Abstract
The Malva genus contains species that reveal therapeutic properties and are mostly important in medicine and the functional food industry. Its breeding, cultivation, and utilization are based on proper germplasm/plant identification, which is difficult using morphological features. For this reason, we applied flow cytometry and inter simple sequence repeat polymerase chain reaction (ISSR-PCR) for fast and accurate species identification. Genome size estimation by flow cytometry was proposed as the first-choice method for quick accession screening. Out of the 12 tested accessions, it was possible to identify six genotypes based on genome size estimation, whereas all species and varieties were identified using ISSR markers. Flow cytometric analyses revealed that Malva species possessed very small (1.45–2.77 pg/2C), small (2.81–3.80 pg/2C), and intermediate (11.06 pg/2C) genomes, but the majority of accessions possessed very small genomes. Additionally, this is the first report on genome size assessment for eight of the accessions. The relationships between the investigated accessions showed the presence of two clusters representing malvoid and lavateroid group of species. Flow cytometry and ISSR molecular markers can be effectively used in the identification and genetic characterization of Malva species.
1. Introduction
Malva L. (mallow) is the genus within the Malvaceae Juss. family, which includes 25–40 species and several hybrids [1,2,3]. This genus contains herbaceous annual, biennial, and perennial species that are native to regions of Africa, Asia, and Europe [4,5]. Malva species contain a lot of mucilage, malvin, flavonoids, terpenoids, polysaccharides, and vitamin A. Therefore, leaves, flowers, roots, and seeds as well as the whole herb are used in traditional phytotherapy, medicine, horticulture, and functional food industry [6,7,8,9,10]. Some species (e.g., M. neglecta Wallr. M. parviflora L. and M. sylvestris L.) are consumed as vegetables in Turkey, Morocco, Egypt, and Mexico as well [11,12,13,14,15]. In medicine, mallow species are used in the treatment of respiratory, urinary, and digestive problems as they have high bactericidal, antiulcerogenic, anti-inflammatory, hepatoprotective, and antidiabetic activities [16,17,18,19,20,21,22]. M. neglecta, M. parviflora, M. sylvestris, and M. verticillata L. are the most investigated species in the genus and at the same time the most used ones in the industry [9]. Moreover, some mallow species also have the capacity to accumulate heavy metals and are potentially used for phytoremediation [23,24,25,26,27,28].
The Malva genus is morphologically very diverse, but some species are hardly distinguishable based on morphological features [29]. Several studies have been conducted to clarify the taxonomic affiliation of Malva species using different features, such as molecular data (nuclear ribosomal DNA (rDNA), internal transcribed spacer (ITS) region, intron–exon splice junction (ISJ), and inter simple sequence repeat polymerase chain reaction (ISSR) markers) [3,30,31,32], differentiation of seed and seed coat structure [33,34], morphology of pollen grains [4,5], epidermal structures and stem hairs [6,35,36], and plant morphological traits [34,37,38].
The variability in mallow species is due, at least in part, to hybridization. Natural crossings between M. pusilla Sm. and M. neglecta, M. alcea L., and M. moschata L. as well as M. sylvestris and M. neglecta were found in Europe [1]. However, no evidence for hybridization among M. pusilla plants was observed in the Canadian collection [39]. Kristofferson [40] studied artificial Malva crosses and found that inter- and intra-section hybridization between species was also possible but extremely rare. Hybrids obtained in this way (e.g., M. pusilla × M. neglecta, M. parviflora × M. neglecta) had low fertility and produced unfilled seeds or fruits without seeds, indicating that the formation of viable hybrid forms is not a common phenomenon in Malva genus. Ray [3] stated that hybridization or polyploidy is probably a factor in the evolution of these species, but this aspect has not been investigated so far.
The taxonomy and systematics of the Malva genus are still unclear and very complicated. Taxonomic doubts have appeared because of the high level of homoplasty in morphological traits that are usually used as diagnostic features [29]. Ray [30] pinpointed that this genus is closely related to Lavatera L. and that some species included in Lavatera are more closely related to M. sylvestris than some other species included in the Malva genus. Based on the flower structure, Dalby [1] divided the Malva genus into two sections: Bismalva (with M. alcea, M. excisa Rchb., and M. moschata) and Malva (M. neglecta, M. pusilla, M. sylvestris, and M. verticillata). Additionally, within the Malva section, two subsections were distinguished: Planocentrae (with M. neglecta and M. pusilla) and Cenocentrae (with M. verticillata) [41]. A different classification based on ITS molecular markers as well as fruit morphology and seed structure was reported by Ray [3,30], and two groups were distinguished: malvoid and lavateroid. Additionally, species of the section Bismalva were placed in one group with the species of the genus Lavatera. The malvoid group included several Lavatera species and at least nine Malva species (M. sylvestris, M. nicaeensis All., M. verticillata, M. parviflora, M. pusilla, M. neglecta, M. aegyptia L., M. cretica Cav., and M. stipulacea Cav.). The second group consisted of about 10 Lavatera species and three Malva species (M. alcea, M. hispanica L., and M. tournefortiana L.). Only M. moschata was placed outside of the above groups. A similar division was proposed by Escobar Garcia et al. [29] based on five ITS molecular markers (matK plus trnK, ndhF, trnL-trnF, and psbA-trnH). Section Bismalva (M. alcea, M. tournefortiana, and M. moschata) were included in the lavateroid clade, while M. sylvestris, M. verticillata, M. parviflora, and M. neglecta were clustered in the malvoid clade. M. cretica ssp. althaeoides was placed outside of these two clades, together with annual Althaea, while M. aegyptia and M. trifida Cav. were placed in the M. aegyptia clade. These genetic relationships and the classification of Malva species were also confirmed by Celka et al. [31] and Lo Bianco et al. [42] based on ITS and ISSR molecular markers along with seed image analysis.
Most of the Malva species are polyploids with the base chromosome number n = x = 7 [3,43]. Numerous species are hexaploids, where the chromosome number is in the range of 40 to 44, and a few species possess higher numbers of chromosomes (e.g., M. alcea, 2n = 78 or 84; M. verticillate, 2n = 76, 84, 112, 120, 126; and M. crispa L., 2n = 84, 112, 120 [3,44]). Only one reported species (M. hispanica) possesses 2n = 24 chromosomes [3], and this species could be considered as tetraploid.
This study aimed to evaluate genetic diversity and relationships between selected Malva accessions through the use of genome size estimation and ISSR molecular markers. The genetic characteristics of the selected species/varieties are intended to support the correct identification of the species/varieties, which could be of particular importance for its taxonomy, breeding, and germplasm collection. In addition, this is the first report on 2C DNA content (genome size) for eight of the Malva accessions. Moreover, no such research on the combined nuclear DNA content and ISSR markers for Malva species characterization has been reported so far.
2. Materials and Methods
2.1. Plant Material
Twelve Malva accessions were used in this study (Table 1). The seeds of 11 species were obtained from GRIN-ARS-USDA (Germplasm Resources Information Network, Agricultural Research Service, United States Department of Agriculture) gene bank, while seeds of M. sylvestris var. mauritiana (L.) Boiss. were collected from the Medicinal and Cosmetic Plant Garden of the Nicolaus Copernicus University, Collegium Medicum, Bydgoszcz, Poland. The seeds were sown in pots with 1:2 sand and commercial humus mixture in a growth chamber for 16/8 h at 26/18 °C (day/night).

Table 1.
List of the investigated Malva accessions.
2.2. Genome Size Estimation
Leaves of Malva accessions were prepared for nuclear DNA content analysis as previously described by Jedrzejczyk and Rewers [45] with some modifications. Plant tissue (about 0.5–1.0 cm2) was chopped in 1 mL of nuclei isolation buffer (200 mM TRIS, 4 mM MgCl2•6H2O, 0.5% (v/v) Triton X-100) with the addition of propidium iodide (PI, 50 μg/mL) and ribonuclease A (RNase A, 50 μg/mL). The prepared suspension of nuclei was analyzed using a CyFlow SL Green (Partec GmbH, Münster, Germany) flow cytometer. The nuclear DNA content of 5000–7000 nuclei was measured for each sample on a linear scale. The fluorescence intensity histograms (CV = 5.28%–6.57%) were evaluated using FloMax software (Partec GmbH, Münster, Germany). Leaves of Zea mays L. CE-777 (5.43 pg/2C; [46]) and Glycine max (L.) Merr. “Polanka” (2.50 pg/2C; [47]) were used as internal standards (Table 2). For each Malva accession, six randomly selected plants were analyzed. Genome size was determined using the linear relationship between the mean ratio of the 2C peak positions of the Malva accessions and the internal standard on the histogram of fluorescence intensities. The obtained 2C DNA values (pg) were converted to megabase pairs (Mbp) of nucleotides using the following formula: 1 pg = 978 Mbp [48]. The results were analyzed using a one-way analysis of variance (ANOVA) and a Duncan’s post-hoc test (p < 0.05; Statistica v. 13.3, StatSoft, Poland).

Table 2.
Nuclear DNA content of the Malva accessions.
2.3. Genomic DNA Extraction
Total DNA was isolated from 0.12 g of fresh mallow leaves from three randomly selected plants per accession using a GeneJet Plant Genomic DNA Purification Mini Kit (Thermo Scientific, Waltham, MA, USA). After DNA extraction, the quality and quantity of DNA samples were assessed by spectrophotometry, and the quality was additionally assessed by 1% (w/v) agarose gel electrophoresis.
2.4. ISSR-PCR Amplification
Twenty-five ISSR primers (Genomed, Warsaw, Poland) were examined, out of which 13 generated stable and clear band patterns and were used for PCR amplifications (Table 3). The PCR reactions were carried out in a total volume of 12.5 μL containing 30 ng of genomic DNA template, 0.1 U/µL Taq DNA polymerase, 4 mM MgCl2, 0.5 mM of each dNTP, 10 µM ISSR primer, and sterile deionized water. Reactions were performed using T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA) and the following conditions: an initial denaturation at 94 °C for 5 min, then 1 min denaturation at 94 °C (40 cycles), followed by 1 min annealing at 48.5–67.2 °C (depending on the ISSR primer, Table 3), 2 min elongation at 72 °C, and 7 min of final extension at 72 °C. The PCR products were separated on 1.5% (w/v) of agarose gel electrophoresis. The size of the obtained bands was estimated by comparison to a 3000 bp DNA ladder (GenoPlast Biochemicals, Rokocin, Poland). The gels were visualized and archived via GelDoc XR+ (Bio-Rad, Hercules, CA, USA).

Table 3.
The inter simple sequence repeat polymerase chain (ISSR) primers applied in the molecular analyses of Malva accessions.
2.5. ISSR Marker Analysis
The ISSR amplicons were scored as presence (1) or absence (0) of bands in the form of a binary matrix. Only reproducible and clear bands were counted. Monomorphic and polymorphic bands were calculated for each ISSR primer. The polymorphic information content (PIC) was determined using the following equation: PIC = 1 − p2 − q2, where p is the band frequency and q is no-band frequency [50]. Genetic distances were calculated for all accessions according to Nei and Li [51]. Cluster analysis was performed with the UPGMA algorithm (unweighted pair group method with arithmetic average) and the Treecon ver. 3.1 software [52]. To estimate the statistical support of the dendrogram branches, the data were bootstrapped using 2000 replications, and values higher than 50% were mapped (Figure 1).
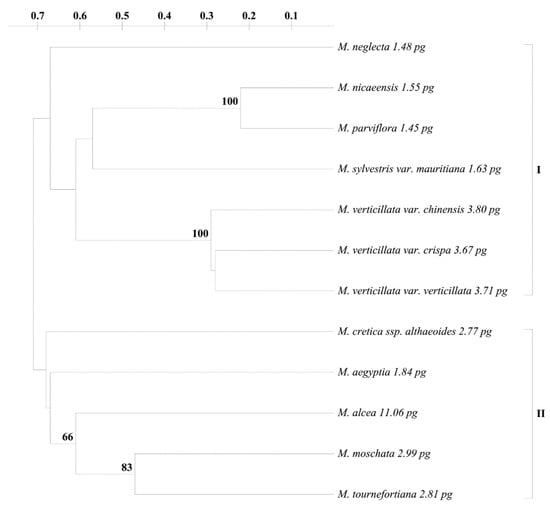
Figure 1.
The UPGMA (unweight pair group method with arithmetic average) dendrogram computed using genetic distance matrix based on ISSR markers. Only bootstrap values > 50% are indicated. Scale indicates genetic distance, and the values in pg represent genome size (2C) of the particular species.
3. Results
3.1. Genome Size
The genome size of the Malva accessions was different and ranged from 1.45 pg/2C in M. parviflora to 11.06 pg/2C in M. alcea (Table 2 and Figure 2), which corresponds to 1418 and 10,817 Mbp, respectively. The mean nuclear DNA content (2C) determined for all Malva accessions reached 3.23 pg/2C, which relates to 3159 Mbp. In three varieties of M. varticillata, the differences between the smallest and the highest 2C DNA content was 0.13 pg. The obtained data of genome size for all accessions were separated into three groups according to the classification of Soltis et al. ([49]; Table 2). Six accessions were classified into a group with very small genomes, with 2C DNA content ranging from 1.45 (M. parviflora) to 2.77 (M. cretica ssp. althaeoides) pg/2C, meaning an almost 2-fold difference. The group with small genomes included five accessions, with genome size ranging from 2.81 (M. tournefortiana) to 3.80 (M. verticillata var. chinensis (Mill.) S. Y. Hu) pg/2C, meaning a nearly 1.4-fold variation. The group with intermediate genomes was represented by one species (M. alcea) with the highest genome size (11.06 pg/2C) observed within the studied accessions.
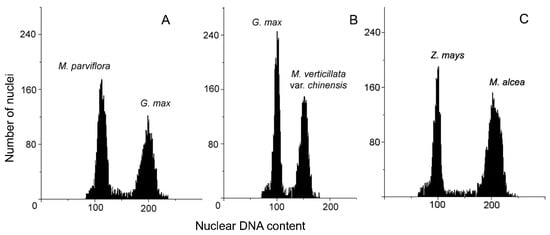
Figure 2.
Selected histograms of 2C DNA content of M. parviflora (A), M. verticillata var. chinensis (B), and M. alcea (C).
3.2. ISSR Markers
Twelve Malva genotypes were examined with 13 ISSR primers, which revealed unambiguous and reproducible band patterns. The primers yielded 243 polymorphic loci, with the size of the bands ranging from 212 (ISSR-1) to 2392 (ISSR-7) bp. The percentage of polymorphism amounted to 100% for all investigated primers (Table 3). The number of bands varied from 15 (ISSR-5 and ISSR-44) to 25 (ISSR-6), with an average of 19 bands per primer. The PIC values differed from 0.350 (ISSR-50) to 0.466 (ISSR-42), with an average of 0.395 (Table 3). The ISSR-4 and ISSR-42 primers revealed to be useful to distinguish all the investigated species, while the ISSR-5 and ISSR-6 primers enabled the discrimination of species and also M. verticillata varieties (Figure 3).
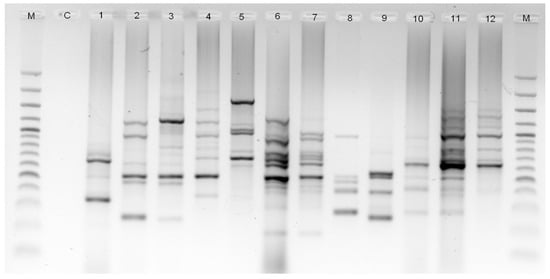
Figure 3.
Banding profiles generated by ISSR-PCR using ISSR-6 primer. DNA ladder (M); negative control (C), 1, M. aegyptia; 2, M. alcea; 3, M. cretica ssp. althaeoides; 4, M. moschata; 5, M. neglecta; 6, M. nicaeensis; 7, M. parviflora; 8, M. sylvestris var. mauritiana; 9, M. tournefortiana; 10, M. verticillata var. chinensis; 11, M. verticillata var. crispa; 12, M. verticillata var. verticillata.
The genetic distance was calculated to reflect the relationships between the tested accessions. The close relationship between Malva accessions was expressed by a low degree of the genetic distance. The values of genetic distance between the studied accessions ranged from 0.22 to 0.81 (Table 4), with the lowest being detected between M. parviflora and M. nicaeensis, while the highest was found between M. sylvestris var. mauritiana and M. alcea. The UPGMA algorithm clustered accessions into two groups (bootstrap values below 50%; Figure 1). The first group (marked as I) contained all M. verticillata accessions as well as M. sylvestris var. mauritiana, M. parviflora, M. nicaeensis, and M. neglecta. The second group (II) represented five species: M. cretica ssp. althaeoides, M. aegyptia, M. alcea, M. moschata, and M. tournefortiana.

Table 4.
Genetic distance between Malva accessions.
4. Discussion
Distinguishing Malva species from each other and closely related genera is mainly based on morphological analysis. However, within the genus, some species (e.g., M. parviflora, M. pusilla, M. nicaeensis, and M. neglecta) are regularly misidentified based on morphological features [38]. Moreover, the phylogenetic relationships and taxonomic organization of the Malva genus are still unclear. Therefore, molecular analysis can be useful in species identification and study of the genetic relationships between Malva species/subspecies/varieties.
Flow cytometry was used for nuclear DNA content estimation as a first step of Malva species identification and genetic characterization. To the best of our knowledge, this is the first study to determine the 2C DNA content for eight of the studied accessions. The 2C DNA content measured for all accessions ranged from 1.45 to 11.06 pg, with more than half the accessions (54%) possessing very small genomes. Most of the studied Malva accessions are hexaploids with small and numerous chromosomes, which makes chromosome counting challenging, so the published numbers (40–44) could be incorrect [3]. This means that it is possible several hexaploid species have the same chromosome number. In this study, the hexaploid level was observed for all accessions with very small genomes (M. aegyptia, M. cretica ssp. althaeoides, M. neglecta, M. nicaeensis, M. parviflora, and M. sylvestris var. mauritiana) and for two accessions with small genomes (M. moschata and M. tournefortiana). Until now, the genome size has been reported for only five mallow species: M. parviflora, M. neglecta, M. sylvestris, M. moschata, and M. alcea. 2C DNA content of M. parviflora was estimated previously by Feulgen microdensitometry (1.05 pg; [53]) and was lower than that determined here by flow cytometry, probably due to the use of different methods. Nuclear DNA content of M. neglecta (1.48 pg/2C) was close to 2C DNA content reported by both Bou Dagher-Kharrat et al. ([54]; 1.26 pg) and Bai et al. ([55]; 1.60 pg), which can suggest the same ploidy level. The genome size calculated for M. sylvestris var. mauritiana was similar to 2C DNA of M. sylvestris reported by Siljak-Yakovlev et al. ([56]; 1.5 pg). Higher values of 2C DNA content (2.90 and 3.0 pg) of this species were reported formerly by Caccarelli et al. [57] and Wendel et al. [58], which can indicate the occurrence of plants with higher ploidy level. The genome size obtained for M. moschata (2.99 pg/2C) was comparable to the 2C DNA value estimated by Pustahija et al. ([59]; 2.71 pg), which demonstrates the same ploidy level for this accession. The genome size of M. alcea (11.06 pg/2C) was 4-fold higher than that reported by Pustahija et al. ([59]; 2.65 pg/2C). According to Ray [3], M. alcea is a dodecaploid with 78 or 84 chromosomes, so it can be suggested that different cytotypes can occur among this species. However, this supposition should be confirmed by determining the number of chromosomes. Within the M. verticillata varieties, the 2C DNA content was similar and ranged from 3.71 (M. verticillata var. verticillata) to 3.80 (M. verticillata var. chinensis) pg. However, for this species, different chromosome numbers (76,84,112,120,126) have also been reported [3]. Thus, the chromosome number and ploidy level for these accessions should be clarified by chromosome recounting.
Differences in genome size allowed half of the studied mallow accessions to be identified. M. neglecta (1.48 pg/2C) could not be distinguished from M. parviflora (1.45 pg/2C), M. verticillata var. crispa (3.67 pg/2C) could not be distinguished from M. verticillata var. verticillata (3.71 pg/2C), and M. cretica ssp. althaeoides (2.77 pg/2C) could not be distinguished from M. tournefortiana (2.81 pg/2C) based on flow cytometric analysis. Therefore, to identify all studied accessions and to investigate genetic relationships between them, ISSR molecular markers were applied. The usefulness of these markers in species identification and detection of genetic diversity have been proven in studies of several other herbal genera (e.g., Trigonella L., Thymus L., Ocimum L., Origanum L., Mentha L.; [45,60,61,62,63]). Until now, molecular studies using ISSR markers conducted in the Malva genus have only included a few species [31,32]. All primers used in ISSR-PCRs for the Malva genus revealed 100% polymorphism between all accessions. Therefore, it was possible to identify all tested species. Moreover, for M. verticillata taxon, it was possible to distinguish all studied varieties. The usefulness of most of the used ISSR primers was also confirmed in Ocimum, Origanum, and Mentha identification [45,62,63].
The systematics of the Malva genus and closely related genera is complicated. Moreover, the relationships obtained from molecular studies do not confirm traditional classification [3,29]. So far, only molecular analysis relying on rDNA ITS sequences and ISSR markers have shed light on taxonomical relationships between Malva species [3,29,30,31]. Phylogenetic analyses of rDNA ITS sequences indicated the presence of two well-supported clusters within the mallow species (malvoid and lavateroid clades), which is consistent with the presented data. In this study, malvoid clade (I) was represented by five species: M. neglecta, M. nicaeensis, M. parviflora, M. sylvestris var. mauritiana, and M. verticillata. Moreover, within this cluster, two subgroups were detected. The first subgroup contained all M. verticillata varieties, while the second subgroup included two species: M. nicaeensis and M. parviflora. An earlier study reported that the two latter species are difficult to identify using morphological features [39]. Our work also confirmed a close relationship between M. parviflora and M. nicaeensis. Both M. sylvestris var. mauritiana and M. neglecta were placed outside of the subgroups. The lavateroid group (II) consisted of five species: M. cretica ssp. althaeoides, M. aegyptia, M. alcea, M. moschata, and M. tourneforiana. Within this group M. alcea together with M. moschata and M. tournefortiana created one subcluster, which confirms earlier phylogenetic analysis [29]. M. aegyptia and M. cretica ssp. althaeoides were placed outside of this subgroup. An earlier phylogenetic study revealed problems in the affiliation of those two species [29], while seed image analysis grouped them into separate Bibracteolatae sections [42]. Within the investigated genotypes, M. sylvestris var. mauritiana and M. alcea were the most genetically diverse. Nevertheless, the phylogenetic data presented in this study mostly agree with the division of mallow species proposed by Escobar Garcia et al. [29] on malvoid and lavateroid sections of the Malva genus. The data are also consistent with the study of Celka et al. [31], although the authors proposed the affiliation of the two clusters to Malva and Bismalva sections.
5. Conclusions
This study proved the potential of flow cytometric genome size determination and ISSR marker analyses in the identification and evaluation of genetic relationships between Malva species/varieties. Out of the 12 studied accessions, it was possible to identify six genotypes using flow cytometry, whereas all species and varieties were identified by the ISSR-PCR method. This is the first report about the application of these two techniques to estimate the genetic variation and genetic characterization of Malva accessions. The results of this study can be useful in mallow breeding programs, planning of conservation strategies, germplasm collection, and taxonomy of the genus.
Author Contributions
Conceptualization I.J.; data curation, I.J. and M.R.; formal analysis, I.J. and M.R.; investigation, I.J.; methodology, I.J. and M.R.; project administration, I.J.; software, I.J; supervision, I.J.; validation, I.J. and M.R.; writing—original draft, I.J. and M.R.; writing—review and editing, I.J. and M.R. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
The authors are grateful to the GRIN-ARS-USDA gene bank (USA) and the Medicinal and Cosmetic Plant Garden of the Nicolaus Copernicus University, Collegium Medicum, Bydgoszcz, Poland, for providing Malva accessions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dalby, D.H.; Malva, L. Flora Europea. Rosaceae to Umbelliferae; Tutin, T.G., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Walters, S.M., Weeb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1968; Volume 2, pp. 249–251. [Google Scholar]
- Abedin, S. Malvaceae. In Flora of West Pakistan 130; Nasir, E., Ali, S.I., Eds.; University of Karachi: Karachi, Pakistan, 1979; pp. 1–98. [Google Scholar]
- Ray, M.F. Systematics of Lavatera and Malva (Malvaceae, Malveae)—A new perspective. Plant Syst. Evol. 1995, 198, 29–53. [Google Scholar] [CrossRef]
- El Naggar, S.M. Pollen Morphology of Egyptian Malvaceae: An assessment of taxonomic value. Turk. J. Bot. 2004, 28, 227–240. [Google Scholar]
- Shaheen, N.; Khan, M.A.; Yasmin, G.; Hayat, M.Q.; Ali, S. Taxonomic implication of palynological characters in the genus Malva, L., family Malvaceae from Pakistan. Am. Eurasian J. Agric. Environ. Sci. 2009, 6, 716–722. [Google Scholar]
- Akçin, Ö.E.; Özbucak, T.B. Morphological, anatomical and ecological studies on medicinal and edible plant Malva neglecta Wallr. (Malvaceae). Pak. J. Biol. Sci. 2006, 9, 2716–2719. [Google Scholar] [CrossRef][Green Version]
- DellaGreca, M.; Cutillo, F.; D’Abrosca, B.; Fiorentino, A.; Pacifico, S.; Zarrelli, A. Antioxidant and radical scavenging properties of Malva sylvestris. Nat. Prod. Commun. 2009, 4, 893–896. [Google Scholar] [CrossRef]
- Boual, Z.; Kemassi, A.; Khelil, A.O.E.H.; Michaud, P.; Hadj, M.D.O.E. Partial characterization of water soluble polysaccharides extracted from one Saharian medicinal plant: Malva aegyptiaca L. Proceedings International Conference on Biology. Environ. Chem. 2011, 24, 420–424. [Google Scholar]
- Azab, A. Malva: Food, medicine and chemistry. Eur. Chem. Bull. 2017, 6, 295–320. [Google Scholar] [CrossRef]
- Fakhfakh, N.; Abdelhedi, O.; Jdir, H.; Nasri, M.; Zouari, N. Isolation of polysaccharides from Malva aegyptiaca and evaluation of their antioxidant and antibacterial properties. Int. J. Biol. Macromol. 2017, 105, 1519–1525. [Google Scholar] [CrossRef]
- Badawi, M.A.; El-Sahhar, K.F. Effect of Gibberellin on Some Vegetative Characters of Malva parviflora; Research Bulletin, Faculty of Agriculture Ain Shams University: Cairo, Egypt, 1978; p. 16. [Google Scholar]
- Baytop, T. Dictionary of Turkish Plant Names; Ankara Ataturk Culture, Language and History Institution Press: Ankara, Turkey, 1994; p. 578. [Google Scholar]
- Tanji, A.; Nassif, F. Edible weeds in Morocco. Weed Technol. 1995, 9, 617–620. [Google Scholar] [CrossRef]
- Ranhotra, G.S.; Gelroth, J.A.; Leinen, S.D.; Vinas, M.A.; Lorenz, K.J. Nutritional profile of some edible plants from México. J. Food Compos. Anal. 1998, 11, 298–304. [Google Scholar] [CrossRef]
- Doan, D.; Baolar, S.; Ay, G.; Mert, H.H. The use of wild edible plants in Western and Central Anatolia (Turkey). Econ. Bot. 2004, 58, 684–690. [Google Scholar]
- Baytop, T. The Medicinal and Poisonous Plants of Turkey; Istanbul University Press: Istanbul, Turkey, 1963; p. 1039. [Google Scholar]
- Asimgil, A. Medicinal Plants; Times Press: Istanbul, Turkey, 1993. [Google Scholar]
- Çakilcioğlu, U.; Sengun, M.T.; Türkoglu, I. An ethnobotanical survey of medicinal plants of Yazıkonak and Yurtbaşı districts of Elazığ province, Turkey. J. Med. Plants Res. 2010, 4, 567–572. [Google Scholar]
- Gasparetto, J.C.; Ferreira Martins, C.A.; Hayashi, S.S.; Otuky, M.F.; Pontarolo, R. Ethnobotanical and scientific aspects of Malva sylvestris L.: A millennial herbal medicine. J. Pharm. Pharmacol. 2012, 64, 172–189. [Google Scholar] [CrossRef] [PubMed]
- Hussain, L.; Ikram, J.; Rehman, K.; Tariq, M.; Ibrahim, M.; Akash, M.S.H. Hepatoprotective effects of Malva sylvestris L. against paracetamol-induced hepatotoxicity. Turk. J. Biol. 2014, 38, 396–402. [Google Scholar] [CrossRef]
- Mirghiasi, S.M.; Akhzari, M.; Vassaf, M.; Akbari, A.; Baghi, S.M. The effect of Malva neglecta on the reduction of inflammatory agents in patients with osteoarthritis. Mol. Biol. 2015, 4, 135. [Google Scholar] [CrossRef]
- Keyrouz, E.; El Feghali, P.A.R.; Jaafar, M.; Nawas, T. Malva neglecta: A natural inhibitor of bacterial growth and biofilm formation. J. Med. Plants Res. 2017, 11, 380–386. [Google Scholar] [CrossRef][Green Version]
- Del Rio, M.; Font, R.; Almela, C.; Velez, D.; Montoro, R.; De Haro Bailon, A. Heavy metals and arsenic uptake by wild vegetation in the Guadiamar river area after the toxic spill of the Aznalcóllar mine. J. Biotechnol. 2002, 98, 125–137. [Google Scholar] [CrossRef]
- Tlustos, P.; Szakova, J.; Hruby, J.; Hartman, I.; Najmanova, J.; Nedelnik, J.; Pavlikova, D.; Batysta, M. Removal of As, Cd, Pb, and Zn from contaminated soil by high biomass producing plants. Plant Soil Environ. 2006, 52, 413–423. [Google Scholar] [CrossRef]
- Ibrahim, M.M.; Alsahli, A.A.; El-Gaaly, G. Evaluation of phytoremediation potential of six wild plants for metal in a site polluted by industrial wastes: A field study in Riyadh, Saudi Arabia. Pak. J. Bot. 2013, 42, 571–576. [Google Scholar]
- Ahmed, Y.M.; Mahmood, A.B.; Ibrahim, H.J. Measuring the accumulation of copper and cadmium in the vegetative parts of the plant and the root of Malva parviflora as a result of irrigation with sewage in city of Kirkuk. Int. J. Curr. Res. Aca. Rev. 2016, 4, 149–154. [Google Scholar] [CrossRef]
- Rahbar, A.; Farjadfard, S.; Leili, M.; Kafaei, R.; Haghshenas, V.; Ramavandi, B. Experimental data of biomaterial derived from Malva sylvestris and charcoal tablet powder for Hg2+ removal from aqueous solutions. Data Brief 2016, 8, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Salahandish, R.; Ghaffarinejad, A.; Norouzbeigi, R. Rapid and efficient lead (II) ion removal from aqueous solutions using Malva sylvestris flower as a green biosorbent. Anal. Methods 2016, 8, 2515–2525. [Google Scholar] [CrossRef]
- Escobar García, P.; Schönswetter, P.; Fuertes Aguilar, J.; Nieto Feliner, G.; Schneeweiss, G.M. Five molecular markers reveal extensive morphological homoplasy and reticulate evolution in the Malva alliance (Malvaceae). Mol. Phylogenet. Evol. 2009, 50, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.F. New combinations in Malva (Malvaceae: Malveae). Novon 1998, 3, 288–295. [Google Scholar] [CrossRef]
- Celka, Z.; Szczecińska, M.; Sawicki, J. Genetic relationships between some of Malva species as determined with ISSR and ISJ markers. Biodivers. Res. Conserv. 2010, 19, 23–32. [Google Scholar] [CrossRef]
- Celka, Z.; Szczecińska, M.; Sawicki, J.; Shevera, M.V. Molecular studies did not support the distinctiveness of Malva alcea and M. excise (Malvaceae) in Central and Eastern Europe. Biologia 2012, 67, 1088–1098. [Google Scholar] [CrossRef]
- El Naggar, S.M. Systematic implications of seed coat morphology in Malvaceae. Pak. J. Biol. Sci. 2001, 4, 822–828. [Google Scholar] [CrossRef]
- Celka, Z.; Drapikowska, M.; Buczkowska, K.; Bączkiewicz, A.; Marciniak, J. Morphological variability of Malva alcea L. populations from Poland. In Environmental Changes and Biological Assessment; Kočáarek, P., Plášek, V., Malachová, K., Eds.; Scripta Facultatis Rerum Naturalium Universitatis Ostraviensis: Ostrava, Czech Republic, 2006; Volume 163, pp. 159–165. [Google Scholar]
- Celka, Z.; Szkudlarz, P.; Biereżnoj, U. Morphological variation of hairs in Malva alcea L. (Malvaceae). Biodivers. Res. Conserv. 2006, 3, 258–261. [Google Scholar]
- Celka, Z.; Drapikowska, M.; Jusik, S.; Olejnik, N.; Shevera, M.V.; Szkudlarz, P. Morphological variability of hairs in Malva alcea L. (Malvaceae) populations from Central and Eastern Europe, and consideration of the status of Malva excisa Rchb. Pak. J. Bot. 2015, 47, 467–476. [Google Scholar]
- Celka, Z.; Drapikowska, M.; Ogrodowicz, K.; Shevera, M.V.; Szkudlarz, P. Differentiation of petals in the Malva alcea populations from the region of Central and Eastern Europe. Biodivers. Res. Conserv. 2007, 5, 17–24. [Google Scholar]
- Michael, P.J.; Steadman, K.J.; Plummer, J.A. The biology of Australian weeds 52. Malva parviflora L. Plant Prot. Q. 2009, 24, 2–9. [Google Scholar]
- Makowski, R.M.D.; Morrison, I.N. The biology of Canadian weeds. 91. Malva pusilla Sm. (= M. rotundifolia L.). Can. J. Plant Sci. 1989, 69, 861–879. [Google Scholar] [CrossRef]
- Kristofferson, K.B. Species crossing in Malva. Hereditas 1926, 7, 233–354. [Google Scholar] [CrossRef]
- Olyanitskaya, L.G.; Tzvelev, N.N. Malvaceae. In Flora Europae Orientalis; Tzvelev, N.N., Ed.; ‘Mir i Semia-XCV’: St. Petersburg, Russia, 1996; Volume 9, pp. 231–255. [Google Scholar]
- Lo Bianco, M.; Grillo, O.; Escobar Garcia, P.; Mascia, F.; Venora, G.; Bacchetta, G. Morpho-colorimetric characterization of Malva alliance taxa by seed image analysis. Plant Biol. 2017, 19, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Adamkiewicz, E.; Bijok, K. Badania kariologiczne nad trzema gatunkami rodzaju Malva, L. Acta Soc. Bot. Pol. 1971, 2, 395–400. [Google Scholar]
- Góralski, G.; Judasz, A.; Gacek, P.; Grabowska-Joachimiak, A.; Joachimiak, A.J. Polyploidy, alien species and invasiveness in Polish angiosperms. Plant Syst. Evol. 2014, 300, 225–238. [Google Scholar] [CrossRef][Green Version]
- Jedrzejczyk, I.; Rewers, M. Genome size and ISSR markers for Mentha, L. (Lamiaceae) genetic diversity assessment and species identification. Ind. Crops Prod. 2018, 120, 171–179. [Google Scholar] [CrossRef]
- Lysak, M.A.; Dolezel, J. Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia 1998, 52, 123–132. [Google Scholar] [CrossRef]
- Doležel, J.; Dolezelova, M.; Novak, F.J. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol. Plant. 1994, 36, 351–357. [Google Scholar] [CrossRef]
- Doležel, J.; Bartoš, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S.; Bennett, M.D.; Leitch, I.J. Evolution of genome size in the Angiosperms. Am. J. Bot. 2003, 90, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Ghislain, M.; Zhang, D.; Fajardo, D.; Huamán, Z.; Hijmans, R.J. Marker-assisted sampling of the cultivated Andean potato Solanum phureja collection using RAPD markers. Genet. Resour. Crop Evol. 1999, 46, 547–555. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; De Wachter, Y. TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 1994, 10, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Bidak, L.; Brandham, P.E. Intraspecific uniformity of chromosome number and nuclear DNA quantity in two Egyptian weedy species, Malva parviflora (Malvaceae) and Trigonella stellata (Leguminosae). Kew Bull. 1995, 50, 595–599. [Google Scholar] [CrossRef]
- Bou Dagher-Kharrat, M.; Abdel-Samad, N.; Douaihy, B.; Bourge, M.; Fridlender, A.; Siljak-Yakovlev, S.; Brown, S.C. Nuclear DNA C-values for biodiversity screening: Case of the Lebanese flora. Plant Biosyst. 2013, 147, 1228–1237. [Google Scholar] [CrossRef]
- Bai, C.; Alverson, W.S.; Follansbee, A.; Waller, D.M. New reports of nuclear DNA content for 407 U.S. plant species. Ann. Bot. 2012, 110, 1623–1629. [Google Scholar] [CrossRef]
- Siljak-Yakovlev, S.; Pustahija, F.; Solic, E.M.; Bogunic, F.; Muratovic, E.; Basic, N.; Catrice, O.; Brown, S.C. Towards a genome size and chromosome number database of Balkan flora: C-values in 343 taxa with novel values for 242. Adv. Sci. Lett. 2010, 3, 190–213. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Morosi, L.; Cionini, P.G. Chromocenter association in plant cell nuclei: Determinants, functional significance, and evolutionary implications. Genome 1998, 41, 96–103. [Google Scholar] [CrossRef]
- Wendel, J.F.; Cronn, R.C.; Johnston, J.S.; Price, H.J. Feast and famine in plant genomes. Genetica 2002, 115, 37–47. [Google Scholar] [CrossRef]
- Pustahija, F.; Brown, S.C.; Bogunic, F.; Bašic, N.; Muratovic, E.; Ollier, S.; Hidalgo, O.; Bourge, M.; Stevanovic, V.; Siljak-Yakovlev, S. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil 2013, 373, 427–453. [Google Scholar] [CrossRef]
- Dangi, R.S.; Lagu, M.D.; Choudhary, L.B.; Ranjekar, P.K.; Gupta, V.S. Assessment of genetic diversity in Trigonella foenum–graecum and Trigonella caerulea using ISSR and RAPD markers. BMC Plant Biol. 2004, 4, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Rahimmalek, M.; Bahreininejad, B.; Khorrami, M.; Tabatabaei, B.E. Genetic variability and geographic differentiation in Thymus daenensis subsp. daenensis, an endangered medicinal plant, as revealed by inter simple sequence repeat (ISSR) markers. Biochem. Genet. 2009, 47, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Rewers, M.; Jedrzejczyk, I. Genetic characterization of Ocimum genus using flow cytometry and inter-simple sequence repeat markers. Ind. Crops Prod. 2016, 91, 142–151. [Google Scholar] [CrossRef]
- Jedrzejczyk, I. Study on genetic diversity between Origanum, L. species based on genome size and ISSR markers. Ind. Crops Prod. 2018, 126, 201–207. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).