Poaceae with PGPR Bacteria and Arbuscular Mycorrhizae Partnerships as a Model System for Plant Microbiome Manipulation for Phytoremediation of Petroleum Hydrocarbons Contaminated Agricultural Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of the Native Bacteria of Contaminated Soil
2.2. Mesocosm Experiment
- (1)
- The first set of mesocosms was formed by 50 pots of O. miliaceum (20 seeds per pot) in contaminated soil without the inoculum of endophytic consortium strains selected (Group 1).
- (2)
- The second set of mesocosms consisted of 50 pots of O. miliaceum (20 seeds per pot) in contaminated soil supplemented by the endophytic consortium strains selected (4.0 × 1011–1.0 × 1012 CFU mL−1 g of soil) (Group 2).
- (3)
- The control set consisted of 50 pots of O. miliaceum (20 seeds per pot) with select endophytic consortium strains selected (4.0 × 1011–1.0 × 1012 CFU mL−1 g of soil) in commercial non-contaminated soil (Control Group).
2.3. Plant Analysis
Stress Marker and Antioxidant Enzyme Activity
2.4. Chlorophyll Content
2.5. Biomass
2.6. Quantification of Petroleum Hydrocarbons
2.7. Soil Analysis
2.7.1. Quantification of Petroleum Hydrocarbons
2.7.2. Dehydrogenase Activity
2.8. Statistical Analysis
3. Results
3.1. Screening of the Isolated Hydrocarbon Degraders’ Bacteria
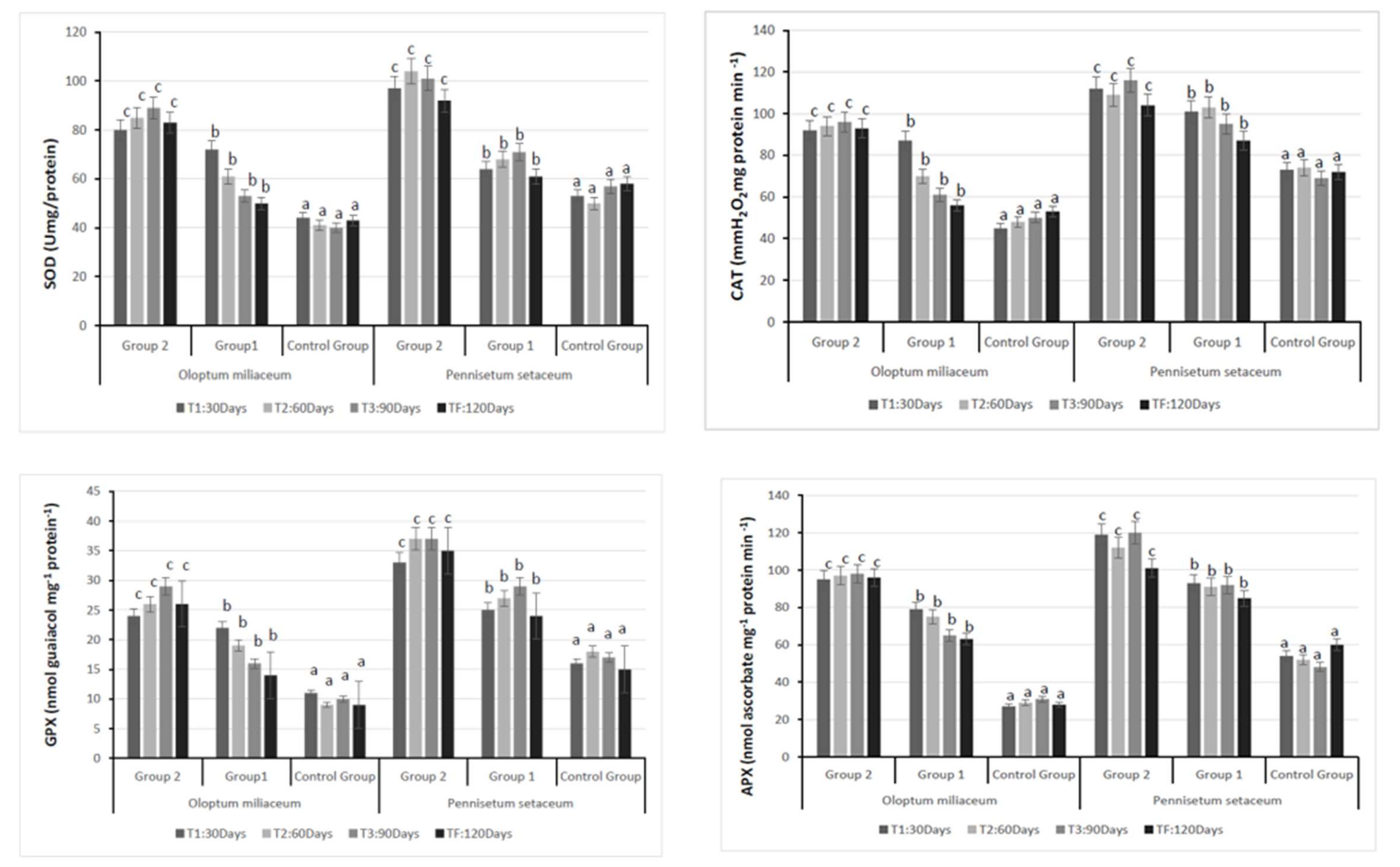
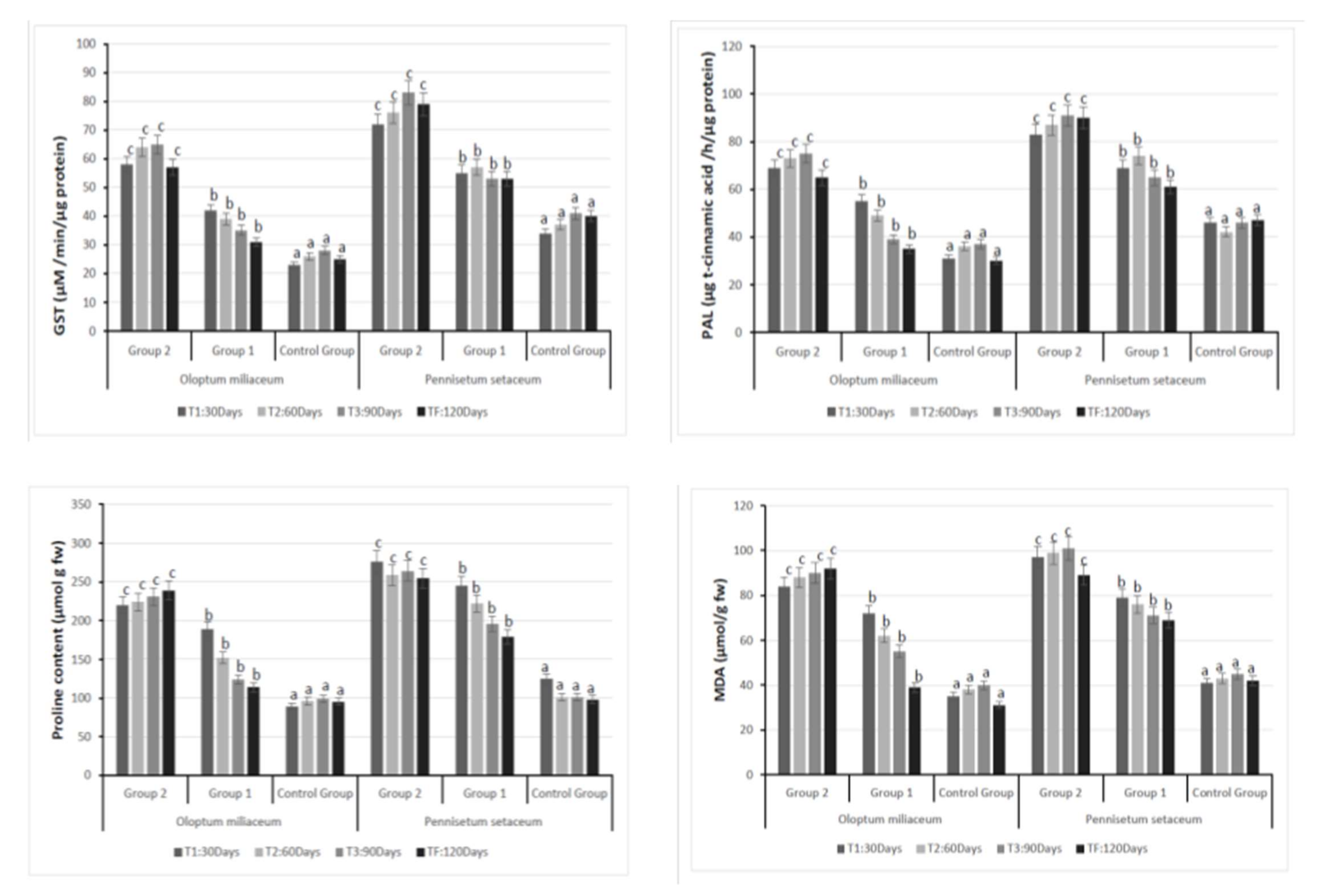
3.2. Plant Analysis: Antioxidant Enzyme Activity and Stress Marker
3.3. Biomass
3.4. Chlorophyll Content
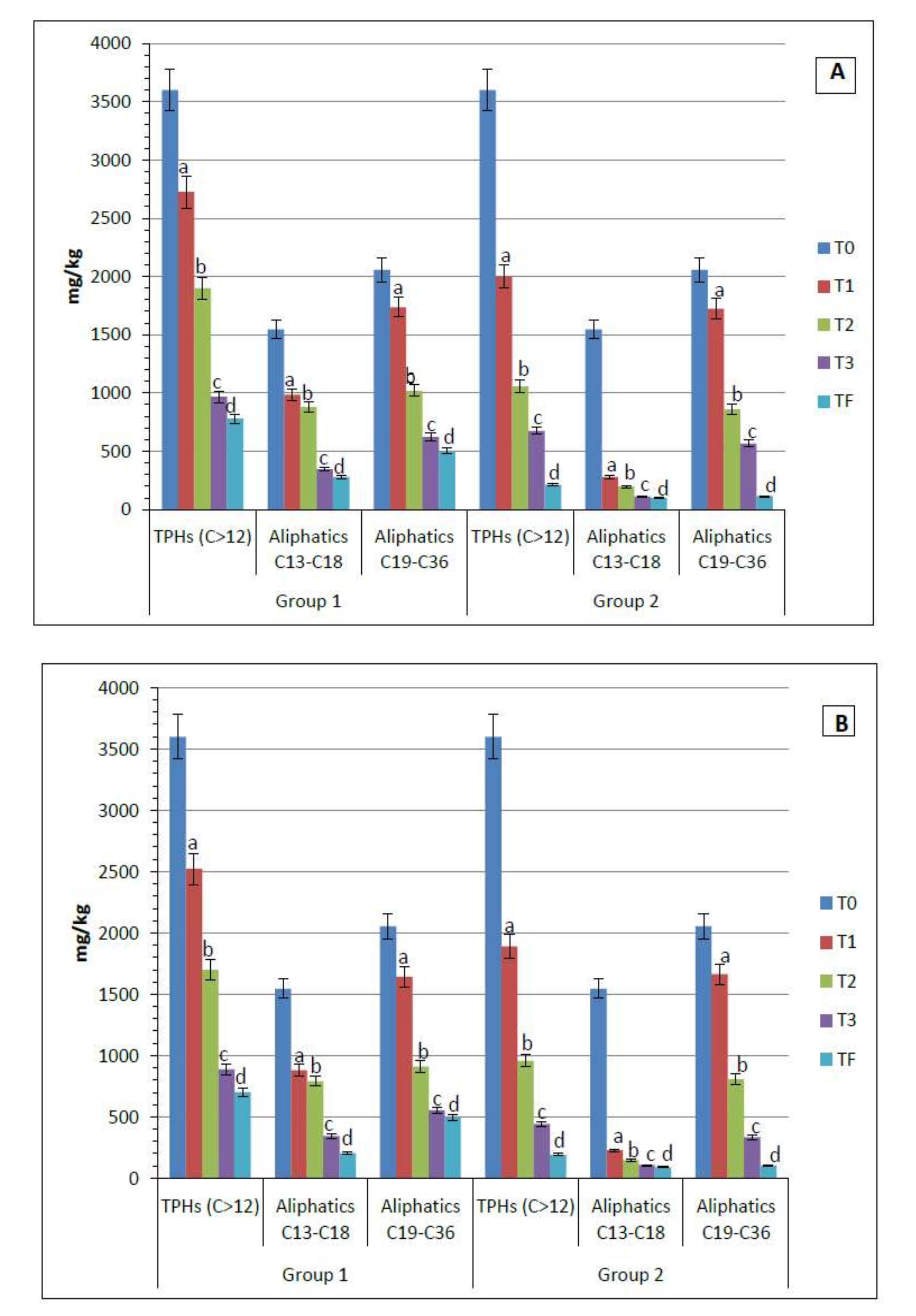
3.5. The Adsorption of TPHs by O. miliaceum and P. setaceum Tissue (Roots and Leaves)
3.6. Soil Analysis: Quantification of Petroleum Hydrocarbons and Dehydrogenase Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Huang, X.D.; El-Alawi, Y.; Gurska, J.; Glick, B.R.; Greenberg, B.M. A multi-process phytoremediation system for decontamination of persistent total petroleum hydrocarbons (TPHs) from soils. Microchem. J. 2005, 81, 139–147. [Google Scholar] [CrossRef]
- Hong, S.H.; Ryu, H.W.; Kim, J.; Cho, K.S. Rhizoremediation of diesel-contaminated soil using the plant growth-promoting rhizobacterium Gordonia sp. S2RP-17. Biodegradation 2011, 22, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, W.; Wang, B.; Wang, Q.; Luo, Y.; Franks, A.E. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, K.E.; Huang, X.D.; Glick, B.R.; Greenberg, B.M. Phytoremediation and rhizoremediation of organic soil contaminants: Potential and challenges. Plant Sci. 2009, 176, 20–30. [Google Scholar] [CrossRef]
- Barrutia, O.; Garbisu, C.; Epelde, L.; Sampedro, M.C.; Goicolea, M.A.; Becerril, J.M. Plant tolerance to diesel minimizes its impact on soil microbial characteristics during rhizoremediation of diesel-contaminated soils. Sci. Total Environ. 2011, 409, 4087–4093. [Google Scholar] [CrossRef]
- Guo, H.; Yao, J.; Cai, M.; Qian, Y.; Guo, Y.; Richnow, H.H.; Blake, R.E.; Doni, S.; Ceccanti, B. Effects of petroleum contamination on soil microbial numbers, metabolic activity and urease activity. Chemosphere 2012, 87, 1273–1280. [Google Scholar] [CrossRef]
- Taiwo, A.M. Composting as A Sustainable Waste Management Technique in Developing Countries. Artic. J. Environ. Sci. Technol. 2011. [Google Scholar] [CrossRef]
- Guarino, C.; Spada, V.; Sciarrillo, R. Assessment of three approaches of bioremediation (Natural Attenuation, Landfarming and Bioagumentation—Assistited Landfarming) for a petroleum hydrocarbons contaminated soil. Chemosphere 2017, 170, 10–16. [Google Scholar] [CrossRef]
- Guarino, C.; Sciarrillo, R. Effectiveness of in situ application of an Integrated Phytoremediation System (IPS) by adding a selected blend of rhizosphere microbes to heavily multi-contaminated soils. Ecol. Eng. 2017, 99, 70–82. [Google Scholar] [CrossRef]
- Ma, X.; Hovy, E. End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, ACL 2016—Long Papers, Berlin, Germany, 7–12 August 2016; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA; Volume 2, pp. 1064–1074.
- Gkorezis, P.; Daghio, M.; Franzetti, A.; Van Hamme, J.D.; Sillen, W.; Vangronsveld, J. The interaction between plants and bacteria in the remediation of petroleum hydrocarbons: An environmental perspective. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Hussain, I.; Puschenreiter, M.; Gerhard, S.; Sani, S.G.A.S.; Khan us din, W.; Reichenauer, T.G. Differentiation between physical and chemical effects of oil presence in freshly spiked soil during rhizoremediation trial. Environ. Sci. Pollut. Res. 2019, 26, 18451–18464. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.X.; Yu, J.; Zhao, H.M.; Cheng, Y.T.; Mo, C.H.; Cai, Q.Y.; Li, Y.W.; Li, H.; Wong, M.H. Efficient phytoremediation of organic contaminants in soils using plant–endophyte partnerships. Sci. Total Environ. 2017, 583, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Wani, P.A.; Omozele, A.B. CR (VI) Removal by Indigenous Klebsiella Species PB6 Isolated from Contaminated Soil under the Influence of Various Factors. Curr. Res. Bacteriol. 2015, 8, 62–69. [Google Scholar] [CrossRef]
- Compant, S.; Saikkonen, K.; Mitter, B.; Campisano, A.; Mercado-Blanco, J. Editorial special issue: Soil, plants and endophytes. Plant Soil 2016, 405, 1–11. [Google Scholar] [CrossRef]
- Ijaz, A.; Imran, A.; Anwar ul Haq, M.; Khan, Q.M.; Afzal, M. Phytoremediation: Recent advances in plant-endophytic synergistic interactions. Plant Soil 2016, 405, 179–195. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Khaksar, G.; Treesubsuntorn, C.; Thiravetyan, P. Effect of endophytic Bacillus cereus ERBP inoculation into non-native host: Potentials and challenges for airborne formaldehyde removal. Plant Physiol. Biochem. 2016, 107, 326–336. [Google Scholar] [CrossRef]
- Bisht, S.; Pandey, P.; Kaur, G.; Aggarwal, H.; Sood, A.; Sharma, S.; Kumar, V.; Bisht, N.S. Utilization of endophytic strain bacillus sp. SBER3 for biodegradation of polyaromatic hydrocarbons (PAH) in soil model system. Eur. J. Soil Biol. 2014, 60, 67–76. [Google Scholar] [CrossRef]
- Kukla, M.; Płociniczak, T.; Piotrowska-Seget, Z. Diversity of endophytic bacteria in Lolium perenne and their potential to degrade petroleum hydrocarbons and promote plant growth. Chemosphere 2014, 117, 40–46. [Google Scholar] [CrossRef]
- Afzal, M.; Yousaf, S.; Reichenauer, T.G.; Sessitsch, A. The Inoculation Method Affects Colonization and Performance of Bacterial Inoculant Strains in the Phytoremediation of Soil Contaminated with Diesel Oil. Int. J. Phytoremediation 2012, 14, 35–47. [Google Scholar] [CrossRef]
- Palmroth, M.R.T.; Pichtel, J.; Puhakka, J.A. Phytoremediation of subarctic soil contaminated with diesel fuel. Bioresour. Technol. 2002, 84, 221–228. [Google Scholar] [CrossRef]
- Kim, J.; Kang, S.-H.; Min, K.-A.; Cho, K.-S.; Lee, I.-S. Rhizosphere Microbial Activity During Phytoremediation of Diesel-Contaminated Soil. J. Environ. Sci. Health Part A 2006, 41, 2503–2516. [Google Scholar] [CrossRef] [PubMed]
- Liste, H.H.; Felgentreu, D. Crop growth, culturable bacteria, and degradation of petrol hydrocarbons (PHCs) in a long-term contaminated field soil. Appl. Soil Ecol. 2006, 31, 43–52. [Google Scholar] [CrossRef]
- Yoshitomi, K.J.; Shann, J.R. Corn (Zea mays L.) root exudates and their impact on 14C-pyrene mineralization. Soil Biol. Biochem. 2001, 33, 1769–1776. [Google Scholar] [CrossRef]
- Tesar, M.; Reichenauer, T.G.; Sessitsch, A. Bacterial rhizosphere populations of black poplar and herbal plants to be used for phytoremediation of diesel fuel. Soil Biol. Biochem. 2002, 34, 1883–1892. [Google Scholar] [CrossRef]
- Basumatary, B.; Bordoloi, S. A study on degradation of heavy metals in crude oil-contaminated soil using Cyperus rotundus. In Phytoremediation: Management of Environmental Contaminants; Springer International Publishing: Cham, Switzerland, 2016; Volume 4, pp. 53–60. ISBN 9783319418117. [Google Scholar]
- Adam, G.; Duncan, H. Influence of diesel fuel on seed germination. Environ. Pollut. 2002, 120, 363–370. [Google Scholar] [CrossRef]
- Dominguez-Rosado, E.; Pichtel, J. Transformation of Fulvic Substances in the Rhizosphere during Phytoremediation of Used Motor Oil. J. Environ. Sci. Health Part A 2004, 39, 2369–2381. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Saravana Babu, S. Assessment Studies on River Pollution by Textile Dyes and Heavy Metals around Bhavani-Komaraplalayam, Tamilnadu, India. Int. J. Curr. Res. Biosci. Plant Biol. 2015, 2, 210–213. [Google Scholar]
- De Oliveira, D.M.; Finger-Teixeira, A.; Rodrigues Mota, T.; Salvador, V.H.; Moreira-Vilar, F.C.; Correa Molinari, H.B.; Craig Mitchell, R.A.; Marchiosi, R.; Ferrarese-Filho, O.; Dantas dos Santos, W. Ferulic acid: A key component in grass lignocellulose recalcitrance to hydrolysis. Plant Biotechnol. J. 2015, 13, 1224–1232. [Google Scholar] [CrossRef]
- Pignatti, S.; Guarino, R.; La Rosa, M. Flora d’Italia; Ed. Agricole: Milan, Italy, 2017; Volume 21, ISBN 9788850652426. [Google Scholar]
- Guarino, C.; Paura, B.; Sciarrillo, R. Enhancing phytoextraction of HMs at real scale, by combining salicaceae trees with microbial consortia. Front. Environ. Sci. 2018, 6, 1–11. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Wyszkowski, M.; Ziólkowska, A. Role of compost, bentonite and calcium oxide in restricting the effect of soil contamination with petrol and diesel oil on plants. Chemosphere 2009, 74, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Al-Baldawi, I.A.; Abdullah, S.R.S.; Anuar, N.; Suja, F.; Mushrifah, I. Phytodegradation of total petroleum hydrocarbon (TPH) in diesel-contaminated water using Scirpus grossus. Ecol. Eng. 2015, 74, 463–473. [Google Scholar] [CrossRef]
- Escalante-Espinosa, E.; Gallegos-Martínez, M.E.; Favela-Torres, E.; Gutiérrez-Rojas, M. Improvement of the hydrocarbon phytoremediation rate by Cyperus laxus Lam. inoculated with a microbial consortium in a model system. Chemosphere 2005, 59, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Chaîneau, C.H.; Yepremian, C.; Vidalie, J.F.; Ducreux, J.; Ballerini, D. Bioremediation of a crude oil-polluted soil: Biodegradation, leaching and toxicity assessments. Water. Air. Soil Pollut. 2003, 144, 419–440. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Lotfinasabasl, S.; Gunale, V.R.; Rajurkar, N.S. Petroleum Hydrocarbons Pollution in Soil and its Bioaccumulation in mangrove species, Avicennia marina from Alibaug Mangrove Ecosystem, Maharashtra, India. Int. J. Adv. Res. Technol. 2013, 2. [Google Scholar]
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of plant growth by ACC deaminase-producing soil bacteria. In New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research; Springer: Dordrecht, The Netherlands; pp. 329–339. ISBN 9781402067761.
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Glick, B.R. Phytoremediation. In Beneficial Plant-Bacterial Interactions; Springer International Publishing: Cham, Switzerland, 2015; pp. 191–221. [Google Scholar]
- Joner, E.J.; Hirmann, D.; Szolar, O.H.J.; Todorovic, D.; Leyval, C.; Loibner, A.P. Priming effects on PAH degradation and ecotoxicity during a phytoremediation experiment. Environ. Pollut. 2004, 128, 429–435. [Google Scholar] [CrossRef]
- Namkoong, W.; Hwang, E.Y.; Park, J.S.; Choi, J.Y. Bioremediation of diesel-contaminated soil with composting. Environ. Pollut. 2002, 119, 23–31. [Google Scholar] [CrossRef]
- Kaimi, E.; Mukaidani, T.; Miyoshi, S.; Tamaki, M. Ryegrass enhancement of biodegradation in diesel-contaminated soil. Environ. Exp. Bot. 2006, 55, 110–119. [Google Scholar] [CrossRef]
- Kaimi, E.; Mukaidani, T.; Tamaki, M. Effect of rhizodegradation in diesel-contaminated soil under different soil conditions. Plant Prod. Sci. 2007, 10, 105–111. [Google Scholar] [CrossRef]
- Kaimi, E.; Mukaidani, T.; Tamaki, M. Screening of twelve plant species for phytoremediation of petroleum hydrocarbon-contaminated soil. Plant Prod. Sci. 2007, 10, 211–218. [Google Scholar] [CrossRef]
- Chakraborty, J.; Das, S. Molecular perspectives and recent advances in microbial remediation of persistent organic pollutants. Environ. Sci. Pollut. Res. 2016, 23, 16883–16903. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, W.W. Rhizosphere processes and management in plant-assisted bioremediation (phytoremediation) of soils. Plant Soil 2009, 321, 385–408. [Google Scholar] [CrossRef]
- Shekhar, S.; Sundaramanickam, A.; Balasubramanian, T. Biosurfactant Producing Microbes and their Potential Applications: A Review. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1522–1554. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Jhaveri, D.J.; Marshall, V.M.; Bauer, D.C.; Edson, J.; Narayanan, R.K.; Robinson, G.J.; Lundberg, A.E.; Bartlett, P.F.; Wray, N.R.; et al. A comparative study of techniques for differential expression analysis on RNA-seq data. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Juwarkar, A.A.; Singh, S.K.; Mudhoo, A. A comprehensive overview of elements in bioremediation. Rev. Environ. Sci. Biotechnol. 2010, 9, 215–288. [Google Scholar] [CrossRef]
- Zhang, T.; Lan, R.; Chan, C.-F.; Law, G.-L.; Wong, W.-K.; Wong, K.-L. In vivo selective cancer-tracking gadolinium eradicator as new-generation photodynamic therapy agent. Proc. Natl. Acad. Sci. USA 2014, 111, E5492–E5497. [Google Scholar] [CrossRef]

| Bacterial Isolates | Production IAA | Siderophores Release | Exopolysaccharides Production | Production of Ammonia |
|---|---|---|---|---|
| Acetobacterium woodii | - | - | - | - |
| Achromobacter marplatensis | - | - | - | - |
| Achromobacter spanius | + | - | - | - |
| Achromobacter xylosoxidans | ++ | - | - | ++ |
| Comamonas koreensis | +++ | +++ | +++ | +++ |
| Comamonastesto steroni | ++ | ++ | ++ | ++ |
| Comamonas aquatica | ++ | ++ | ++ | + |
| Ochrobactrum anthropi | +++ | +++ | +++ | +++ |
| Pseudomonas aeruginosa | +++ | +++ | +++ | +++ |
| Pseudomonas aeruginosa PA7 | ++ | ++ | ++ | + |
| Pseudomonas stuzeri | ++ | ++ | + | + |
| Pseudomonas mendocina NK-01 | + | - | + | - |
| Pseudomonas resinovorans | - | - | - | - |
| Pseudomonas resinovorans NBRC 106553 | - | - | - | - |
| Pseudomonas fluorescens | +++ | +++ | +++ | +++ |
| Pseudomonas fluorescens Pf0-1 | ++ | ++ | ++ | ++ |
| Pseudomonas fluorescens SBW25 | ++ | ++ | ++ | ++ |
| Pseudomonas fluorescens F113 | + | + | + | + |
| Pseudomonas poae | - | ++ | - | - |
| Pseudomonas putida | +++ | +++ | +++ | +++ |
| Pseudomonas putida H8234 | - | ++ | - | - |
| Pseudomonas putida NBRC 14164 | - | - | - | - |
| Pseudomonas putida GB-1 | ++ | ++ | - | - |
| Pseudomonas putida HB3267 | + | - | - | - |
| Pseudomonas fulva 12-X | - | + | + | - |
| Pseudomonas stuzeri DSM 10701 | + | - | - | ++ |
| Pseudomonas stuzeri DSM 4166 | + | - | - | + |
| Pseudomonas stuzeri A1501 | - | - | - | - |
| Pseudomonas syringaepv.tomato | - | - | ++ | ++ |
| Pseudomonas brassicacearum | + | + | - | - |
| Pseudomonas pertucinogena group | + | + | - | - |
| Rhodococcus opacus B4 | - | ++ | - | - |
| Rhodococcus jostii | - | +++ | - | - |
| Paenibacillus mucilaginosus K02 | ++ | ++ | - | - |
| Paenibacillus mucilaginosus 3016 | ++ | ++ | - | - |
| Paenibacillus polymyxa | +++ | - | - | - |
| Bacilluscereus group | +++ | +++ | +++ | +++ |
| Deinococcus proteolyticus | - | - | - | - |
| Bradyrhizobium japonicum USDA6 | - | ++ | ++ | - |
| Bradyrhizobium japonicum | - | ++ | ++ | - |
| Bradyrhizobium diazoefficiens | - | + | + | - |
| Bradyrhizobium oligotrophicum | - | + | ++ | - |
| Bradyrhizobium oligotrophicum S58 | - | + | ++ | - |
| Nitrobacter hamburgensis | - | - | - | - |
| Nitrobacter hamburgensis X14 | - | - | - | - |
| Rhizobium leguminosaurum | +++ | - | - | - |
| Biomass (Dry Weight (g plant−1)) | ||||
|---|---|---|---|---|
| Roots | Leaves | Roots | Leaves | |
| Oloptum miliaceum | Pennisetum setaceum | |||
| Group 1 | ||||
| T1 | 34.1 ± 9a | 25 ± 5a | 37.3 ± 3a | 27.7 ± 4a |
| T2 | 45.2 ± 5b | 36 ± 10b | 72.1 ± 5b | 49.9 ± 5b |
| T3 | 150 ± 10c | 49.6 ± 5c | 160.3 ± 14c | 69.3 ± 3c |
| TF | 190 ± 5d | 77.8 ± 10d | 201.3 ± 10d | 71.2 ± 5d |
| Group 2 | ||||
| T1 | 34.2 ± 10a | 36.4 ± 6a | 38.5 ± 5a | 41.3 ± 4a |
| T2 | 71.8 ± 11b | 52.6 ± 10b | 88.4 ± 8b | 73.7 ± 5 b |
| T3 | 234 ± 10c | 71.9 ± 6c | 189.8 ± 11c | 91.1 ± 7c |
| TF | 285 ± 15d | 118 ± 4d | 291.6 ± 15d | 97.6 ± 5d |
| Control Group | ||||
| T1 | 50.9 ± 5a | 55 ± 5a | 48.4 ± 5a | 44.7 ± 5a |
| T2 | 98.1 ± 5b | 69 ± 5b | 101.4 ± 10b | 66.7 ± 4b |
| T3 | 273 ± 10c | 89 ± 10c | 245.6 ± 15c | 89.6 ± 5 c |
| TF | 312 ± 10d | 113 ± 8d | 287.7 ± 12d | 101.3 ± 10d |
| Chlorophyll Content (mg g−1 fw) | ||||||
|---|---|---|---|---|---|---|
| Chlorophyll a | Chlorophyll b | Chlorophyll a + b | Chlorophyll a | Chlorophyll b | Chlorophyll a + b | |
| Oloptum miliaceum | Pennisetum setaceum | |||||
| Group 1 | ||||||
| T1 | 0.85 ± 0.01a | 0.15 ± 0.02a | 1.00 ± 0.03a | 0.79 ± 0.01a | 0.19 ± 0.02a | 0.98 ± 0.01a |
| T2 | 0.82 ± 0.01a | 0.13 ± 0.01a | 0.95 ± 0.02a | 0.76 ± 0.02a | 0.16 ± 0.01a | 0.92 ± 0.02a |
| T3 | 0.79 ± 0.02a | 0.11 ± 0.01a | 0.90 ± 0.01a | 0.84 ± 0.01a | 0.18 ± 0.01a | 1.02 ± 0.04a |
| TF | 0.81 ± 0.01a | 0.18 ± 0.01a | 0.99 ± 0.01a | 0.86 ± 0.01a | 0.16 ± 0.01a | 1.02 ± 0.02a |
| Group 2 | ||||||
| T1 | 1.23 ± 0.04a | 0.19 ± 0.01a | 1.42 ± 0.02a | 0.97 ± 0.02 a | 0.22 ± 0.01 a | 1.19 ± 0.05a |
| T2 | 1.37 ± 0.01a | 0.23 ± 0.02a | 1.60 ± 0.05b | 1.18 ± 0.05b | 0.26 ± 0.02a | 1.44 ± 0.04b |
| T3 | 1.45 ± 0.02b | 0.31 ± 0.01b | 1.76 ± 0.05c | 1.27 ± 0.04c | 0.26 ± 0.01a | 1.53 ± 0.03c |
| TF | 1.59 ± 0.01c | 0.33 ± 0.01c | 1.92 ± 0.05d | 1.36 ± 0.03d | 0.30 ± 0.01a | 1.66 ± 0.05d |
| Control Group | ||||||
| T1 | 1.44 ± 0.02a | 0.27 ± 0.02a | 1.71 ± 0.05a | 1.11 ± 0.05a | 0.29 ± 0.02 a | 1.40 ± 0.05a |
| T2 | 1.49 ± 0.02a | 0.30 ± 0.01a | 1.79 ± 0.04b | 1.25 ± 0.03a | 0.32 ± 0.01a | 1.57 ± 0.04b |
| T3 | 1.57 ± 0.03a | 0.31 ± 0.01a | 1.88 ± 0.03b | 1.35 ± 0.04a | 0.37 ± 0.01a | 1.72 ± 0.03c |
| TF | 1.62 ± 0.02a | 0.39 ± 0.01a | 2.01 ± 0.04a | 1.44 ± 0.01a | 0.40 ± 0.02a | 1.84 ± 0.03d |
| Groups | DHA (mg Formazan 100 g soil dw−1 6 h−1) | ||||
|---|---|---|---|---|---|
| T0 | T1 | T2 | T3 | TF | |
| Oloptum miliaceum | |||||
| Group 1 | 3.3 ± 0.05a | 15.8 ± 0.09a | 26.3 ± 1.08 | 60.8 ± 2.21a | 46.3 ± 1.35a |
| Group 2 | 3.5 ± 0.06b | 23.3 ± 1.01b | 32.8 ± 1.11b | 77.5 ± 2.10b | 67.8 ± 1.15b |
| Control Group | 3.5 ± 0.05 | 14.3 ± 0.05 | 35.1 ± 0.05 | 69.3 ± 0.05 | 75.7 ± 0.05 |
| Pennisetum setaceum | |||||
| Group 1 | 3.2 ± 0.05a | 7.6 ± 0.05a | 15.9 ± 0.02a | 64.9 ± 0.03a | 56.7 ± 0.05a |
| Group 2 | 3.6 ± 0.04b | 26.4 ± 0.02b | 30.1 ± 0.02b | 79.3 ± 0.04b | 70.1 ± 0.05b |
| Control Group | 3.5 ± 0.05 | 14.3 ± 0.05 | 35.1 ± 0.05 | 69.3 ± 0.05 | 75.7 ± 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guarino, C.; Marziano, M.; Tartaglia, M.; Prigioniero, A.; Postiglione, A.; Scarano, P.; Sciarrillo, R. Poaceae with PGPR Bacteria and Arbuscular Mycorrhizae Partnerships as a Model System for Plant Microbiome Manipulation for Phytoremediation of Petroleum Hydrocarbons Contaminated Agricultural Soils. Agronomy 2020, 10, 547. https://doi.org/10.3390/agronomy10040547
Guarino C, Marziano M, Tartaglia M, Prigioniero A, Postiglione A, Scarano P, Sciarrillo R. Poaceae with PGPR Bacteria and Arbuscular Mycorrhizae Partnerships as a Model System for Plant Microbiome Manipulation for Phytoremediation of Petroleum Hydrocarbons Contaminated Agricultural Soils. Agronomy. 2020; 10(4):547. https://doi.org/10.3390/agronomy10040547
Chicago/Turabian StyleGuarino, Carmine, Mario Marziano, Maria Tartaglia, Antonello Prigioniero, Alessia Postiglione, Pierpaolo Scarano, and Rosaria Sciarrillo. 2020. "Poaceae with PGPR Bacteria and Arbuscular Mycorrhizae Partnerships as a Model System for Plant Microbiome Manipulation for Phytoremediation of Petroleum Hydrocarbons Contaminated Agricultural Soils" Agronomy 10, no. 4: 547. https://doi.org/10.3390/agronomy10040547
APA StyleGuarino, C., Marziano, M., Tartaglia, M., Prigioniero, A., Postiglione, A., Scarano, P., & Sciarrillo, R. (2020). Poaceae with PGPR Bacteria and Arbuscular Mycorrhizae Partnerships as a Model System for Plant Microbiome Manipulation for Phytoremediation of Petroleum Hydrocarbons Contaminated Agricultural Soils. Agronomy, 10(4), 547. https://doi.org/10.3390/agronomy10040547






