Assessment of 16 Peanut (Arachis hypogaea L.) CSSLs Derived from an Interspecific Cross for Yield and Yield Component Traits: QTL Validation
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Experimental Design and Trial Management
2.3. Rainfall Amount in the Six Environnements
2.4. Harvest and Post-Harvest Management
2.5. Traits Evaluated
2.5.1. Pod and Haulm Yield
2.5.2. Yield Components
2.5.3. Pod and Seed Sizes
2.6. Statistical Analysis
3. Results
3.1. Single-Site Analysis and Comparison between CSSLs and Fleur11
3.1.1. Hundred Pod and Seed Weights (HPW and HSW)
3.1.2. Pod and Seed Length (PL and SL)
3.1.3. Pod and Seed Width (PWI and SWI)
3.1.4. Pod and Haulm Yield (Yield, Hlm) and Pod Maturity (Mat)
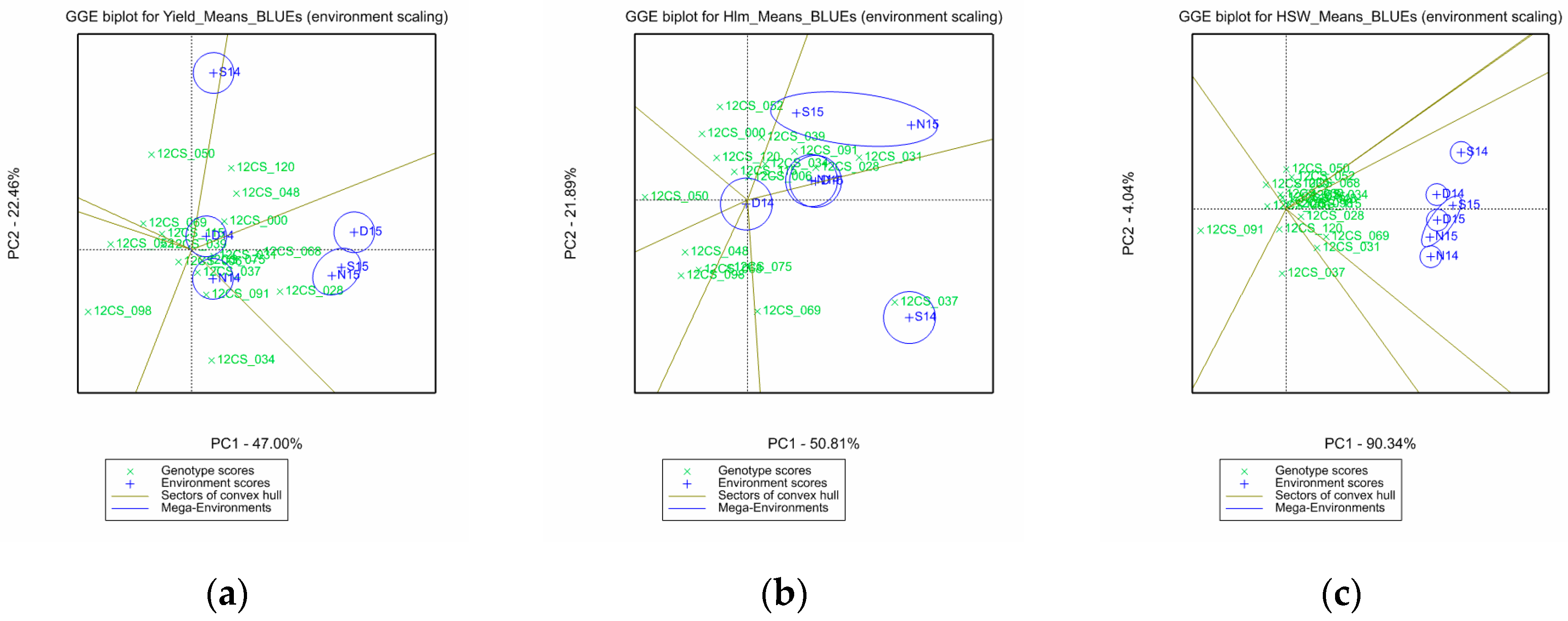
3.2. Mega-Environments, Performance, and Stability of the Genotypes
4. Discussion
4.1. Wild Alleles Contributed Positive Variation to Yield and Yield Related Traits
4.2. CSSLs Are Accurate Populations for QTL Validation and New QTL Discovery
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Davis, J.P.; Dean, L.L. Peanut composition, flavor and nutrition. In Peanuts: Genetics, Processing, and Utilization; Stalker, H.T., Wilson Richard, F., Eds.; Academic Press and AOCS Press: Cambridge, MA, USA, 2016; pp. 289–345. [Google Scholar]
- FAOSTAT. Food and Agriculture Organization -Statistical Database. Available online: www.fao.org/faostat/fr/2017 (accessed on 12 December 2019).
- Husted, L. Cytological Studies an the Peanut, Arachis. Cytologia (Tokyo) 1933, 5, 109–117. [Google Scholar] [CrossRef]
- Husted, L. Cytological Studies an the Peanut, Arachis. II. Cytologia (Tokyo) 1936, 7, 396–423. [Google Scholar] [CrossRef]
- Stebbins, G.L. Self Fertilization and Population Variability in the Higher Plants. Am. Nat. 1957, 91, 337–354. [Google Scholar] [CrossRef]
- Seijo, J.G.; Lavia, G.I.; Fernandez, A.; Krapovickas, A.; Ducasse, D.; Moscone, E.A. Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaensis are the wild diploid progenitors of A. hypogaea (Leguminosae). Am. J. Bot. 2004, 91, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Grabiele, M.; Chalup, L.; Robledo, G.; Seijo, G. Genetic and geographic origin of domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences. Plant Syst. Evol. 2012, 298, 1151–1165. [Google Scholar] [CrossRef]
- Kochert, G.; Halward, T.; Branch, W.D.; Simpson, C.E. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. TAG Theor. Appl. Genet. Theor. Angew. Genet. 1991, 81, 565–570. [Google Scholar] [CrossRef]
- Kochert, G.; Stalker, H.T.; Gimenes, M.; Galgaro, L.; Lopes, C.R.; Moore, K. RFLP and Cytogenetic Evidence on the Origin and Evolution of Allotetraploid Domesticated Peanut, Arachis hypogaea (Leguminosae). Am. J. Bot. 1996, 83, 1282–1291. [Google Scholar] [CrossRef]
- Seijo, G.; Lavia, G.I.; Fernández, A.; Krapovickas, A.; Ducasse, D.A.; Bertioli, D.J.; Moscone, E.A. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 2007, 94, 1963–1971. [Google Scholar] [CrossRef]
- Halward, T.M.; Stalker, H.T.; Larue, E.A.; Kochert, G. Genetic variation detectable with molecular markers among unadapted germ-plasm resources of cultivated peanut and related wild species. Genome 1991, 34, 1013–1020. [Google Scholar] [CrossRef]
- Halward, T.; Stalker, T.; LaRue, E.; Kochert, G. Use of single-primer DNA amplifications in genetic studies of peanut (Arachis hypogaea L.). Plant Mol. Biol. 1992, 18, 315–325. [Google Scholar] [CrossRef]
- He, G.; Prakash, C.S. Identification of polymorphic DNA markers in cultivated peanut (Arachis hypogaea L.). Euphytica 1997, 97, 143–149. [Google Scholar] [CrossRef]
- Subramanian, V.; Gurtu, S.; Rao, R.N.; Nigam, S.N. Identification of DNA polymorphism in cultivated groundnut using random amplified polymorphic DNA (RAPD) assay. Genome 2000, 43, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.N.; Rani, V.; Kojima, T.; Ogihara, Y.; Singh, K.P.; Devarumath, R.M. RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 2001, 44, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Gimenes, M.A.; Lopes, C.R.; Valls, J.F.M. Genetic relationships among Arachis species based on AFLP. Genet. Mol. Biol. 2002, 25, 349–353. [Google Scholar] [CrossRef]
- Milla, S.R.; Isleib, T.G.; Stalker, H.T. Taxonomic relationships among Arachis sect. Arachis species as revealed by AFLP markers. Genome 2005, 48, 1–11. [Google Scholar] [PubMed]
- Cuc, L.M.; Mace, E.S.; Crouch, J.H.; Quang, V.D.; Long, T.D.; Varshney, R.K. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biol. 2008, 8, 55. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Grandillo, S.; Fulton, T.M.; Zamir, D.; Eshed, Y.; Petiard, V.; Lopez, J.; Beck-Bunn, T. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 1996, 92, 213–224. [Google Scholar] [CrossRef]
- Fulton, T.M.; Grandillo, S.; Beck-Bunn, T.; Fridman, E.; Frampton, A.; Lopez, J.; Petiard, V.; Uhlig, J.; Zamir, D.; Tanksley, S.D. Advanced backcross QTL analysis of a Lycopersicon esculentum× Lycopersicon parviflorum cross. Theor. Appl. Genet. 2000, 100, 1025–1042. [Google Scholar] [CrossRef]
- Gur, A.; Zamir, D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2004, 2, e245. [Google Scholar] [CrossRef]
- Causse, M.; Duffé, P.; C Gomez, M.; Buret, M.; Damidaux, R.; Zamir, D.; Gur, A.; Chevalier, C.; Lemaire-Chamley, M.; Rothan, C. A Genetic Map of Candidate Genes and QTLs Involved in Tomato Fruit Size and Composition. J. Exp. Bot. 2004, 55, 1671–1685. [Google Scholar] [CrossRef]
- Xiao, J.; Li, J.; Grandillo, S.; Ahn, S.N.; Yuan, L.; Tanksley, S.D.; McCouch, S.R. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 1998, 150, 899–909. [Google Scholar] [PubMed]
- Moncada, P.; Martínez, C.P.; Borrero, J.; Chatel, M.; Gauch Jr, H.; Guimaraes, E.; Tohme, J.; McCouch, S.R. Quantitative trait loci for yield and yield components in an Oryza sativa×Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 2001, 102, 41–52. [Google Scholar] [CrossRef]
- Thomson, M.J.; Tai, T.H.; McClung, A.M.; Lai, X.-H.; Hinga, M.E.; Lobos, K.B.; Xu, Y.; Martinez, C.P.; McCouch, S.R. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2003, 107, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Septiningsih, E.M.; Prasetiyono, J.; Lubis, E.; Tai, T.H.; Tjubaryat, T.; Moeljopawiro, S.; McCouch, S.R. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2003, 107, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiao, J.; Grandillo, S.; Jiang, L.; Wan, Y.; Deng, Q.; Yuan, L.; McCouch, S.R. QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome 2004, 47, 697–704. [Google Scholar] [CrossRef]
- Shah, M.M.; Gill, K.S.; Baenziger, P.S.; Yen, Y.; Kaeppler, S.M.; Ariyarathne, H.M. Molecular Mapping of Loci for Agronomic Traits on Chromosome 3A of Bread Wheat. Crop Sci. 1999, 39, 1728–1732. [Google Scholar] [CrossRef]
- Börner, A.; Schumann, E.; Fürste, A.; Cöster, H.; Leithold, B.; Röder, M.; Weber, W. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef]
- Huang, X.Q.; Cöster, H.; Ganal, M.W.; Röder, M.S. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor. Appl. Genet. 2003, 106, 1379–1389. [Google Scholar] [CrossRef]
- Chaim, A.B.; Paran, I.; Grube, R.C.; Jahn, M.; Van Wijk, R.; Peleman, J. QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 2001, 102, 1016–1028. [Google Scholar] [CrossRef]
- Rao, G.U.; Ben Chaim, A.; Borovsky, Y.; Paran, I. Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2003, 106, 1457–1466. [Google Scholar] [CrossRef]
- Dwivedi, N.; Kumar, R.; Paliwal, R.; Kumar, U.; Kumar, S.; Singh, M.; Singh, R.K. QTL mapping for important horticultural traits in pepper (Capsicum annuum L.). J. Plant Biochem. Biotechnol. 2015, 24, 154–160. [Google Scholar] [CrossRef]
- Stalker, H.T.; Tallury, S.P.; Ozias-Akins, P.; Bertioli, D.; Bertioli, S.L. The value of diploid peanut relatives for breeding and genomics. Peanut Sci. 2013, 40, 70–88. [Google Scholar] [CrossRef]
- Sharma, S.; Pandey, M.K.; Sudini, H.; Upadhyaya, H.D.; Varshney, R.K. Harnessing Genetic Diversity of Wild Arachis Species for Genetic Enhancement of Cultivated Peanut. Crop Sci. 2017, 57, 1121–1131. [Google Scholar] [CrossRef]
- Fávero, A.P.; Simpson, C.E.; Valls, J.F.M.; Vello, N.A. Study of the Evolution of Cultivated Peanut through Crossability Studies among Arachis ipaënsis, A. duranensis, and A. hypogaea. Crop Sci. 2006, 46, 1546–1552. [Google Scholar] [CrossRef]
- Mallikarjuna, N. Production of hybrids between Arachis hypogaea and A. chiquitana (section Procumbentes). Peanut Sci. 2005, 32, 148–152. [Google Scholar] [CrossRef]
- Mallikarjuna, N.; Hoisington, D. Peanut improvement: Production of fertile hybrids and backcross progeny between Arachis hypogaea and A. kretschmeri. Food Secur. 2009, 1, 457–462. [Google Scholar] [CrossRef]
- Mallikarjuna, N.; Senthilvel, S.; Hoisington, D. Development of new sources of tetraploid Arachis to broaden the genetic base of cultivated groundnut (Arachis hypogaea L.). Genet. Resour. Crop Evol. 2011, 58, 889–907. [Google Scholar] [CrossRef]
- Rami, J.-F.; Leal-Bertioli, S.C.M.; Foncéka, D.; Moretzsohn, M.C.; Bertioli, D.J. Groundnut. In Alien Gene Transfer in Crop Plants; Pratap, A., Kumar, J., Eds.; Springer: New York, NY, USA, 2014; Volume 2, pp. 253–279. ISBN 978-1-4614-9571-0. [Google Scholar]
- Stalker, H.T. Utilizing wild species for peanut improvement. Crop Sci. 2017, 57, 1102–1120. [Google Scholar] [CrossRef]
- Nagy, E.D.; Chu, Y.; Guo, Y.; Khanal, S.; Tang, S.; Li, Y.; Dong, W.B.; Timper, P.; Taylor, C.; Ozias-Akins, P.; et al. Recombination is suppressed in an alien introgression in peanut harboring Rma, a dominant root-knot nematode resistance gene. Mol. Breed. 2010, 26, 357–370. [Google Scholar] [CrossRef]
- Leal-Bertioli, S.C.M.; Cavalcante, U.; Gouvea, E.G.; Ballén-Taborda, C.; Shirasawa, K.; Guimarães, P.M.; Jackson, S.A.; Bertioli, D.J.; Moretzsohn, M.C. Identification of QTLs for Rust Resistance in the Peanut Wild Species Arachis magna and the Development of KASP Markers for Marker-Assisted Selection. G3 Genes Genomes Genet. 2015, 5, 1403–1413. [Google Scholar]
- Leal-Bertioli, S.C.M.; Moretzsohn, M.C.; Roberts, P.A.; Ballén-Taborda, C.; Borba, T.C.O.; Valdisser, P.A.; Vianello, R.P.; Araújo, A.C.G.; Guimarães, P.M.; Bertioli, D.J. Genetic Mapping of Resistance to Meloidogyne arenaria in Arachis stenosperma: A New Source of Nematode Resistance for Peanut. G3 Genes Genomes Genet. 2016, 6, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Rick, C.M.; Chetelat, R.T. Utilization of related wild species for tomato improvement. Acta Hortic. 1995, 21–38. [Google Scholar] [CrossRef]
- Jordan, D.; Butler, D.; Henzell, B.; Drenth, J.; McIntyre, L. Diversification of Australian sorghum using wild relatives, New Directions for a Diverse Planet. In Proceedings of the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004. [Google Scholar]
- Fonceka, D.; Tossim, H.-A.; Rivallan, R.; Vignes, H.; Faye, I.; Ndoye, O.; Moretzsohn, M.C.; Bertioli, D.J.; Glaszmann, J.-C.; Courtois, B.; et al. Fostered and left behind alleles in peanut: Interspecific QTL mapping reveals footprints of domestication and useful natural variation for breeding. BMC Plant Biol. 2012, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D. Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2001, 2, 983–989. [Google Scholar] [CrossRef]
- Foncéka, D. Elargissement de la base génétique de l’arachide cultivée (# Arachis hypogaea#): Applications pour la construction de populations, l’identification de QTL et l’amélioration de l’espèce cultivée; Montpellier SupAgro: Montpellier, France, 2010; Available online: https://www.theses.fr/2010NSAM0023 (accessed on 12 December 2019).
- Fonceka, D.; Tossim, H.-A.; Rivallan, R.; Vignes, H.; Lacut, E.; de Bellis, F.; Faye, I.; Ndoye, O.; Leal-Bertioli, S.C.M.; Valls, J.F.M.; et al. Construction of Chromosome Segment Substitution Lines in Peanut (Arachis hypogaea L.) Using a Wild Synthetic and QTL Mapping for Plant Morphology. PLoS ONE 2012, 7, e48642. [Google Scholar] [CrossRef]
- Van Rossum, B.-J.; van Eeuwijk, F.; Boer, M.; Malosetti, M.; Bustos-Korts, D.; Millet, E.; Paulo, J.; Verouden, M.; Kruijer, W.; Wehrens, R.; et al. statgenSTA: Single Trial Analysis (STA) of Field Trials. R Package version 1.0.4. 2020. Available online: https://rdrr.io/cran/statgenSTA/ (accessed on 12 December 2019).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists; CRC Press: Boca Raton, FL, USA, 2003; ISBN 978-0-429-12272-9. [Google Scholar]
- Breeding Management System | Integrated Breeding Platform | Plant Breeding Software. Available online: https://bmspro.io/1824/breeding-management-system/tutorials/maize-multi-site-gxe-analysis (accessed on 7 April 2020).
- Eshed, Y.; Zamir, D. Introgressions fromLycopersicon pennellii can improve the soluble-solids yield of tomato hybrids. Theor. Appl. Genet. 1994, 88, 891–897. [Google Scholar] [CrossRef]
- McCouch, S. Diversifying selection in plant breeding. PLoS Biol. 2004, 2, e347. [Google Scholar] [CrossRef]
- Gutiérrez, A.G.; Carabalí, S.J.; Giraldo, O.X.; Martínez, C.P.; Correa, F.; Prado, G.; Tohme, J.; Lorieux, M. Identification of a Rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines. BMC Plant Biol. 2010, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Fu, Y.; Zhao, X.; Jiang, L.; Zhu, Z.; Gu, P.; Xu, W.; Su, Z.; Sun, C.; Tan, L. Genomic structure analysis of a set of Oryza nivara introgression lines and identification of yield-associated QTLs using whole-genome resequencing. Sci. Rep. 2016, 6, 27425. [Google Scholar] [CrossRef] [PubMed]
- Swamy, B.P.M.; Sarla, N. Yield-enhancing quantitative trait loci (QTLs) from wild species. Biotechnol. Adv. 2008, 26, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Board, J.E.; Kang, M.S.; Harville, B.G. Path Analyses Identify Indirect Selection Criteria for Yield of Late-Planted Soybean. Crop Sci. 1997, 37. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Ferguson, M.E.; Bramel, P.J.; Chandra, S. Gene diversity among botanical varieties in peanut (Arachis hypogaea L.). Crop Sci. 2004, 44, 1847–1854. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Archer, M.A.; Wayne, R.K. Transgressive segregation, adaptation and speciation. Heredity 1999, 83, 363–372. [Google Scholar] [CrossRef]
- Vega, U.; Frey, K.J. Transgressive segregation in inter and intraspecific crosses of barley. Euphytica 1980, 29, 585–594. [Google Scholar] [CrossRef]
- Nguepjop, J.R.; Tossim, H.-A.; Bell, J.M.; Rami, J.-F.; Sharma, S.; Courtois, B.; Mallikarjuna, N.; Sane, D.; Fonceka, D. Evidence of Genomic Exchanges between Homeologous Chromosomes in a Cross of Peanut with Newly Synthetized Allotetraploid Hybrids. Front. Plant Sci. 2016, 7, 1635. [Google Scholar] [CrossRef]
- Leal-Bertioli, S.; Shirasawa, K.; Abernathy, B.; Moretzsohn, M.; Chavarro, C.; Clevenger, J.; Ozias-Akins, P.; Jackson, S.; Bertioli, D. Tetrasomic Recombination Is Surprisingly Frequent in Allotetraploid Arachis. Genetics 2015, 199, 1093–1105. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Chavarro, C.; Agarwal, G.; Bertioli, D.J.; Leal-Bertioli, S.C.M.; Pandey, M.K.; Vaughn, J.; Abernathy, B.; Barkley, N.A.; et al. Genome-wide SNP Genotyping Resolves Signatures of Selection and Tetrasomic Recombination in Peanut. Mol. Plant 2017, 10, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Beavis, W.D.; Beavis, W.D.; Beavis, W.D.; Beavis, W.D. The Power and Deceit of QTL Experiments: Lessons from Comparative QTL Studies; ScienceOpen: Berlin, Germany, 1994. [Google Scholar]
- Fasoula, V.A.; Harris, D.K.; Boerma, H.R. Validation and Designation of Quantitative Trait Loci for Seed Protein, Seed Oil, and Seed Weight from Two Soybean Populations. Crop Sci. 2004, 44, 1218–1225. [Google Scholar] [CrossRef]
- Keurentjes, J.J.B.; Bentsink, L.; Alonso-Blanco, C.; Hanhart, C.J.; Blankestijn-De Vries, H.; Effgen, S.; Vreugdenhil, D.; Koornneef, M. Development of a Near-Isogenic Line Population of Arabidopsis thaliana and Comparison of Mapping Power With a Recombinant Inbred Line Population. Genetics 2007, 175, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.L.; Zhai, H.Q.; Wan, J.M.; Yasui, H.; Yoshimura, A. Mapping QTL for traits associated with resistance to ferrous iron toxicity in rice (Oryza sativa L.), using japonica chromosome segment substitution lines. Yi Chuan Xue Bao 2003, 30, 893–898. [Google Scholar] [PubMed]
- Sun, D.; Jiang, L.; Zhang, Y.; Cheng, X.; Zhai, H.; Wan, J. Detection of QTL associated with rice stripe resistance in cultivar IR24. Acta Agron Sin. 2007, 33, 25–30. [Google Scholar]
- Irzykowska, L.; Wolko, B. Interval mapping of QTLs controlling yield-related traits and seed protein content in Pisum sativum. J. Appl. Genet. 2004, 45, 297–306. [Google Scholar] [PubMed]
- Timmerman-Vaughan, G.M.; Mills, A.; Whitfield, C.; Frew, T.; Butler, R.; Murray, S.; Lakeman, M.; McCallum, J.; Russell, A.; Wilson, D. Linkage Mapping of QTL for Seed Yield, Yield Components, and Developmental Traits in Pea. Crop Sci. 2005, 45, 1336–1344. [Google Scholar] [CrossRef]
- Ayaz, S.; McKENZIE, B.A.; Hill, G.D.; McNEIL, D.L. Variability in yield of four grain legume species in a subhumid temperate environment. II. Yield components. J. Agric. Sci. 2004, 142, 21–28. [Google Scholar] [CrossRef]
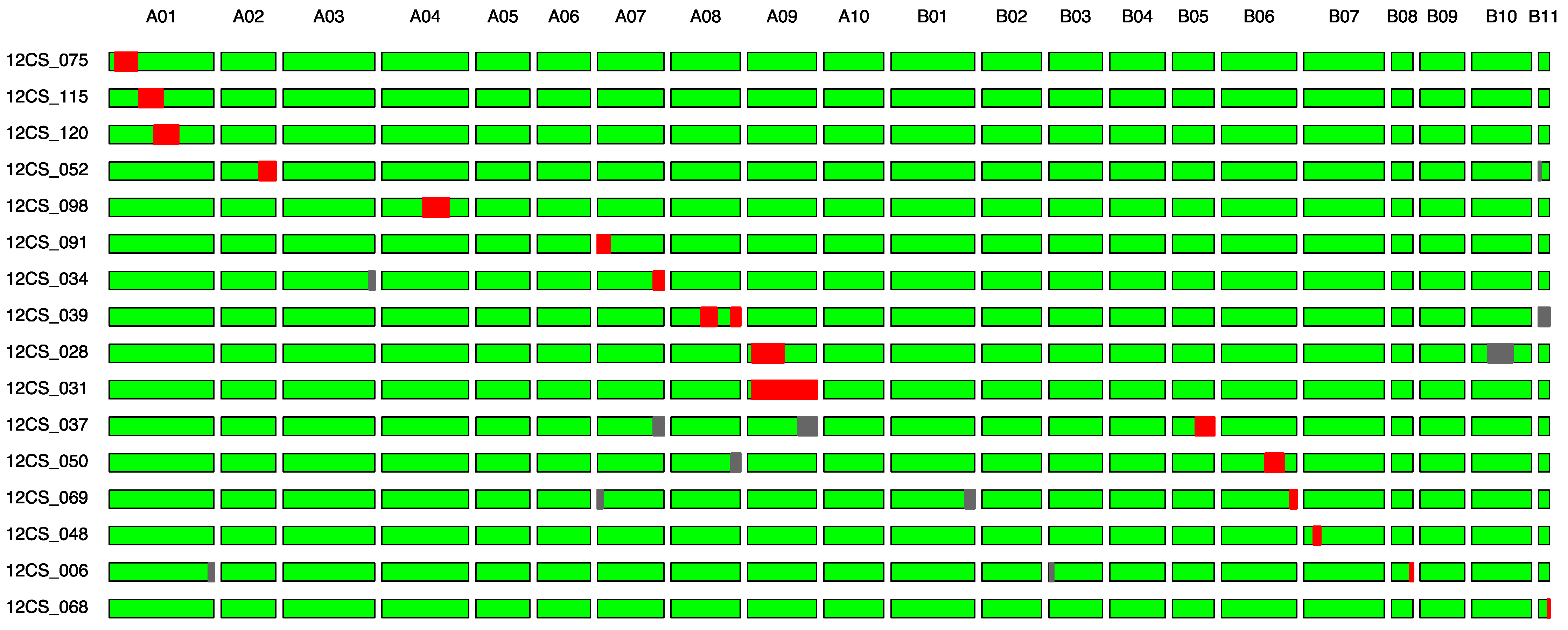


| Lines | Linkage Group | QTLs from AB-QTL Study |
|---|---|---|
| 12CS_075 | A01 | qPN; qPW; qSHW |
| 12CS_115 | A01 | qPN; qPW; qSHW |
| 12CS_120 | A01 | qPN; qPW; qSHW |
| 12CS_052 | A02 | qHW |
| 12CS_098 | A04 | qPH |
| 12CS_091 | A07 | qHSW; qPL; qPWI; qSL; qSWI |
| 12CS_034 | A07 | qHSW; qPL; qPWI; qSL; qSWI |
| 12CS_039 | A08 | qPL; qPWI; qSL |
| 12CS_028 | A09 | qPL; qSL |
| 12CS_031 | A09 | qPL; qSL |
| 12CS_037 | B05 | qHPW; qPWI; qSWI; qSW |
| 12CS_050 | B06 | qPWI, qSWI; qTB; qHW; qPMAT |
| 12CS_069 | B06 | qPWI, qSWI; qTB; qHW; qPMAT |
| 12CS_068 | B11 | qPMAT |
| 12CS_048 | B07 | - |
| 12CS_006 | B08 | - |
| Fleur11 | - | - |
| D14 | D15 | N14 | |||||||||||
| Trait | Term | mean | F | Pr | h2 | Mean | F | Pr | h2 | Mean | F | Pr | h2 |
| Hlm | genotype | 1.92 | 29.16 | <0.001 *** | 0.81 | 2.43 | 7.63 | <0.001 *** | 0.73 | 2.99 | 21.3 | 0.003 ** | 0.55 |
| rep | 0.47 | 0.247 | 3.79 | <0.001 *** | 7.23 | 0.008 ** | |||||||
| HPW | genotype | 111.94 | 12.58 | <0.001 *** | 0.88 | 129.16 | 1.18 | <0.001 *** | 0.91 | 134.10 | 38.66 | <0.001 *** | 0.74 |
| rep | 0.59 | 0.083 | 0.26 | 0.386 | 2.41 | 0.662 | |||||||
| HSW | genotype | 58.95 | 3.72 | <0.001 *** | 0.89 | 56.04 | 2.26 | <0.001 *** | 0.93 | 58.31 | 3.86 | <0.001 *** | 0.87 |
| rep | 0.05 | 0.001 ** | 4.87 | 0.554 | 0.41 | 0.104 | |||||||
| Mat | genotype | 62.20 | 11.41 | <0.001 *** | 0.60 | 89.61 | 14.56 | 0.275 | 0.09 | 88.65 | 15.93 | 0.004 ** | 0.53 |
| rep | 3.09 | 0.974 | 1.25 | 0.77 | 0.79 | 0.277 | |||||||
| PL | genotype | 28.17 | 7.68 | <0.001 *** | 0.92 | 28.77 | 1.16 | <0.001 *** | 0.97 | 28.17 | 14.7 | <0.001 *** | 0.89 |
| rep | 0.51 | 0.885 | 0.08 | 0.090 | 0.69 | 0.13 | |||||||
| PWI | genotype | 11.80 | 3.99 | <0.001 *** | 0.97 | 11.33 | 5.06 | <0.001 *** | 0.90 | 11.76 | 4.1 | <0.001 *** | 0.91 |
| rep | 0.26 | 0.626 | 17.93 | 0.064 | 0.47 | 0.046 * | |||||||
| SL | genotype | 14.45 | 44.78 | <0.001 *** | 0.84 | 14.57 | 17.93 | <0.001 *** | 0.96 | 14.34 | 11.96 | <0.001 *** | 0.93 |
| rep | 7.03 | 0.023 * | 1.11 | 0.54 | 5.36 | 0.287 | |||||||
| SWI | genotype | 8.70 | 22.24 | <0.001 *** | 0.95 | 8.35 | 1.98 | <0.001 *** | 0.94 | 8.69 | 34.39 | <0.001 *** | 0.94 |
| rep | 2.7 | <0.001 *** | 2.55 | 0.687 | 4.07 | 0.452 | |||||||
| Yield | genotype | 1.77 | 4.85 | <0.001 *** | 0.81 | 1.90 | 5.48 | <0.001 *** | 0.88 | 2.65 | 8.05 | <0.001 *** | 0.73 |
| rep | 2.49 | 0.107 | 1.38 | <0.001 *** | 2.49 | 0.954 | |||||||
| N15 | S14 | S15 | |||||||||||
| Trait | Term | mean | F | Pr | h2 | mean | F | Pr | h2 | mean | F | Pr | h2 |
| Hlm | genotype | 3.48 | 8.26 | <0.001 *** | 0.89 | 5.30 | 3.79 | <0.001 *** | 0.79 | 4.60 | 11.39 | 0.002 ** | 0.56 |
| rep | 15.51 | 0.673 | 10.5 | <0.001 *** | 0.95 | <0.001 *** | |||||||
| HPW | genotype | 121.42 | 9.95 | <0.001 *** | 0.79 | 105.20 | 24.28 | <0.001 *** | 0.75 | 111.86 | 15.46 | <0.001 *** | 0.84 |
| rep | 2.74 | 0.545 | 0.62 | 0.627 | 0.37 | 0.563 | |||||||
| HSW | genotype | 52.69 | 7.94 | <0.001 *** | 0.87 | 59.06 | 2.17 | <0.001 *** | 0.96 | 57.56 | 9.36 | <0.001 *** | 0.96 |
| rep | 2.26 | 0.603 | 1.28 | 0.089 | 2.04 | 0.135 | |||||||
| Mat | genotype | 91.61 | 3.94 | 0.296 | 0.15 | 77.71 | 9.4 | <0.001 *** | 0.70 | 77.59 | 5.18 | 0.011 * | 0.46 |
| rep | 0.46 | 0.925 | 0.4 | 0.376 | 0.61 | 0.078 | |||||||
| PL | genotype | 27.21 | 7.53 | <0.001 *** | 0.93 | 27.56 | 8.19 | <0.001 *** | 0.92 | 28.66 | 6.57 | <0.001 *** | 0.97 |
| rep | 1.52 | 0.499 | 3.22 | 0.658 | 2.11 | 0.017 * | |||||||
| PWI | genotype | 10.96 | 27.38 | <0.001 *** | 0.86 | 11.43 | 3.42 | <0.001 *** | 0.98 | 11.31 | 11.86 | <0.001 *** | 0.97 |
| rep | 2.42 | 0.218 | 0.98 | <0.001 *** | 0.42 | 0.392 | |||||||
| SL | genotype | 13.94 | 5.16 | <0.001 *** | 0.87 | 14.38 | 2.32 | <0.001 *** | 0.95 | 14.46 | 6.08 | <0.001 *** | 0.92 |
| rep | 0.21 | 0.040 * | 12.41 | 0.329 | 0.33 | 0.25 | |||||||
| SWI | genotype | 8.04 | 38.13 | <0.001 *** | 0.85 | 8.31 | 12.76 | <0.001 *** | 0.91 | 8.43 | 17.04 | <0.001 *** | 0.94 |
| rep | 0.94 | 0.121 | 1.39 | 0.005 ** | 5.49 | 0.004 ** | |||||||
| Yield | genotype | 2.77 | 9.24 | <0.001 *** | 0.74 | 2.12 | 2.58 | <0.001 *** | 0.71 | 2.55 | 12.03 | <0.001 *** | 0.79 |
| rep | 6.72 | 0.633 | 0.03 | 0.773 | 0.12 | 0.814 | |||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tossim, H.-A.; Nguepjop, J.R.; Diatta, C.; Sambou, A.; Seye, M.; Sane, D.; Rami, J.-F.; Fonceka, D. Assessment of 16 Peanut (Arachis hypogaea L.) CSSLs Derived from an Interspecific Cross for Yield and Yield Component Traits: QTL Validation. Agronomy 2020, 10, 583. https://doi.org/10.3390/agronomy10040583
Tossim H-A, Nguepjop JR, Diatta C, Sambou A, Seye M, Sane D, Rami J-F, Fonceka D. Assessment of 16 Peanut (Arachis hypogaea L.) CSSLs Derived from an Interspecific Cross for Yield and Yield Component Traits: QTL Validation. Agronomy. 2020; 10(4):583. https://doi.org/10.3390/agronomy10040583
Chicago/Turabian StyleTossim, Hodo-Abalo, Joel Romaric Nguepjop, Cyril Diatta, Aissatou Sambou, Maguette Seye, Djibril Sane, Jean-François Rami, and Daniel Fonceka. 2020. "Assessment of 16 Peanut (Arachis hypogaea L.) CSSLs Derived from an Interspecific Cross for Yield and Yield Component Traits: QTL Validation" Agronomy 10, no. 4: 583. https://doi.org/10.3390/agronomy10040583
APA StyleTossim, H.-A., Nguepjop, J. R., Diatta, C., Sambou, A., Seye, M., Sane, D., Rami, J.-F., & Fonceka, D. (2020). Assessment of 16 Peanut (Arachis hypogaea L.) CSSLs Derived from an Interspecific Cross for Yield and Yield Component Traits: QTL Validation. Agronomy, 10(4), 583. https://doi.org/10.3390/agronomy10040583




