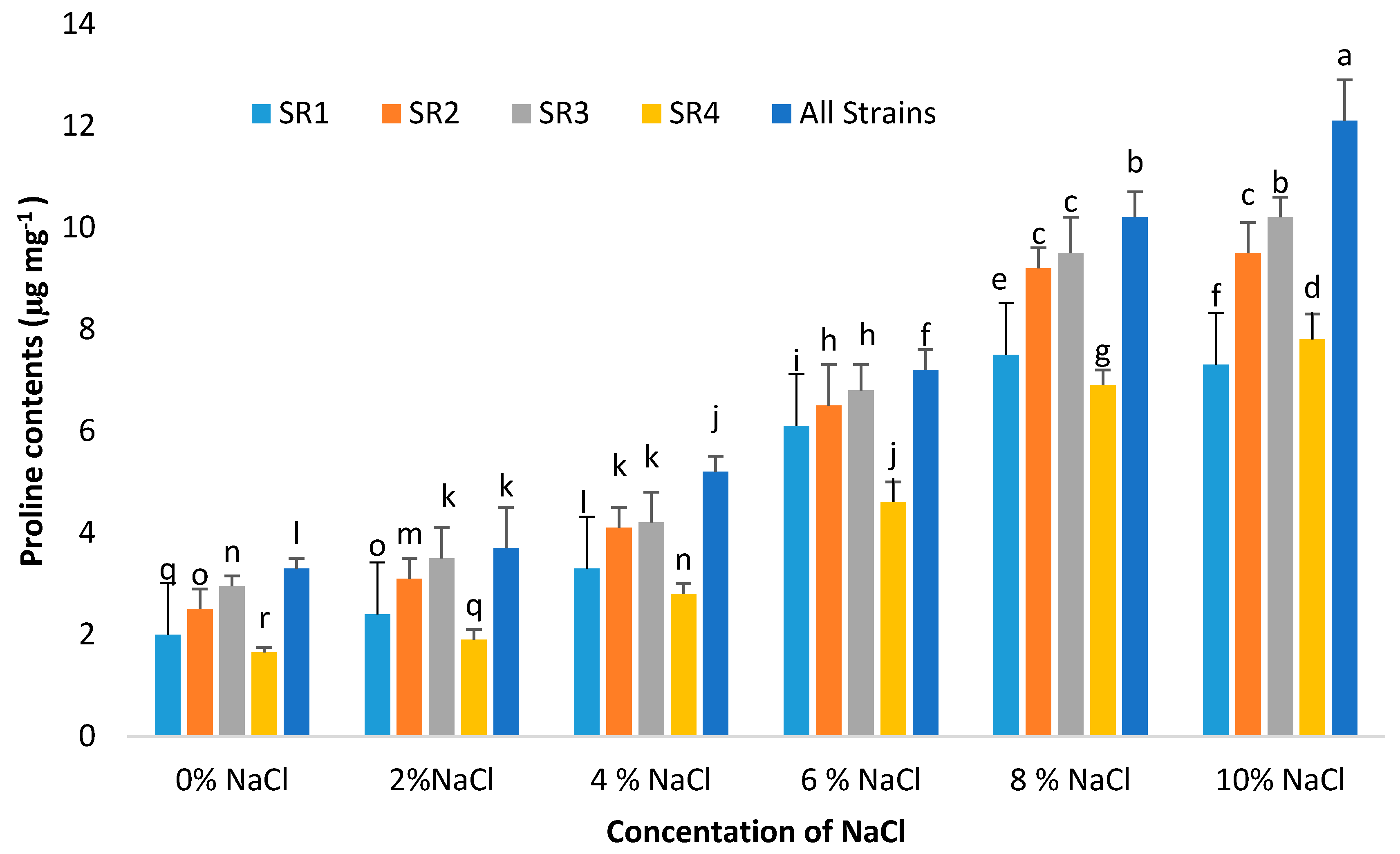
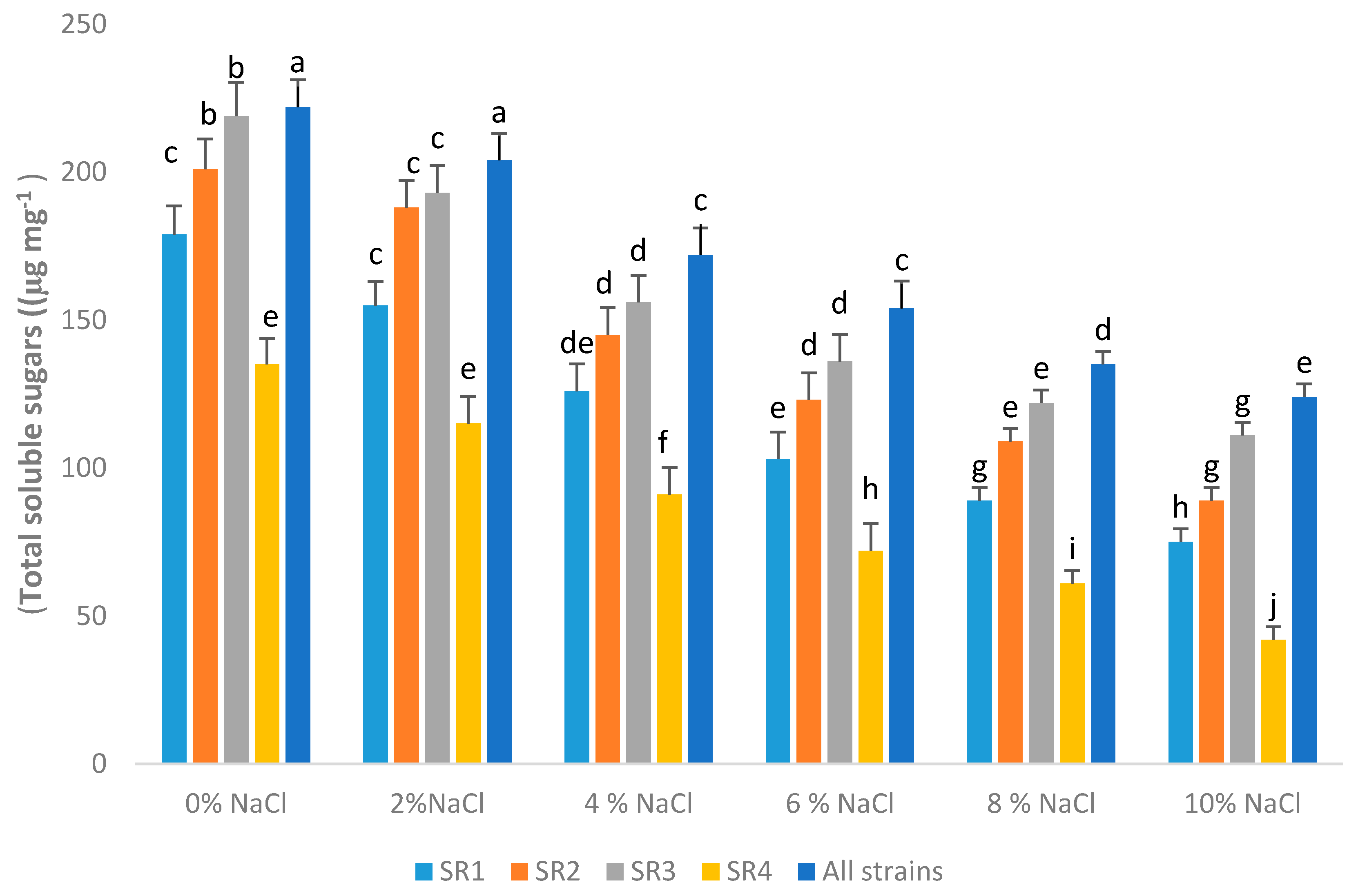
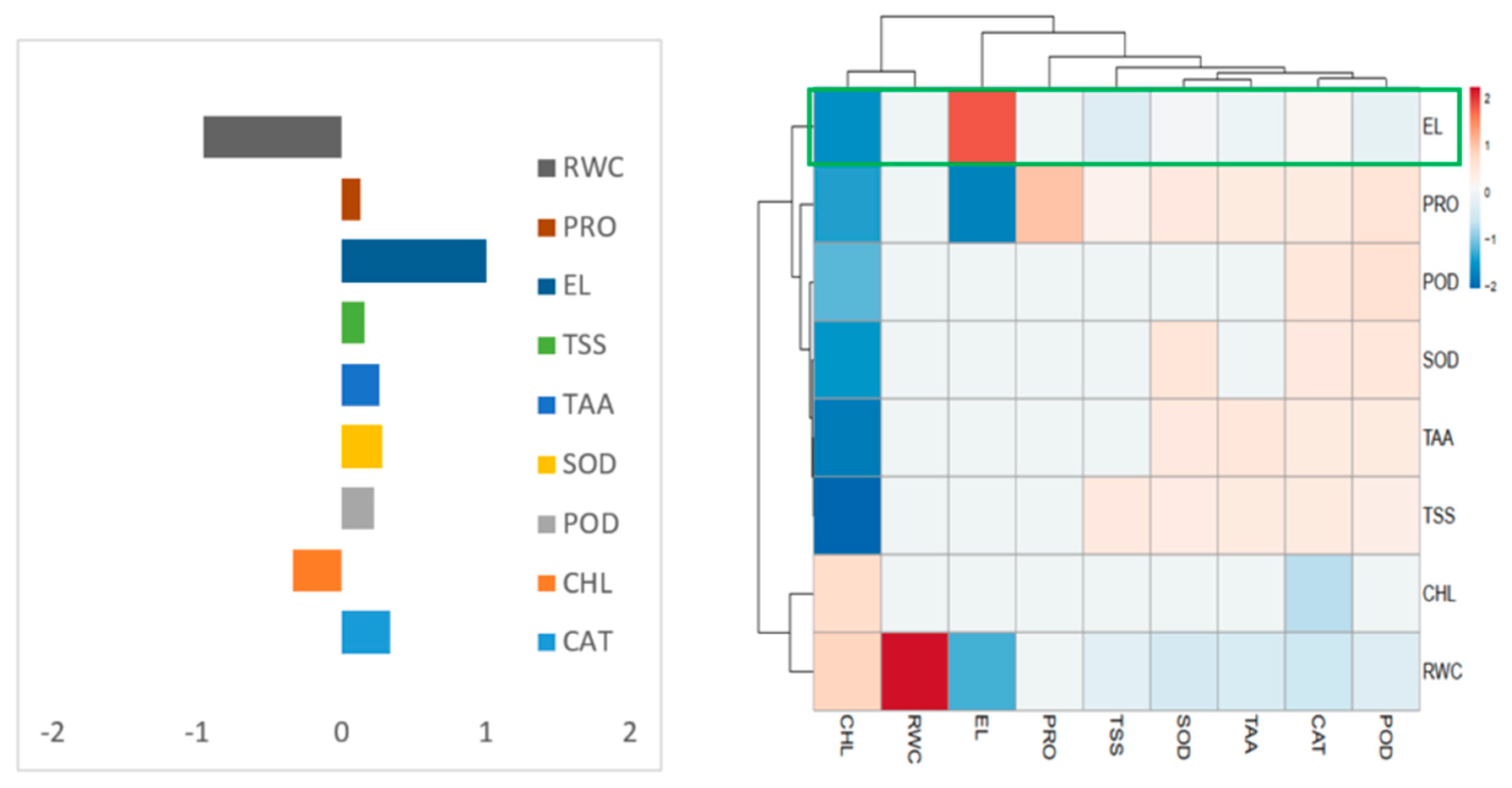
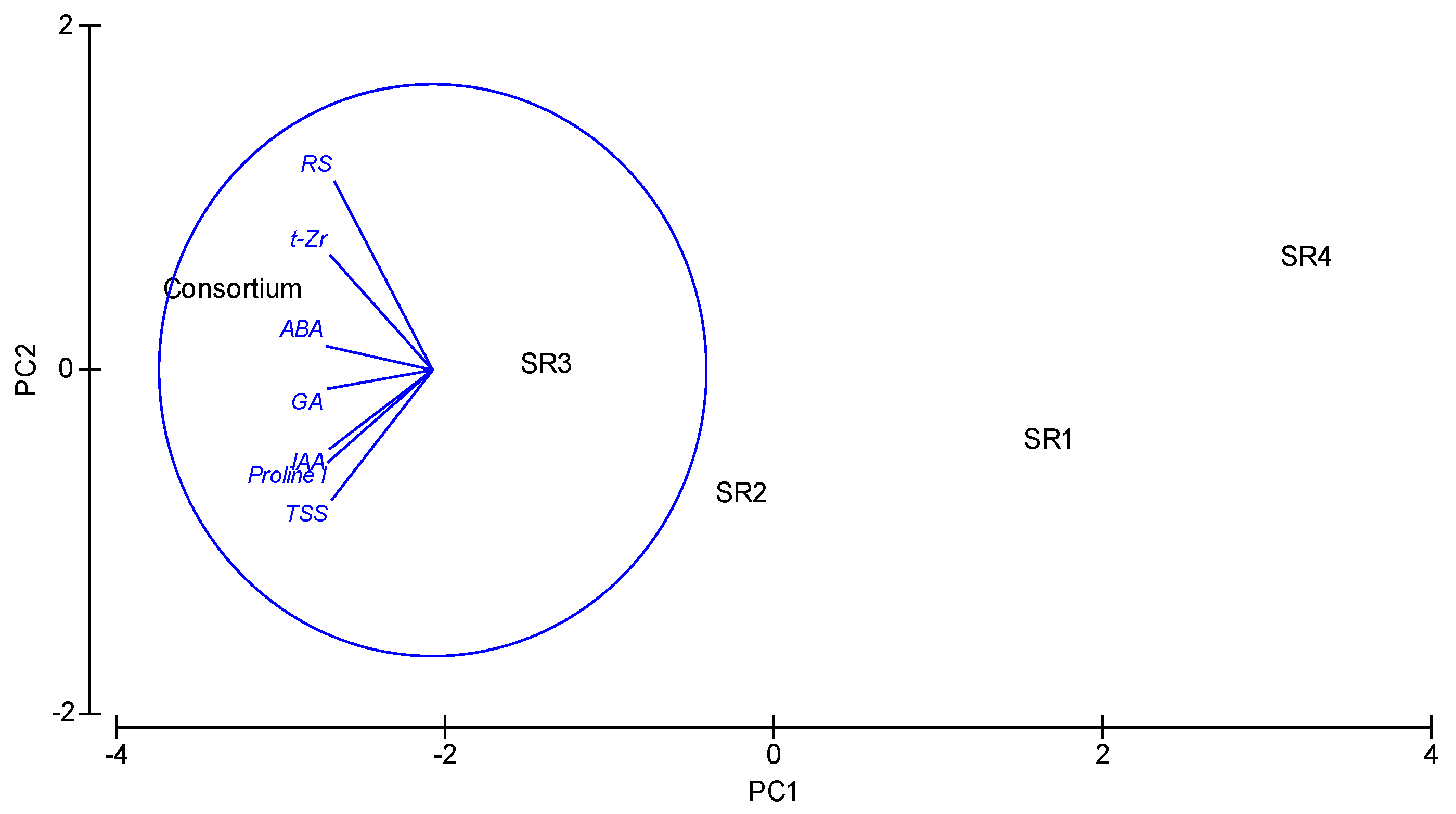
| Host Plant Species | pH | EC (dSm−1) | Soil Texture | SAR (mmol/L) | OC (%) | Macronutrient (meq/L) | Available Nutrients (kg/ha) | PGPR Population (cfu × 105 g−1 of Soil) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CO3 | HCO3 | Cl | Ca+ | Na+ | K+ | N | P | K | |||||||
| Abutilom bidiantum | 7.99 | 0.82 | Sandy clay loam | 39.6 | 0.72 | 3.9 | 15 | 50 | 3 | 82.95 | 0.3 | 240 | 200 | 320 | 69 |
| Maytenus royleanus | 8.12 | 0.85 | Sandy clay loam | 40.3 | 0.54 | 4 | 14 | 48 | 2.5 | 80.6 | 0.4 | 238 | 196 | 325 | 64 |
| Kallar grass | 7.80 | 0.76 | Sandy clay loam | 42.5 | 0.62 | 4.2 | 17 | 47 | 2.8 | 78.9 | 0.5 | 243 | 190 | 315 | 62 |
| Dedonia viscoca | 8.01 | 0.79 | Sandy clay loam | 38.1 | 0.64 | 4.5 | 17.5 | 48 | 3.1 | 81.2 | 0.4 | 237 | 205 | 330 | 65 |
| No | Isolates | Base Pair Length | Similarity (%) | Strain Identification | Accession No. |
|---|---|---|---|---|---|
| 1 | SR1 | 1485 | 98% | Bacillus sp. | KF719179 |
| 2 | SR2 | 1480 | 99% | Azospirillum brasilense | KJ194586 |
| 3 | SR3 | 1482 | 96% | Azospirillum lipoferum | KJ434039 |
| 4 | SR4 | 1263 | 99% | Pseudomonas stutzeri | KJ685889 |
| Treatments | Fresh Biomass (g) | Dry Biomass (g) | Leaf Area (cm2) | |||
|---|---|---|---|---|---|---|
| 0 mM | 150 mM | 0 mM | 150 mM | 0 mM | 150 mM | |
| Control | 10 ± 1g | 7.3 ± 0.4i | 3.3 ± 0.1c | 2.2 ± 0.04d | 140 ± 12e | 120 ± 17f |
| SR1 | 11 ± 0.5f | 8.2 ± 0.9h | 3.5 ± 0.4c | 2.8 ± 0.06d | 150 ± 15d | 130 ± 14f |
| SR2 | 13.2 ± 0.9e | 10.3 ± 0.7g | 4.1 ± 0.1b | 3.5 ± 0.09c | 167 ± 12c | 140 ± 24e |
| SR3 | 16.9 ± 1.2 b | 13.9 ± 1.4d | 4.7 ± 0.5b | 3.8 ± 0.03 c | 177 ± 17b | 147 ± 17e |
| SR4 | 11.50.98f | 7.8 ± 0.54i | 3.6 ± 0.5c | 3.0 ± 0.08d | 160 ± 14c | 135 ± 12f |
| Consortium | 20.3 ± 1.8a | 14.1 ± 1.9c | 5.9 ± 0.2a | 4.3 ± 0.03b | 186 ± 19a | 152 ± 13d |
| Treatments | Percent Water Content | Electrolyte Leakage (%) | ||
|---|---|---|---|---|
| 0 mM | 150 mM | 0 mM | 150 mM | |
| Control | 85 ± 1.5b | 57 ± 0.9e | 33 ± 0.4d | 55 ± 0.5a |
| SR1 | 86.3 ± 1.6b | 60 ± 1d | 30 ± 0.3d | 50 ± 0.45a |
| SR2 | 89 ± 1.9b | 63 ± 1.4d | 26.2 ± 0.25e | 41.3 ± 0.33c |
| SR3 | 95 ± 2.1a | 67 ± 1.7d | 25.7 ± 0.4e | 42.08 ± 0.11c |
| SR4 | 88.7 ± 1.9b | 61 ± 1.15d | 31.2 ± 0.22e | 47.8 ± 0.44b |
| Consortium | 97 ± 2.0a | 70 ± 1.75c | 22.1 ± 0.22f | 35.2 ± 0.23d |
| Chlorophyll a (mg/g Fresh Weight) | Chlorophyll b (mg/g Fresh Weight) | Total Chlorophyll (mg/g Fresh Weight) | Carotenoid (mg/g Fresh Weight) | |||||
|---|---|---|---|---|---|---|---|---|
| Treatments | 0 mM | 150 mM | 0 mM | 150 mM | 0 mM | 150 mM | 0 mM | 150 mM |
| Control | 1.06 ± 0.01d | 0.59 ± 0.01h | 0.27 ± 0.02d | 0.12 ± 0.01h | 1.18 ± 0.10e | 0.86 ± 0.05f | 46.9 ± 0.1f | 65.8 ± 0.15k |
| SR1 | 1.13 ± 0.03b | 0.75 ± 0.03g | 0.29 ± 0.04b | 0.13 ± 0.02g | 1.26 ± 0.09d | 1.01 ± 0.03k | 47.3 ± 0.3e | 67.6 ± 0.5i |
| SR2 | 1.18 ± 0.04c | 0.81 ± 0.02f | 0.32 ± 0.03c | 0.15 ± 0.02f | 1.33 ± 0.7c | 1.13 ± 0.02j | 50.5 ± 0.4c | 69.8 ± 0.4h |
| SR3 | 1.2 ± 0.05c | 0.85 ± 0.04g | 0.33 ± 0.05c | 0.17 ± 0.03f | 1.37 ± 0.8b | 1.18 ± 0.04k | 51.1 ± 0.2b | 69.2 ± 0.5i |
| SR4 | 1.12 ± 0.02b | 0.77 ± 0.03b | 0.28 ± 0.01b | 0.13 ± 0.02g | 1.13 ± 0.6f | 1.05 ± 0.01d | 48.2 ± 0.4d | 68.1 ± 0.6j |
| Consortium | 1.4 ± 0.04a | 0.9 ± 0.02e | 0.35 ± 0.05a | 0.19 ± 0.04e | 1.59 ± 0.5a | 1.25 ± 0.03h | 52.8 ± 0.6a | 70.4 ± 0.8g |
| Treatments | Total Soluble Sugar (µg g−1 FW) | Total Amino Acid (µg g−1 FW) | Proline (µg g−1 FW) | |||
|---|---|---|---|---|---|---|
| 0 mM | 150 mM | 0 mM | 150 mM | 0 mM | 150 mM | |
| Control | 27 ± 2d | 33 ± 5i | 330 ± 10g | 368 ± 20e | 40 ± 03d | 120 ± 5j |
| SR1 | 29 ± 3d | 35 ± 7h | 345 ± 12 f | 379 ± 17e | 44 ± 05d | 128 ± 6i |
| SR2 | 31 ± 5c | 39 ± 6g | 360 ± 15e | 401 ± 27c | 51 ± 3c | 130 ± 7g |
| SR3 | 33 ± 3c | 33 ± 1.0f | 370 ± 24e | 420 ± 25b | 54 ± 3b | 135 ± 9h |
| SR4 | 29 ± 4 d | 36 ± 8h | 350 ± 12f | 387 ± 24.4d | 43 + 3 c | 125 ± 5i |
| Consortium | 39 ± 5 c | 43 ± 5e | 381 ± 10d | 439 ± 15a | 57 ± 4b | 145 ± 7f |
| Treatments | Superoxide Dismutase (EU mg−1 Protein) | Catalase (EU mg−1 Protein) | Peroxidase (EU mg−1 Protein) | |||
|---|---|---|---|---|---|---|
| 0 mM | 150 mM | 0 mM | 150 mM | 0 mM | 150 mM | |
| Control | 0.74 ± 0.06k | 1.83 ± 0.02f | 2.5 ± 0.03h | 4.13 ± 0.02f | 144 ± 3f | 255 ± 5f |
| SR1 | 0.76 ± 0.04j | 1.85 ± 0.01e | 2.7 ± 0.02k | 4.3 ± 0.04e | 148 ± 7j | 260 ± 4d |
| SR2 | 0.8 ± 0.03i | 1.9 ± 0.04c | 3.01 ± 0.04i | 4.7 ± 0.09c | 153 ± 4i | 263 ± 6c |
| SR3 | 0.82 ± 0.05h | 1.91 ± 0.03b | 3.12 ± 0.05h | 4.8 ± 0.10b | 155 ± 6h | 267 ± 7b |
| SR4 | 0.78 ± 0.3j | 1.85 ± 0.4d | 2.6 ± 0.04j | 4.5 ± 0.05d | 150 ± 4j | 257 ± 5d |
| Consortium | 0.86 ± 0.07g | 1.96 ± 0.05a | 3.25 ± 0.05g | 5.05 ± 0.04a | 162 ± 3g | 270 ± 6a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilyas, N.; Mazhar, R.; Yasmin, H.; Khan, W.; Iqbal, S.; Enshasy, H.E.; Dailin, D.J. Rhizobacteria Isolated from Saline Soil Induce Systemic Tolerance in Wheat (Triticum aestivum L.) against Salinity Stress. Agronomy 2020, 10, 989. https://doi.org/10.3390/agronomy10070989
Ilyas N, Mazhar R, Yasmin H, Khan W, Iqbal S, Enshasy HE, Dailin DJ. Rhizobacteria Isolated from Saline Soil Induce Systemic Tolerance in Wheat (Triticum aestivum L.) against Salinity Stress. Agronomy. 2020; 10(7):989. https://doi.org/10.3390/agronomy10070989
Chicago/Turabian StyleIlyas, Noshin, Roomina Mazhar, Humaira Yasmin, Wajiha Khan, Sumera Iqbal, Hesham El Enshasy, and Daniel Joe Dailin. 2020. "Rhizobacteria Isolated from Saline Soil Induce Systemic Tolerance in Wheat (Triticum aestivum L.) against Salinity Stress" Agronomy 10, no. 7: 989. https://doi.org/10.3390/agronomy10070989
APA StyleIlyas, N., Mazhar, R., Yasmin, H., Khan, W., Iqbal, S., Enshasy, H. E., & Dailin, D. J. (2020). Rhizobacteria Isolated from Saline Soil Induce Systemic Tolerance in Wheat (Triticum aestivum L.) against Salinity Stress. Agronomy, 10(7), 989. https://doi.org/10.3390/agronomy10070989