1. Introduction
One of the most important soil components, which is closely related to the activity of microorganisms, is soil organic matter (SOM). SOM consists of all organic substances originating from living organisms in different phases of decomposition. The sources of organic matter in soil are mainly the dead parts of plants and animals, microorganisms and metabolites produced during their growth and decomposition, as well as humic compounds [
1]. Microorganisms play a crucial role in the decomposition of organic matter and the global cycle of elements, and they are involved in the breakdown of complex organic compounds into compounds, which are simple and more easily absorbed by plants. Organic matter (OM) is a key relevant factor responsible for the physical, chemical and biological properties of soil. The content of OM in agricultural soils is gradually decreasing as a result of its intensive use [
2]. It is estimated that during the postindustrial era (since 1850) under the influence of cultivation and soil erosion on a global scale 42–78 gigatonnes (Gt) of carbon have already been lost [
3]. Even a small loss of SOM may result in the deterioration of soil structure and decreases in soil quality. Furthermore, maintaining resources of OM at an appropriate level is important not only due to the production function of soils, but also due to the role of soil in the sequestration of carbon dioxide from the atmosphere, thereby contributing to a reduction in the greenhouse effect [
2].
Exogenous organic matter (EOM) is defined as the whole of the OM that is introduced into the soil in order to improve its fertility, quality and the potential of the land for non-agricultural use [
4,
5]. EOM includes a very wide range of biowastes from various sources [
6]. Sources of EOM include: natural fertilizers (manure, slurry), post-harvest residues, green manures, composted materials from industrial and municipal waste, municipal and industrial waste (sewage sludge, digestate from biogas plants) or waste of animal origin (animal meal) [
7,
8]. These materials are characterized by different physical and chemical properties, differing humification rates and the abilities to produce soil organic matter. Recycling biowaste as an organic fertilizer gives an opportunity to decrease disposal at landfill, and also to reduce greenhouse gas emissions [
9,
10]. The annual production of biowaste in the European Union is estimated at 1.6 billion tons, of which 61% is waste produced by animal breeding, 25% originates from plant residues, 7% is industrial waste and 7% is municipal waste (sewage sludge, biowaste, plant waste). Despite such a large production of biowaste, the most common of all of the organic materials introduced into the soil (97%) is manure and slurry, with only a minor contribution made by industrial waste (2%) and sewage sludge (1%) [
11]. The addition of OM to the soil beneficially affects the degree of aggregation and compaction, water infiltration and water-retention capacity, volumetric density as well as the resistance of the soil to water and wind erosion [
12,
13,
14,
15,
16]. It is commonly known that the use of plant residues, manure and composts obtained from organic waste causes an increase of SOM, which in turn may have a positive effect on crop yields through direct nutrient supply or indirectly through the modification of soil physical properties [
17,
18]. SOM plays one more important function, influencing on the microbial biomass carbon (MBC) [
12]. For this reason, it is necessary to conduct research concerning the impact of different types of EOMs on the functioning and properties of soil. Assessing the impact of various types of EOM amendments should be carried out on the basis of short and long-term experiments to determine the physical, chemical and biological properties of the soil.
The addition of OM may have an influence on soil productivity, the availability of nutrients and processes associated with the mineralization of organic compounds [
19,
20]. Organic wastes, which are used as soil additives constitute a source of energy for microorganisms, and also cause changes in their diversity and activity [
21,
22]. An assessment of the effect EOM additives on soil microbiome diversity may be performed on the basis of genetic, metabolic and enzymatic activity assays [
19,
23] the results of which may be used as an indicator of the shifts that occur due to the influence of applied waste [
24,
25]. Unfortunately, the use of organic waste as a soil fertilizer also carries a certain risk as well as producing significant benefits. Organic fertilizers have been used for a relatively long period of time, but new types of waste are still being produced, for example digestate [
26], which requires appropriate levels of control and monitoring. The development of laboratory techniques, including molecular biology methods, allows for the detailed analyses of the microbial properties of organic waste. Furthermore, the range and types of potential EOM risk depend on the raw material from which they were prepared. The main threats resulting from EOM application are: (i) chemical pollution: heavy metals, pesticides, polycyclic aromatic hydrocarbons (PAHs), phthalates, volatile organic compounds, phenolic compounds [
26,
27]; (ii) contamination with pathogenic and phytopathogenic microorganisms and toxins [
19]; (iii) an unbalanced proportion of C and N [
26]. All of these threats have given rise to the necessity to conduct detailed research monitoring the impact of applied EOM on soils suitable for agriculture. The assessment of soil biological activity as well as the genetic and functional diversity of the soil microbiome is an important approach in the context of shifts occurring in the soil environment and it could also be a biological indicator of its ecological status [
22,
28,
29].
The effect of amendments with organic matter sources on the microbiological properties could be dependent on the type and doses of organic material. Therefore, the overall objective of the present research was to perform a study to explain shifts in the soil microbiome after the application of different types and doses of tested EOMs (compost from sewage sludge (Cof); industrial compost from households wastes (Cdw), animal meal (Man); digestate from a biogas fry factory (Dgf); digestate from biogas plant from the sugar factory (Dgs); and digestate from an agricultural biogas plant (Dga)). The goal of the study was to determine the influence of different types and doses of EOM incorporated into the soil on its ecological status by assessing the changes in microbial functional activity and genetic diversity. Moreover, the purpose of the implementation of this study was the necessity for rational disposal locally of wastes and organic residues which are rich sources of OM and could be an alternative to mineral fertilizers.
2. Materials and Methods
2.1. Study Site, Experimental and Pot Design and Soil-Sampling Strategy
The experiment was conducted from late spring to early autumn in a greenhouse, under natural illumination. Soil to set up the experiment was collected from 0–30 cm depth of a
Luvisol located at Lower Silesia (Poland) (49°54′26.7″ N, 19°12′57.2″ N). Several samples were taken at random from a surface area of 100 m
2, mixed and then homogenized after sieving (≤2 mm). The soil had a pH H
2O value of 5.78 and the soil C
org content was 0.77% [
30]. To control and minimize the heterogeneity of soil to set up a pot experiment we avoided collecting soils that are too wet to minimize soil compaction and avoided collecting soils that are too dry to aid in separating soil from roots and other residues. The pot experiment was set up in a greenhouse in the Institute of Soil Science and Plant Cultivation (IUNG) in Pulawy. The experiment was performed in four biological replications for each treatment and control in a plastic pots that were filled with 3.5 kg of soil. Six different sources of EOM were applied at 2 rates giving in total 12 treatments. EOM sources are described in detail below. The dose of EOM for each treatment was determined after considering the nitrogen required for each dose and the dry matter content and they accounted for 50% (50% N from EOM and 50% from mineral N) and 100% (100% N from EOM) of the total requirement for nitrogen fertilization. In wheat cultivation, the required nitrogen dose is 140 kg ha
−1 of the soil, therefore doses of EOM for each treatment were calculated taking into consideration this value of nitrogen as a total 100% dose of N. Moreover, soil without EOM additions was used as a control object, in which full nitrogen fertilization rate was supplied from mineral fertilization with NH
4NO
3 (100% from mineral N, EOM dose 0%). The calculated doses of EOM were entirely well mixed with the soil, the soil with additives was homogenized through intensive mixing, immediately filling the particular pots. Seeds of the spring wheat variety of Tybalt were sown. This variety has been chosen due to its characteristics: excellent resistance to mildew, great flexibility in drilling date, high yields, morphology of plant: shorts plants with stiff straw, the strong position of this variety in Western Europe, and also its recommendation for UK, France and Germany.
Soil samples were collected after harvesting, 90 days after the application of EOM. The experiment was carried out to allow time for decomposition of used organic materials and taking into account the duration of the vegetative stage of spring wheat (after 90 days a spring wheat is heading into the development stage). Fresh soil samples were taken from each pot separately, and the experiment was designed for four biological replications for each of tested treatments. Directly after collection, the soil samples were immediately used for measurements or stored at 4 °C for dehydrogenases activity (DHA) and community-level physiological profiling (CLPP) analyses or at −80 °C for DNA extraction.
2.2. Sources of Exogenous Organic Matter
The organic wastes tested in the experiment were residues from local factories e.g., a sugar factory, a fry factory, or they were made from organic, compostable waste originating from the Polish–Czech borderland region. The reasons of this choice was rational waste management at the place of its generation.
2.3. Characteristics of the Tested Exogenous Organic Matter (EOM)
Compost (Cof)—organic fertilizer, produced by the homogenization and composting of materials containing degradable organic substances, sewage sludge from the treatment of waste waters, sawdust, biodegradable garden and park waste, soil, mouldings of medicinal plants (wastes from solvent extraction), lime sludge.
Compost (Cdw)—organic fertilizer, made from separated biodegradable wastes from households such as kitchen wastes (food, vegetable and fruit wastes), garden and park waste (grass, leaves, weeds, soil) and other biodegradable wastes (wood ash, sawdust etc.), wood wastes and sewage sludge from communal sewage disposal plants.
Animal meal (Man)—organic fertilizer produced from animal by-products (classified as category 2 and 3 materials) by techniques that comply with Regulation (EC) No 1069/2009 of the European Parliament and Council.
Digestate (Dgf)—digestate from a biogas plant fed by wastes from a fry factory. The main ingredients are potato peelings, misshapen and defective fries and excess sludge.
Digestate (Dgs)—digestate from a biogas plant fed with 100% sugar beet pulp. The biogas plant is located next to the sugar factory in Poland.
Digestate (Dga)—digestate from an agricultural biogas plant fed with silage maize (70% of the input) and slurry (30% of the input).
Each organic residue was thoroughly homogenized and analysed before use (
Table 1).
2.4. Community-Level Physiological Profiling (CLPP) Analysis
The characterization of the CLPPs was based on the metabolic properties of the soil microbiome obtained using Biolog ECO plates [
31] for the bacterial community, Biolog FF plates for the aerobic fungal community, and Biolog AN plates for the anaerobic community. The Biolog system is a useful technique, which is used to assess the shifts in the microbial community functional diversity of the soil. Portions of the sieved soil (1 g) were shaken with 99 mL of sterile saline peptone water for 20 min at 20 °C and subsequently incubated at 4 °C for 30 min. Then the soil microorganism’s suspension was inoculated into each well of the ECO plates (120 µL), FF and AN plates (100 µL) and incubated at 27 °C. AN plates were incubated under anaerobic conditions, in special jars with a gas mixture containing 5% H
2, 10% CO
2, 85% N
2, gas was injected into each jar using an Anoxomat apparatus (MART Microbiology B.V., USA). The microbial functional profile was recorded by measuring the absorbance (590 nm) during 240 incubation hours at 24 h intervals. The most consistent readings allowing good resolution among the treatments came from the 120 incubation hour Biolog Plates and these were used in the analyses. Based on absorbance measurements the average well colour development (AWCD) [
32] or average well density development (AWDD) [
33] were determined. These parameters are factors of the overall metabolic activity of environmental microorganisms. Moreover, biodiversity indices such as Shannon (H) and Simpson (S) were calculated following [
34].
2.5. Terminal Restriction Fragment Length Polymorphism Analysis (T-RFLP) of Ammonia-Oxidizing Archaea (AOA)
The relative abundance of AOA within the total DNA community of treatments was characterized through a T-RFLP analysis for relative quantification of individual terminal restriction fragments within the polymerase chain reaction (PCR) products. The T-RFLP approach is recommended as a reliable technique for environmental research to show similar general community patterns and offers a solid alternative for comprehensive assessment of microbial communities [
35,
36]. The functional
amoA gene coding the α-subunit of the ammonia monooxygenase was used as a biomarker. Genomic DNA was extracted from a 0.5 g of soil sample using a FastDNA SPIN Kit for Faeces (MP Biomedicals, Solon, OH, USA) according to the manufacturer’s protocol.
PCR was performed in a total volume of 30 μL containing 4 ng of DNA template, 15 μL RedTaq
® ReadyMix™ PCR Reaction Mix (Sigma-Aldrich, St. Louis, MO, USA). The concentration of the primers used for PCR was 0.5 μM. Forward primers: amoA 19F—labelled with 6-carboxyfluorescein- FAM on 5′ end (5′-ATG GTC TGG CTW AGA CG-3′) and reverse primer: amo 643R—(5′-TCC CAC TTW GAC CAR GCG GCC ATC CA-3′) [
37]. The amplification reaction was performed as follows: 5 min at 92 °C, 35 cycles of 45 sec at 92 °C, 30 sec at 59 °C, 1 min at 72 °C, 7 min at 72 °C. The PCR products (700 bp) were purified with ExoSAP-IT
® PCR Products Purification Kit for ABI (Affymetrix Inc., Santa Clara, CA, USA). The total volume of the restriction mixture was 10 μL and it contained approximately 50 ng DNA, 0.6 μL of buffer Tango (Fermentas
® International, Burlington, ON, Canada), and 0.6 μL of restriction enzyme (10 U/μL)
AluI or
Csp6I (Fermentas
® International, Burlington, ON, Canada), the mixture was incubated at 37 °C for 2 h, subsequently, the reaction was terminated by incubation at 65 °C for 20 min. 1 μL of digested product was mixed with 9 μL of deionized formamide and 0.5 μL of DNA fragment length standard (GS-600LIZ, ABI) (Applied Biosystems, Foster City, CA, USA) and placed on a 96-well plate. The samples were denatured at 94 °C for 3 min and then snap-cooled with ice. The fluorescently labelled terminal restriction fragments (T-RFs) were run through an ABI 3130 xl capillary sequencer (Applied Biosystems, Foster City, CA, USA) in the GeneScan mode. T-RFLP data were analysed using GeneMaper
® Software v.4.0 (Applied Biosystems, Foster City, CA, USA).
2.6. Enzymatic Activity
DHA was determined according to the Thalmann method [
38] with TTC (triphenyl tetrazolium chloride) used as a substrate, in Tris-HCl buffer (pH 7.4). Incubation was conducted for 96 h at 30 °C. Enzymatic activity was determined spectrophotometrically at 485 nm, with reference to methanol.
2.7. Statistical Analysis
Statistical analysis was performed with Statistica Software (version 13. software, StatSoft Inc., Tulsa, OK, USA, 2011). Data (AWCD/AWDD/DHA/H/S) were evaluated by analysis of variance (ANOVA) and following the significant differences between treatment were estimated by the post-hoc Tukey HSD (honestly significant differences) test at the p < 0.05 significance level.
The most frequently used form of analysis and comparison of the obtained genetic and metabolic profiles is statistical cluster analysis (the determination of the relationship between the populations using a dendrogram) [
39]. Therefore, multivariate cluster analysis was used to assess the level of substrates utilization in Biolog Plates. Moreover, to show potential patterns in the dataset of each treatment and control objects, data visualizations were presented as heat maps. Moreover, dendrograms were prepared on the basis of cluster analysis from the normalized metabolic and genetic data and representing the scaled similarity (%) on the axis (Ward’s method and the unweighted pair group method with arithmetic mean, UPGMA) and the boundary marked according to Sneath’s criteria (restrictive 33% and less restrictive 66%). The data were also evaluated following multidimensional scaling to detect additional relationships between treatments. Differences between treatments were analyzed by principal component analysis (PCA) based on the results of community level physiological profiling and T-RFLP data.
3. Results
The analysis of the functional diversity of the soil microorganisms included the following communities: bacteria, filamentous fungi and anaerobic microorganisms. The calculated results are presented in
Table 2.
The highest values of AWCD/AWDD and Shannon index were recorded for fungal communities (based on FF plates) while rather low activity was observed for anaerobic communities (based on AN plates). The opposite results were obtained for the Simpson index, indicating in general the highest values for anaerobic and the lowest for fungal communities. However, only in fungal communities did the values of S index differ significantly between treatments, with the highest value after the addition of Cof 50. The functional activity calculated based on the FF plates was very diverse for treatments with EOMs (exception Man). However, in 50% doses a significant reduction in diversity was observed. A slight decrease in activity was recorded for treatments with EOMs in 100% doses. With reference to the activity of bacterial communities (based on ECO plates) the same trend as for fungal communities was observed; the higher activity was observed for treatments with EOMs with doses of 100% compared to the 50% dose, but this change was not statistically significant. Furthermore, the lowest microbial activity was noted for anaerobic communities (with EOM at a dose of 50% as well as a dose of 100%). In order to obtain more information about the similarities or differences between the microbial communities inhabiting the tested soil, clustering analyses were performed. When the bacterial communities were taken into consideration (
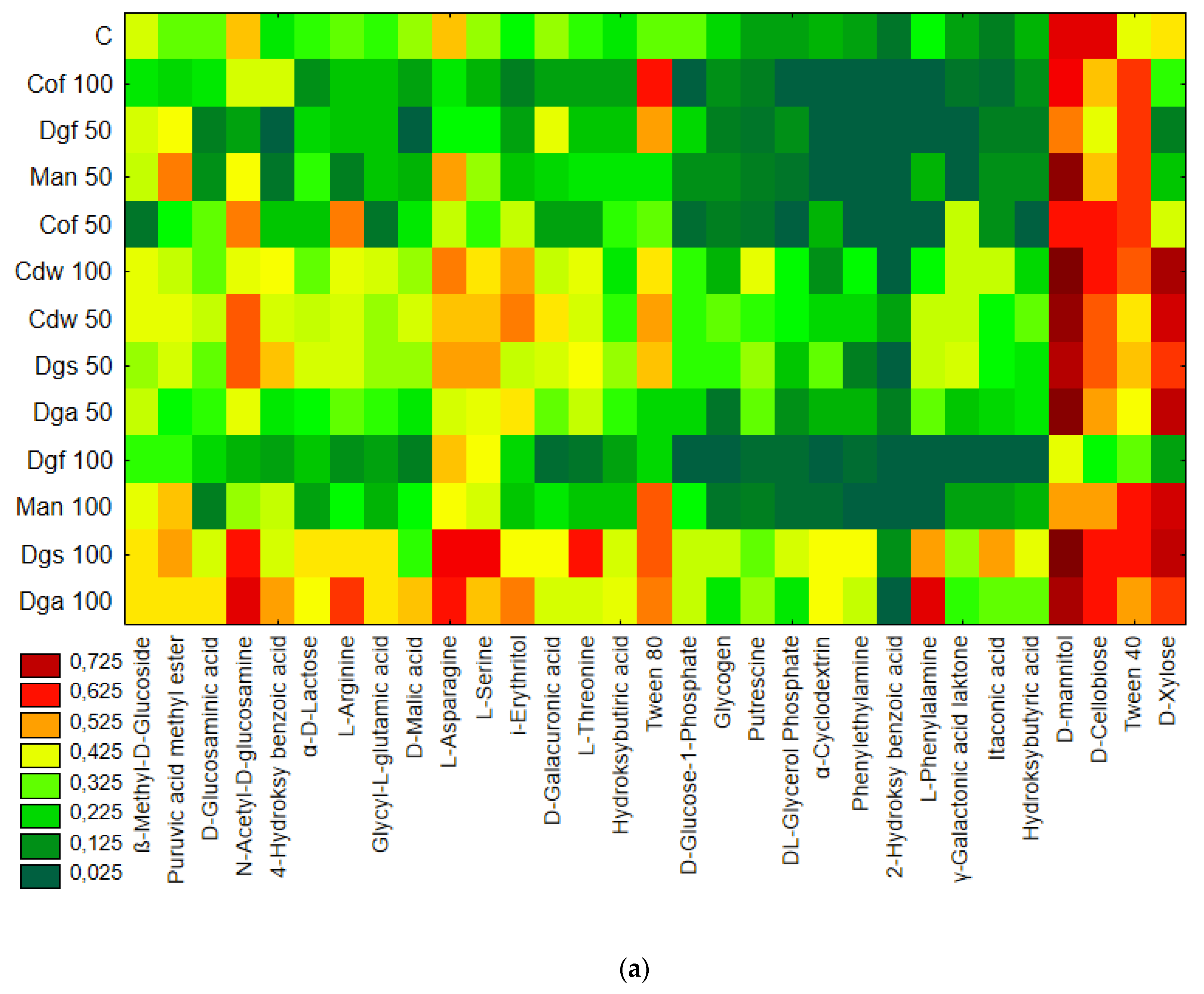
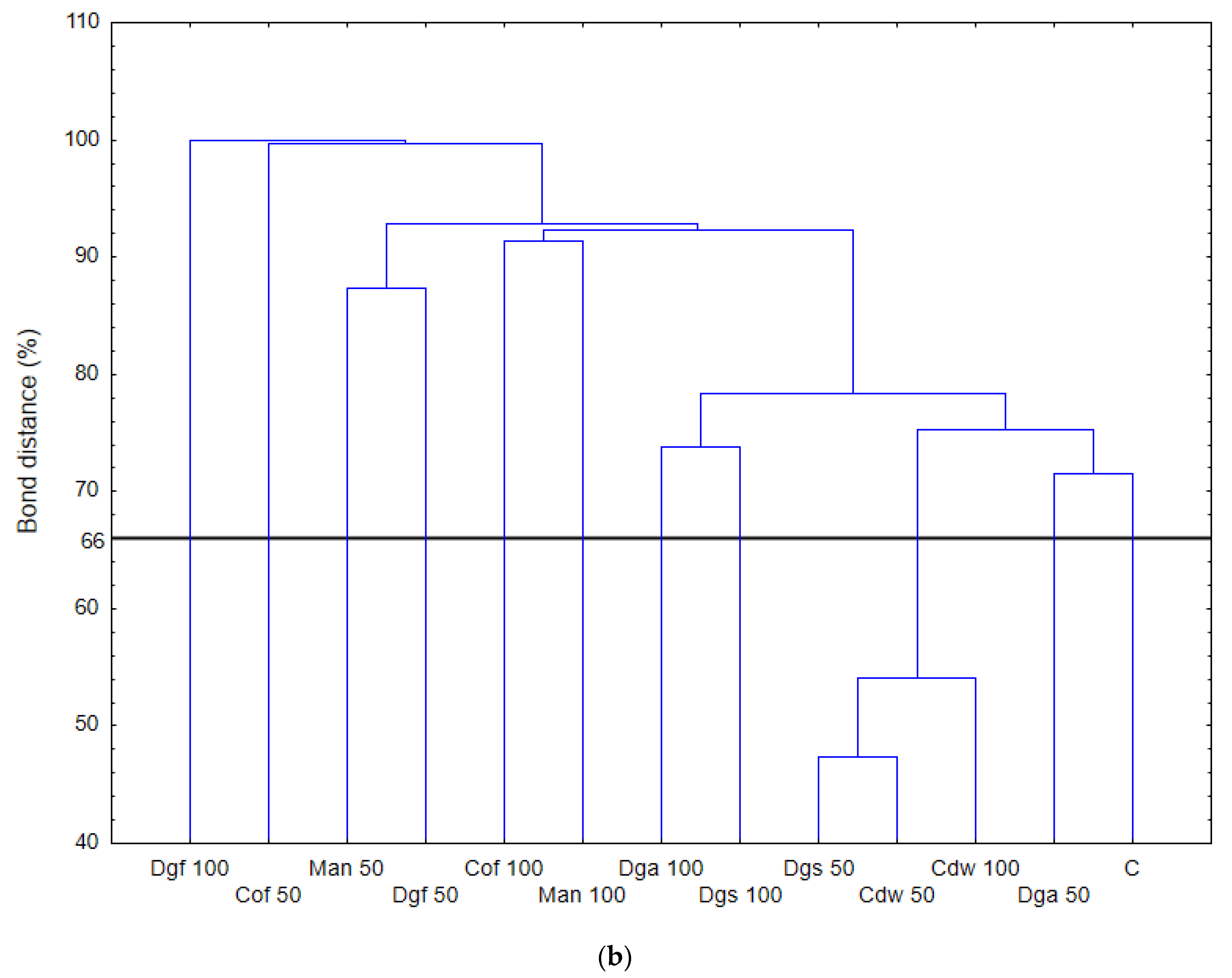
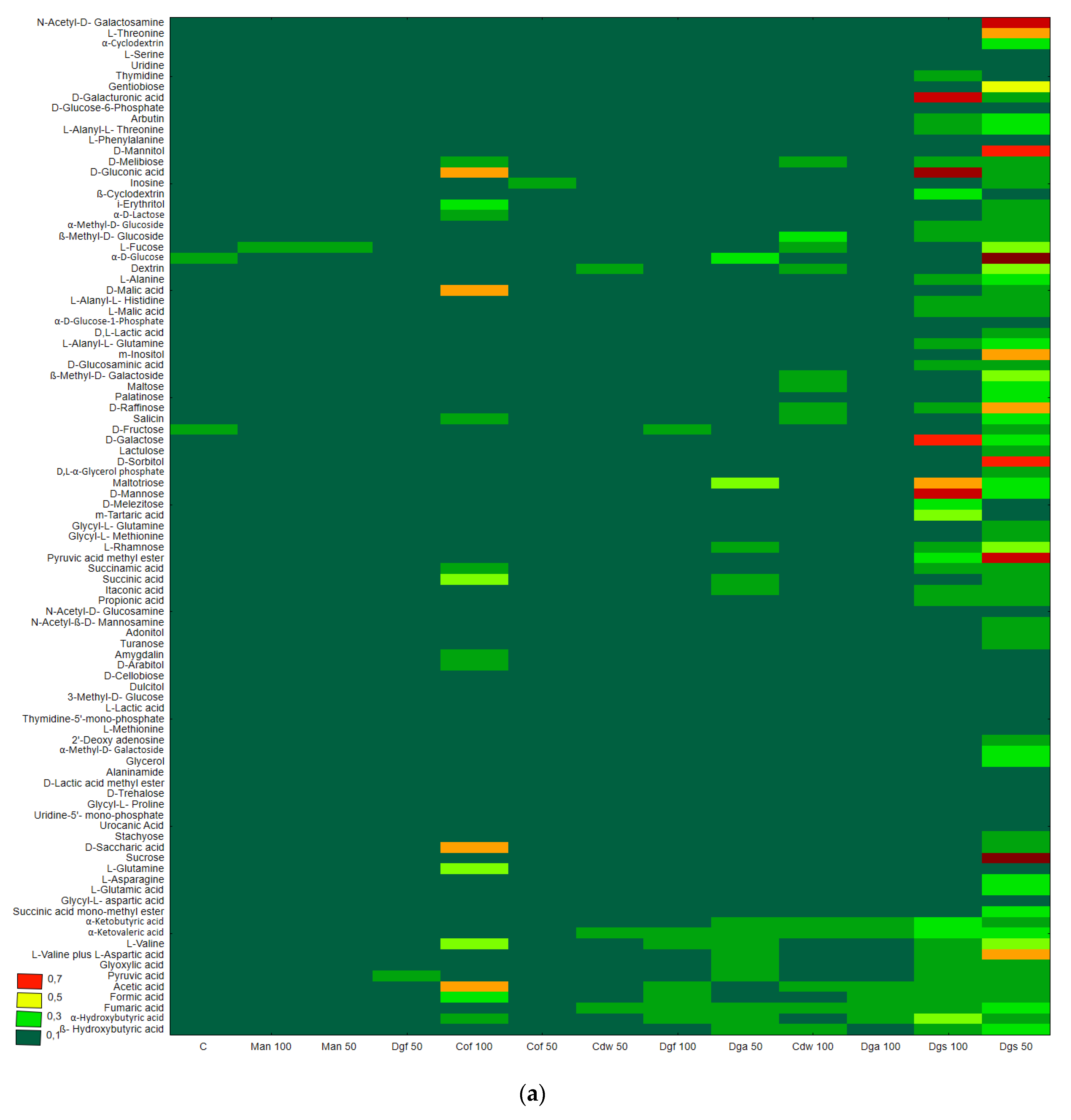
Figure 1), the heat map and dendrogram revealed that almost all carbon substrates were utilized by bacteria in the tested soil.
The metabolic profile was obtained on the basis of the degree of use of carbon substrates from particular groups of compounds in tested soil with different types of EOM in different doses, it may be observed that the level of metabolized carbon sources is very similar. This observation suggests that the proportion of different groups of bacteria in all tested soil may be similar. Data showed (
Figure S1) that the most intensively metabolized group of compounds were carbohydrates (35–40%), while the least intensively used compounds belonged to the group of amines and amides (<7%). An analysis of the utilization rate of individual carbon compounds showed the very intensive use of D-mannitol (AWCD ~ 0.72), D-cellobiose, Tween 40 and D-Xylose. Other substrates whose level of utilization was relatively high were L-asparagine, Tween 80, i-erythritol, L-serine and N-acetyl-D-glucosamine. In contrast the following substrates: glycogen, putrescine, phenylethylamine, 2-hydroxybenzoic acid, hydroxybutyric acid, were catabolized at a very low level. Cluster analysis and the dendrogram of carbon utilization of C-substrates on FF plates are shown at
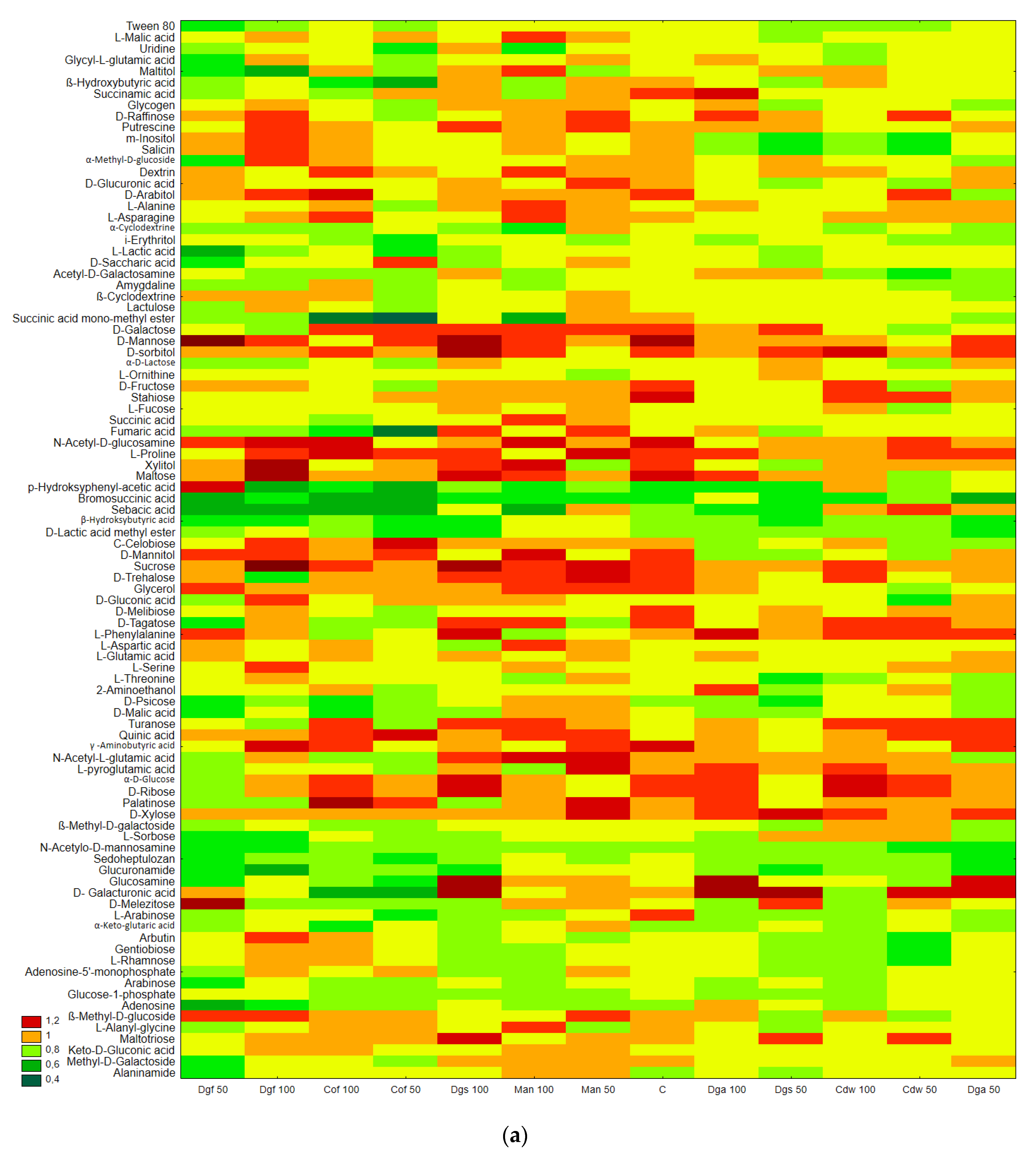
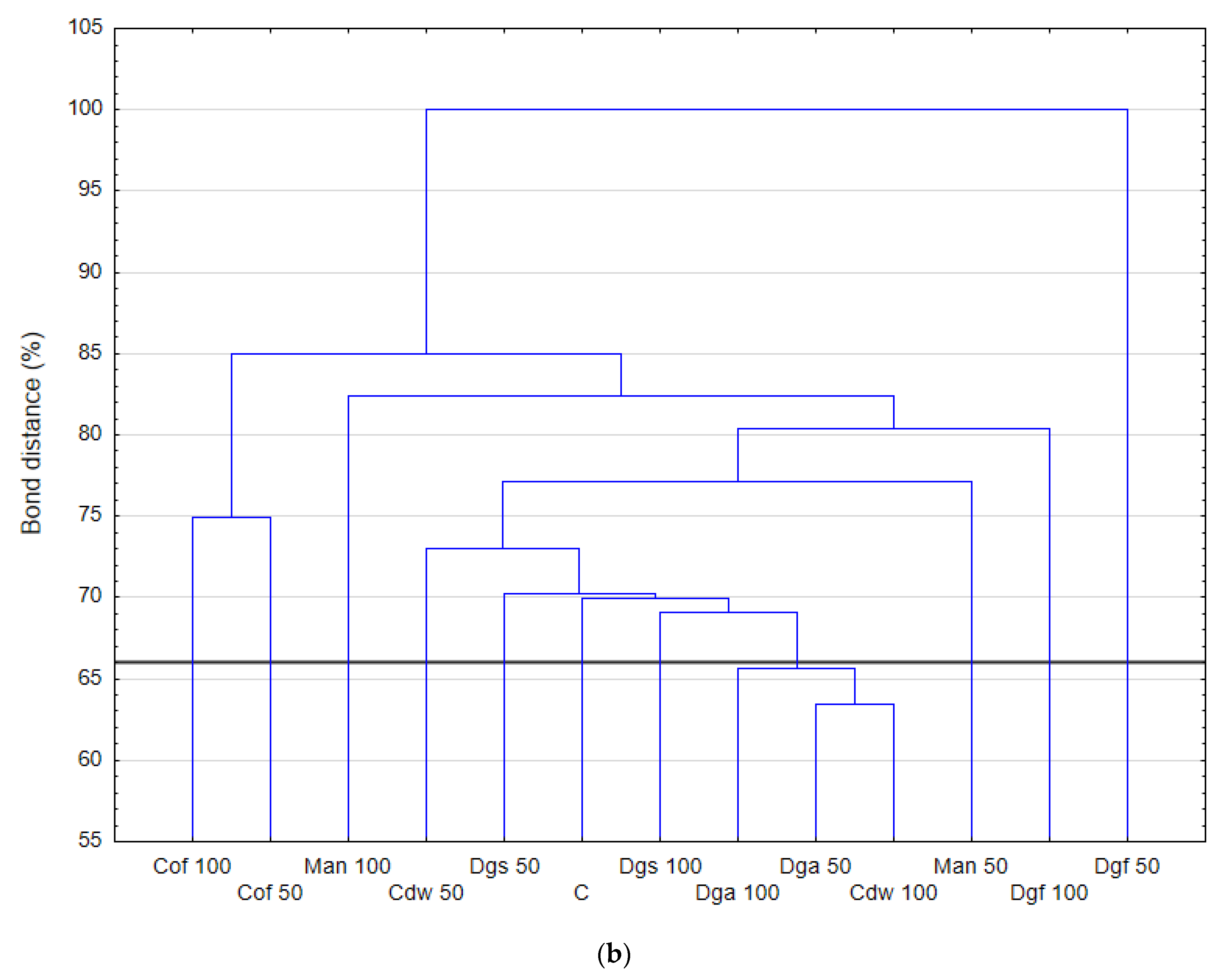
Figure 2.
The results correspond with the ECO plate, i.e., the most intensively utilized group of C-substrates were carbohydrates, distinct differences between the control and tested soil were not observed. Moreover, the highest rate of catabolism for an individual substrate was observed for carbohydrates: Mannose, Xylose, Sucrose, Xylitol, D-sorbitol. A separate analysis for anaerobic communities (AN plates) (
Figure 3) revealed quite a different metabolic profile of anaerobic microorganisms in the tested soil.
Almost all of the tested soils were characterized by very low levels of carbon sources utilization (only a few C-substrates were catabolized). Only the community of microorganisms from soil with the addition of Dgs in a 50% dose was capable of metabolizing more than a dozen C-substrates (e.g., N-acetyl-D-galactosamine, α-D-glucose, sucrose, pyruvic acid methyl ester, lactulose).
To explain the relationship between the EOM treatments and the carbon substrate utilization variables a PCA was performed separately for each group of tested microorganisms, which is presented in
Figure 4a–c.
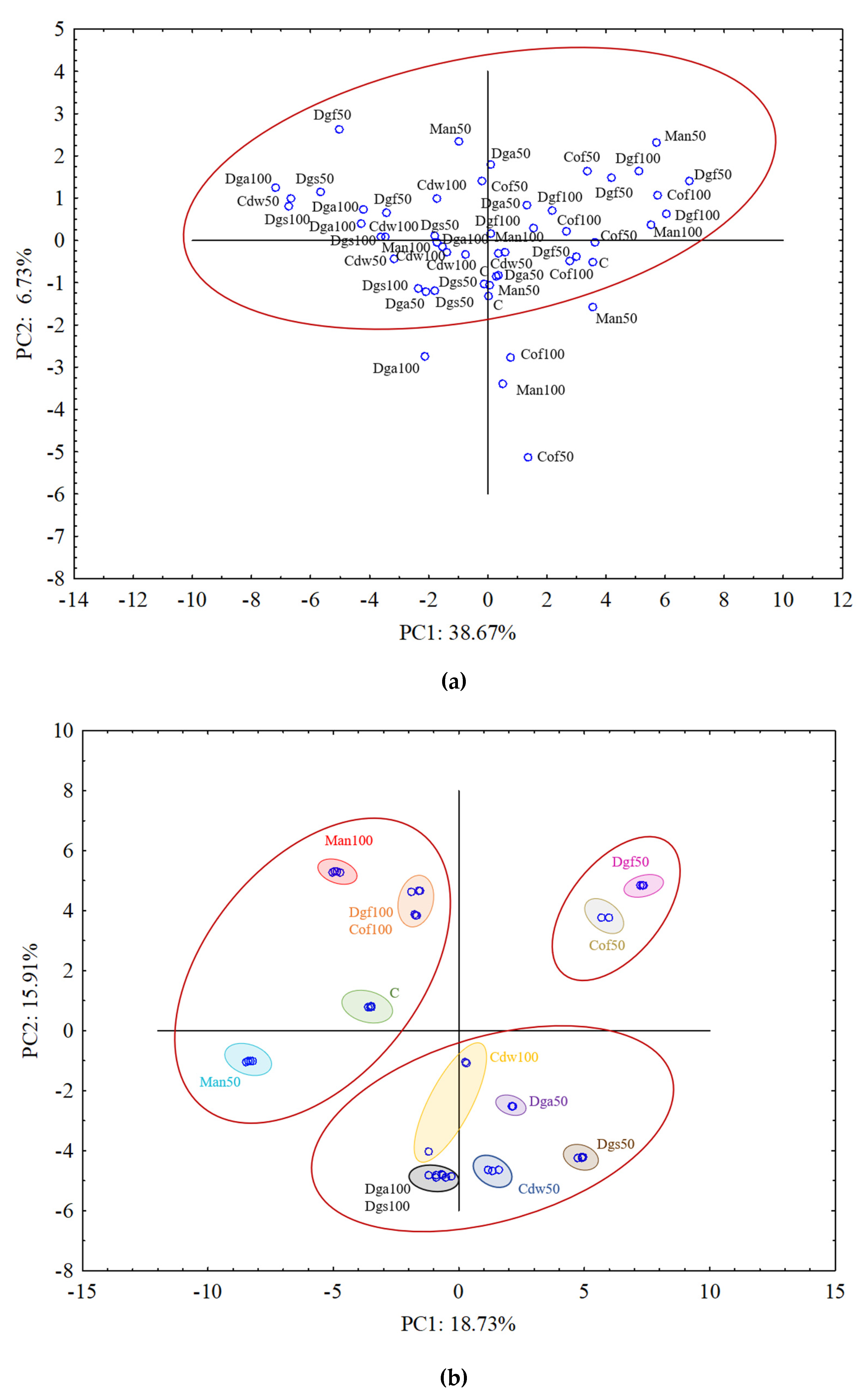
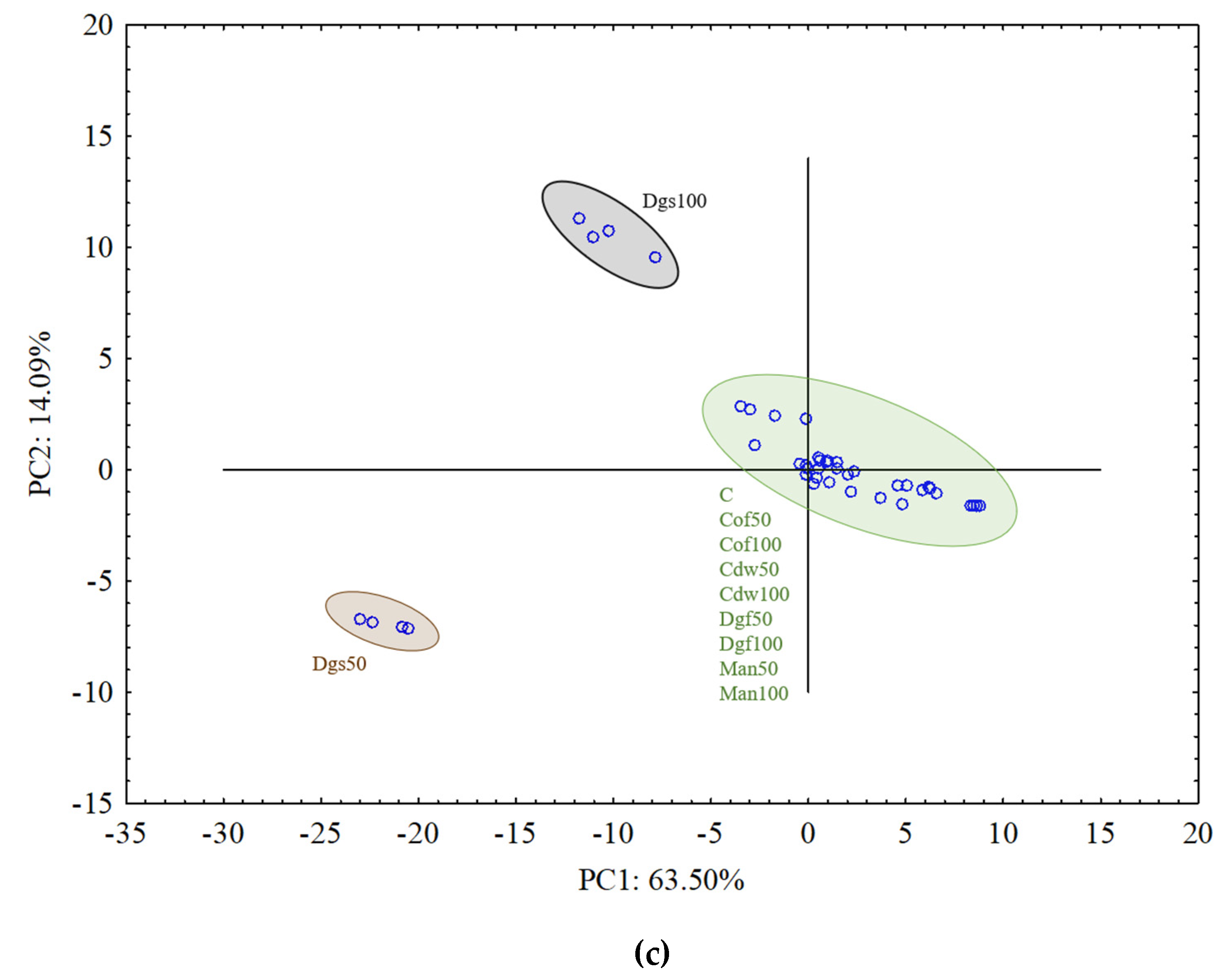
For aerobic bacterial community (ECO plates) the results from all treatments grouped rather together, and in general they did not indicate differences between treatments (
Figure 4a). The results of fungal community on FF plates indicated grouping particular treatments separately and creating three bigger clusters. PCA grouped the following treatments: C, Man 50, Man 100, Dgf 100 and Cof 100 as first cluster, Dga 100, Dga 50, Dgs 100, Dgs 50, Cdw 100 and Cdw 50 as second cluster, Dgf 50 and Cof 50 as the third one (
Figure 4b). For anaerobic microorganisms in AN plates Dgs 100 and Dgs 50 treatments were grouped separately, and the rest of thetreatments grouped together in one cluster (
Figure 4c). This grouping resulted from the similarity in metabolic potential of particular microbes group in the soil. The first (PC1) and the second (PC2) principal components explained 38.67% and 6.73%, 18.73% and 15.91%, 63.50% and 14.09% of the variability for bacteria, fungi and anaerobic microorganisms, respectively.
The evaluation of genetic diversity was performed based on terminal restriction fragment length polymorphism analyses of
amoA gen, which is coding to produce the α-subunit of ammonium monooxygenase. Results are presented on
Figure 5 and as a (
Figures S2 and S3).
The presented results indicated the differences in the intensity and quality of the occurrence of restriction fragments. The high value of the total peak area results from the presence of many restriction fragments, or from the presence of a peak characterized by a very high intensity. In this analysis, the relative abundance of T-RF was calculated based on the peak areas. In the T-RFLP electropherograms obtained both small and large peaks are visible. By analysing their height based on relative fluorescent units (Rfu), it may be concluded that among the observed fragments, there are important components and also components with a lesser importance that represent the ammonia-oxidizing archaeal communities observed in the particular treatments. The intensity of the peaks depends on the intensity of the fluorescence signal received from the restriction fragments. In addition, our own research shows that each experimental treatment was characterized by a different peak profile obtained from the restrictions of DNA isolated from soil. The obtained profiles of all of the tested treatments were characterized by a large number of fragments of different sizes, indicating the presence of many species belonging to the group of ammonia-oxidizing archaea in the environment. The observed differences in the T-RFs profiles obtained indicate a different quantitative and qualitative composition of the examined group of microorganisms in the tested soil samples.
The
Figure S2 and Figure S3 illustrate the results of a cluster analysis of the tested soil samples based on the relative abundance of the restriction fragments obtained after performing restriction. with the
AluI and
Csp6l enzymes, respectively.
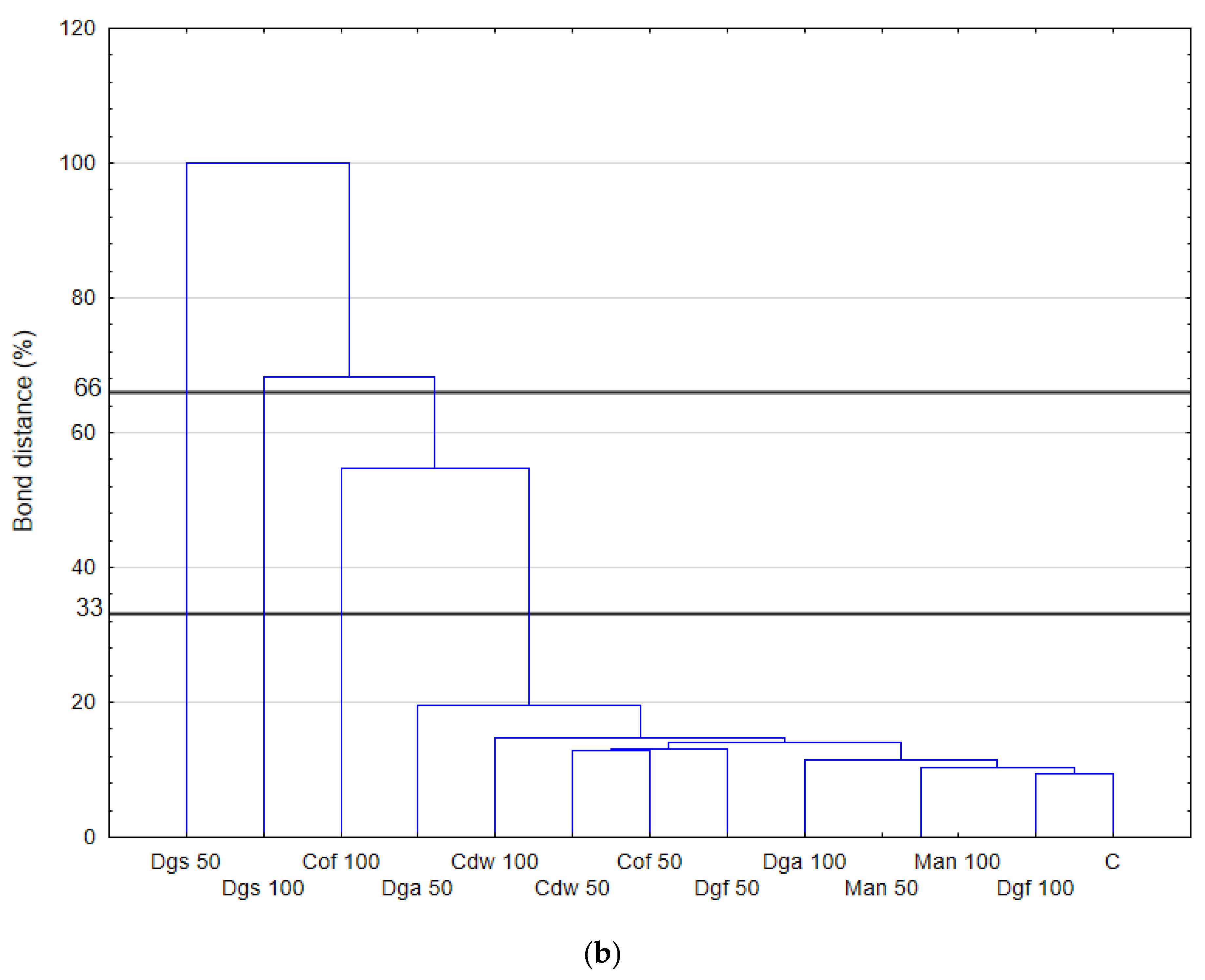
The differences between the treatments were only slightly visible for the treatments grouped in clusters with the criterion below 33%. The treatments: Cof 50, Man 50, Cof 100, Man 100 are grouped into one cluster with the criterion below 33%. A separate cluster contains treatments with Dgs 50, Dga 50, Cdw 50, Dgs 100, Dga 100 and Cdw 100. A restriction profile consisting of specific DNA fragments of various lengths was obtained for each soil treatment.
The restriction profile is characterized by the various lengths of the restriction fragments as well as their relative abundance. The restriction profile with
AluI (
Figure S2) digestion contained from 1 to 20 DNA fragments, the size of these fragments ranged from 55 to 560 base pairs. Among the fragments obtained, there were fragments present in most of the soil treatments (restriction fragments of approximately (± 3bp) 57, 80, 85, 105, 111, 149, 163, 176, 253, 553 bp) and those that were only present on an exceptional basis. A characteristic fragment of 253/254 bp was observed in almost all treatments; however, its intensity was different in various soils. The restriction profile obtained as a result of digestion with the
Csp6l enzyme (
Figure S3) was characterized by the presence of fragments from only 1 to 10, usually 6-8 different fragments, the size range varied from 54/55 to 562 bp. The fragments found in most of the soil samples had a length of (±3bp): 54, 70, 189, 197, 292 and 562 bp.
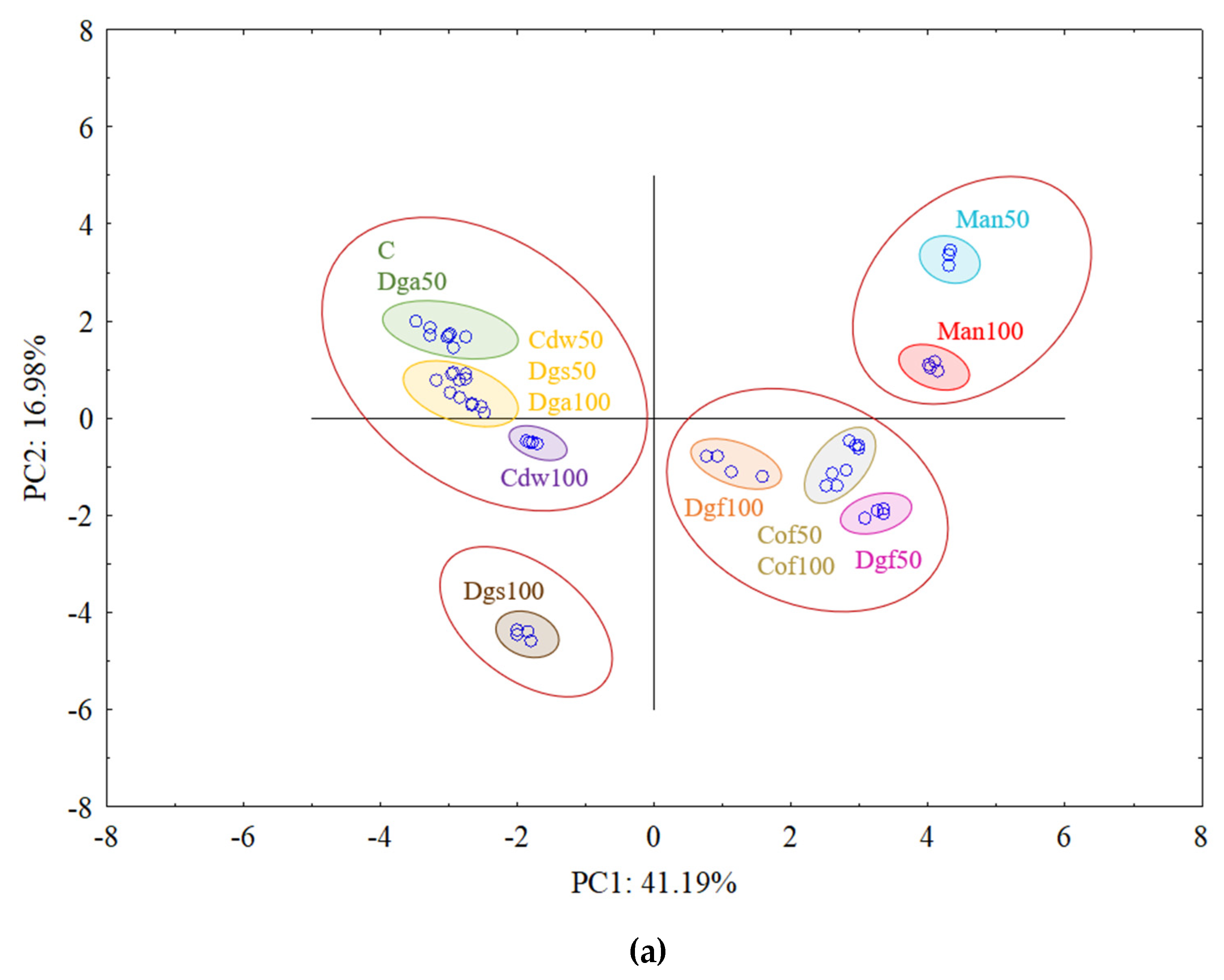
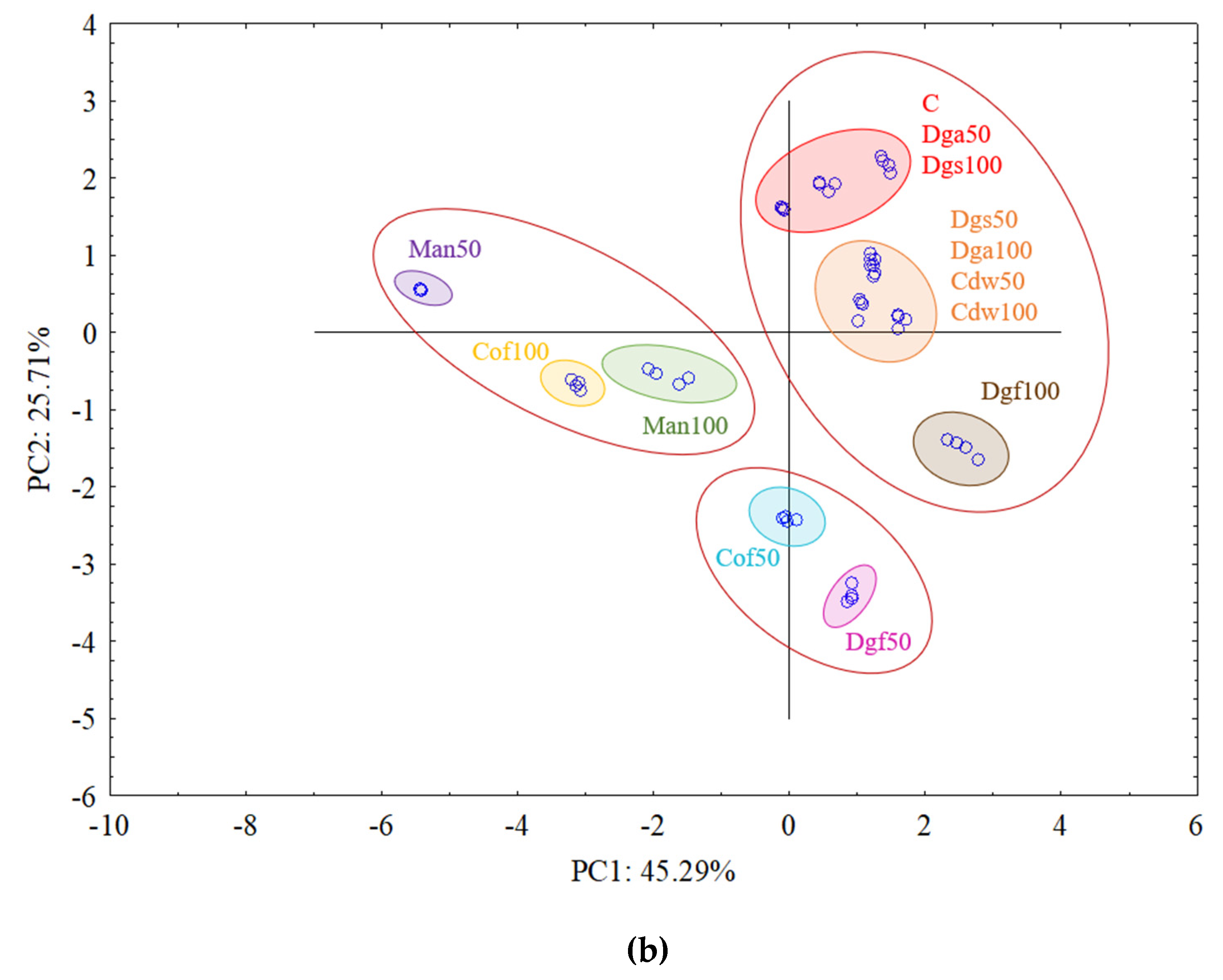
The first (PC1) and the second (PC2) principal components respectively explained 41.19% and 16.98% of the variability for
AluI restriction enzyme and 45.29 and 25.71% of the variability for
Csp6l (
Figure 5). The results of principal component analysis (PCA) indicated the relationship between the EOM treatments and the genetic profile (T-RFs) variables. PCA based on
AluI grouped Man 50 and Man 100 together, Dgf 100, Dgf 50, Cof 100 and Cof 50 together, Dgs 100 as separate cluster and the rest of treatments (C, Dga 100, Dga 50, Cdw 50, Dgs 100, Dgs 50) together. However, for
Csp6l we observed 3 separate clusters with the following treatments in first one Cof 50, Dgf 50, second Man 100, Man 50, Cof 100 and third group with C, Dga 100, Dga 50, Dgs 100, Dgs 50, Cdw 100 and Cdw 50.
To evaluate how much diversity changed in treatments compared to control the T-RFLP results with using
AluI and
Csp6l restriction enzymes were presented on
Figure S4. The results for
AluI indicated increase of diversity of selected T-RFs with the size 69/70, 85, 105/106, 111/112, 149, 176, 186, 253 and 341, where diversity was higher in the ranged from 140% to 650% after EOM application. This increase was observed mainly after incorporation of Dgs and Cdw exogenous organic matter in both doses. Moreover, the highest increase of diversity was noted for T-RF 162/163 in almost all treatments compared to control soil. This increase ranged from 609% in Cof 50 to 7186% in Dgs 100 in comparison to the control. For
Csp6l the most intensive increase of diversity was observed for T-RF 68/69 and ranged from 232% in Dgf 100 to 1563% in Cof 100. However, in the treatments Cdw 100 and Dgs 50 we observed the highest number of TRFs with significant increase compared to the control.
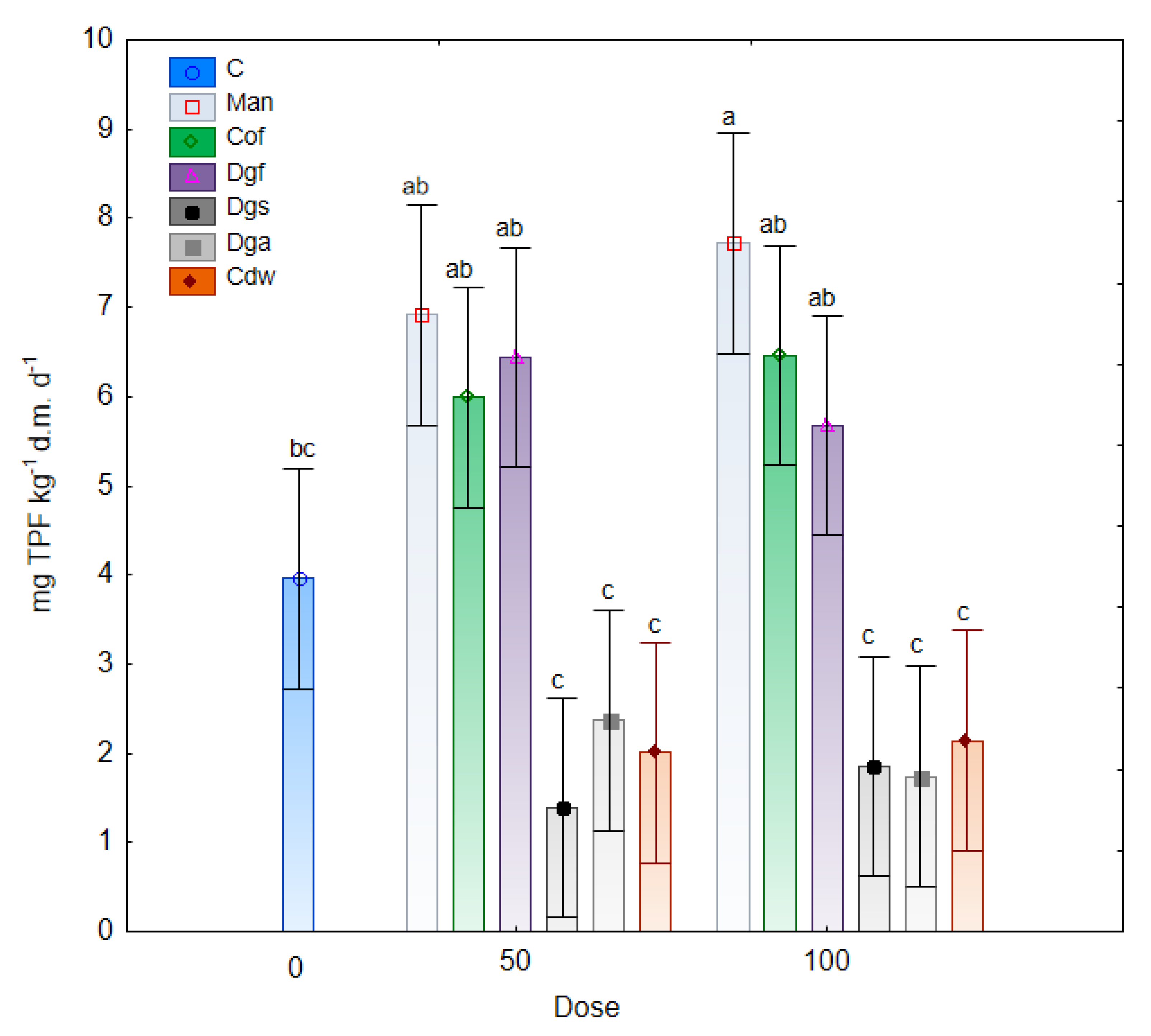
Changes in dehydrogenases activity under the influence of fertilization with different varieties of exogenous organic matter are illustrated in
Figure 6. The results of the study indicated an increase in the activity of DHA in the tested soil with the addition of Man, Cof and Dgf compared to the control soil. Furthermore, a significantly lower enzymatic activity was noted in treatments with the addition of Dgs, Dga and Cdw (at a dose of 50% and 100%). Moreover, significant correlations were observed between dehydrogenase activities and the AWCD of bacteria and anaerobic microbes and non-significant correlation was noted between DHA and AWDD of fungal communities (
Table 3).
4. Discussion
The research conducted in recent years, which has been focused on exploring the possibility of agricultural management of various types of waste from plants or animals, has given rise to the opportunity to use organic waste as fertilizers. This approach is also beneficial not only for the reduction in the amount of waste which should be stored or disposed of, but also because of the possibility of replacing or reducing the use of mineral fertilizers. Appropriate organic waste management contributes to the improvement of the environment and limits the degradation process caused by the overuse of mineral fertilizers [
40]. However, considering all of the ecological and ecotoxicological aspects, the relevant issue is to learn about the impact of organic waste, EOM sources, on the functional diversity of soil microorganisms. The preservation of the natural biodiversity of microorganisms and their balance in the soil environment is the basis of many processes (the prevention of erosion, soil formation, the circulation of elements and others) [
41]. Due to the fact that the soil microbiome is very dynamic and responds rapidly to changes occurring in the environment, the assessment of their condition may be valuable in the assessment of ecotoxicity [
42]. The Biolog system has been used for environmental research by many researchers [
20,
43], allowing for the monitoring of changes in the population of microorganisms in the soil and other ecosystems under the influence of various factors. The assessment of the impact of fertilizer (organic or mineral) addition on the population of microorganisms is important due to the functions they perform in the soil [
44]. The results of our study show, that all of the tested wastes affected the functional diversity of soil microbial communities, including bacteria, fungi and anaerobes in a characteristic way. The highest degree of microorganisms activity is connected with the introduction of organic matter into the soil, which through the supply of C and N sources stimulated the activity of microorganisms. Furthermore, EOMs also provide a large amount of cellulose and lignin-rich substances. The increased metabolic activity of fungi may be associated with their important function in the first stages of the breakdown of macromolecular substances such as lignin or cellulose [
45]. The results of the research show that the use of organic fertilizers (including compost and digestate) in properly balanced doses increased the diversity of microorganisms, on the other hand, the use of doses that are too large, may cause a decrease in the diversity and activity of microorganisms. The results presented show that even a complete replacement of mineral fertilization with the addition of appropriately calculated doses of EOM did not cause a significant decrease in the activity of microorganisms. Therefore, the use of natural fertilizers may be used as a substitute for mineral fertilization, which generates higher costs and, over time, results in a deterioration of the soil condition. The tested doses (50% and 100%) of EOMs, referring to the total N content, had a similar effect on the microbiological soil properties. When comparing the two fertilization strategies, the total nitrogen dose applied to wheat crops (140 kg ha
−1) was delivered to the soil and only the source of the original delivered N was different. The communities of soil microorganisms consist of the aerobic microorganisms, but the anaerobic population cannot be excluded. In soil, anaerobic conditions may occur locally or periodically, in the event of excess water. In situations of oxygen deficiency, the biological decomposition of organic matter conducted by aerobic microorganisms is either terminated or limited. Under anaerobic conditions it is possible to observe the anaerobic decomposition of organic matter carried out by anaerobic bacteria-reducing compounds such as sulphate or nitrates. The anaerobic decomposition processes of organic matter are slower and less energy efficient than those requiring oxygen. The activity of soil anaerobic microorganisms is negligible compared to its aerobic activity; however, the significance of those processes is so great that the determination of the metabolic profile of anaerobes is valuable in the overall assessment of soil microbial activity. Moreover, the introduction of waste material into the soil may cause the occurrence of periodic anaerobic conditions. Our results, regarding metabolic activity based on the level of carbon substrates utilization on Biolog AN Plates demonstrated that the activity of microorganisms in anaerobic conditions was very low. However, it should be mentioned that cultivation of anaerobic microorganisms from environmental samples in Biolog plates is difficult and thus only fragmentary in literature [
43,
46]. The results observed as low activity of microorganisms in anaerobic conditions can be a consequence of their sensitivity to the presence of oxygen in the soil, especially after soil collection from the 0–30 cm layer, and it shows that the main decomposition processes in the soil environment are dependent on oxygen. The biochemical activity resulting from anaerobic activity in the distribution of organic matter plays a complementary role. Because most studies on anaerobic microbes using Biolog Plates are performed in aquatic and other than soil environmental samples [
43,
46], the presented results can serve a valuable reference for soil microbial ecologists deciding to use Biolog Plates.
Ammonia-oxidizing archaea (AOA) is a group of organisms widely distributed in the soil, moreover a significant body of research indicates their advantage over ammonia-oxidizing bacteria (AOB) [
47]. Furthermore, many research indicate the significant role of AOA in the nitrification process, and the genetic profile obtained on the basis of the soil population of AOA is an indicator, which is often used in the determination of biodiversity. EOM application influenced the occurrence of changes in the genetic diversity of AOA, whereas the literature reports that soil pH has the greatest impact on AOA populations. McAndrew and Malhi [
48] reported that the addition of nitrogen fertilizers to the soil may result in the acidification of the environment. A significant reduction in soil pH is also frequently observed during the use of mineral nitrogen fertilization (N, NP, NK, NPK) [
49]. He et al. [
50] presented results indicating a reduction in AOA diversity in the soil as a consequence of lowering the soil pH which is caused by the use of nitrogen fertilizers. On the other hand, the addition of organic fertilizers and organic matter resulted in an increase in the AOA population on account of the greater bioavailability of C [
51]. In this paper, we present results which indicate a positive effect of using the tested EOM on the diversity of the AOA population. In the tested soil, unique restriction fragments were observed which were not detected in the control soil. Moreover, a distinguished group of fragments was observed in all tested treatments as well as in the control soil. The presence of restriction fragments of the same length in most samples may indicate the existence of a certain species belonging to common AOA. However, the observed changes in the abundance of these fragments may be related to the sensitivity of the species, and they represent changes in pH that may be connected with exogenous organic matter application.
Another, relevant parameter which is pivotal in the assessment of soil quality is enzymatic activity. The evaluation of dehydrogenases activity is used in soil condition assessment after the application of organic waste [
52,
53]. It is obvious that the activities of enzymes are correlated with other biochemical and microbiological parameters, such as microbial biomass content, the number of individual groups of soil microorganisms, soil pH and respiration [
54]. The highest degree of soil enzymatic activity is associated with a fairly high content of organic matter and its decomposition rate. The correlation of soil enzymatic activity with the value of the carbon-to-nitrogen ratio (C/N ratio) indicates the importance of exogenous organic matter addition to the soil [
55]. The dehydrogenases activities in treatment with Man, Cof and Dgf supplement in doses of 50% and 100% were higher than 5.5 mg TPF kg
−1 d.m. d
−1 which were significantly (
p < 0.05) higher than in treatments with Dgs, Dga, Cdw (< 2.5 mg TPF kg
−1 d.m. d
−1). The higher DHA activity in the treatment with Mb and Dg is connected with low C/N ratio and affected on carbon and nitrogen cycling in the soil [
56].
Furthermore, the C/N ratio of amendments material reflects the ratio of its decomposition. The ratio of C/N on 24/1 level is ideally balanced for soil microorganisms, when the organic materials is characterized by this ratio the soil microorganisms consume it relatively quickly and no excess carbon or nitrogen is left over. The EOM which were characterized by a lower C/N ratio cause the excess nitrogen to be left in the soil. This surplus nitrogen in the soil will be available for growing plants, or for soil microorganisms to decompose residues that characterized by higher C/N ratio. Peigne et al. [
57] emphasize that the lower C/N ratio in mineralized material the faster the decomposition of organic matter and this releases plant nutrients. Sources of EOM with a very high carbon to nitrogen ratio (such as compost (Cdw) and compost (Cof)) could cause a immobilizing free N by microorganisms and then results lower availability for plant uptake [
58]. Digestate (Dgf, Dgs, Dga) and meat bone meal (Mb) has a low C/N ratio and they are predictable as effective organic fertilizers.
Summarizing, it is worth mentioning that application of EOMs to the soil can be a source of substrates for microbes growing. For different groups of tested microorganisms we observed changes in metabolic of genetic profiles. Bacterial communities had similar metabolic profile as control soil after incorporation of Dgs 50, Cdw 50, Cdw 100 and Dga 50. Fungal communities in control soil were the closest to Cdw 50 and Dgs 50 treatments. However, anaerobic microbes had different metabolic profiles only after application of Dgs 50, Dgs 100 and Cof 100. The rest of treatments had a similar metabolic pattern to the control soil. Genetic profile was the closest to the control soil in treatments with the addition of Dga 50 and Dgs 100.