Pear Flower Cluster Quantification Using RGB Drone Imagery
Abstract
1. Introduction
- (i)
- Examine if the UAV-based top-view is representative for the flower clusters present in the entire tree.
- (ii)
- Evaluation of the pixel-based classification algorithm for segmentation of the flower pixels from the background pixels.
- (iii)
- Evaluation of density-based clustering to group flower pixels for estimating the number of flower clusters.
- (iv)
- Comparison of the accuracy of the 2D- and 3D-based pixel and object-based methods for flower cluster quantification.
- (v)
- Comparison of the accuracy of all methods on individually segmented trees (tree-level) versus per three segmented trees (plot-level) to evaluate the importance and difficulty of tree delineation in the orchard environment.
2. Materials and Methods
2.1. Study Area and Flower Cluster Reference Data
2.2. RGB Drone Data Acquisition
2.2.1. RGB Drone Data for Flower Cluster Estimation
2.2.2. RGB Drone Data for Training the Pixel-Based Classifier
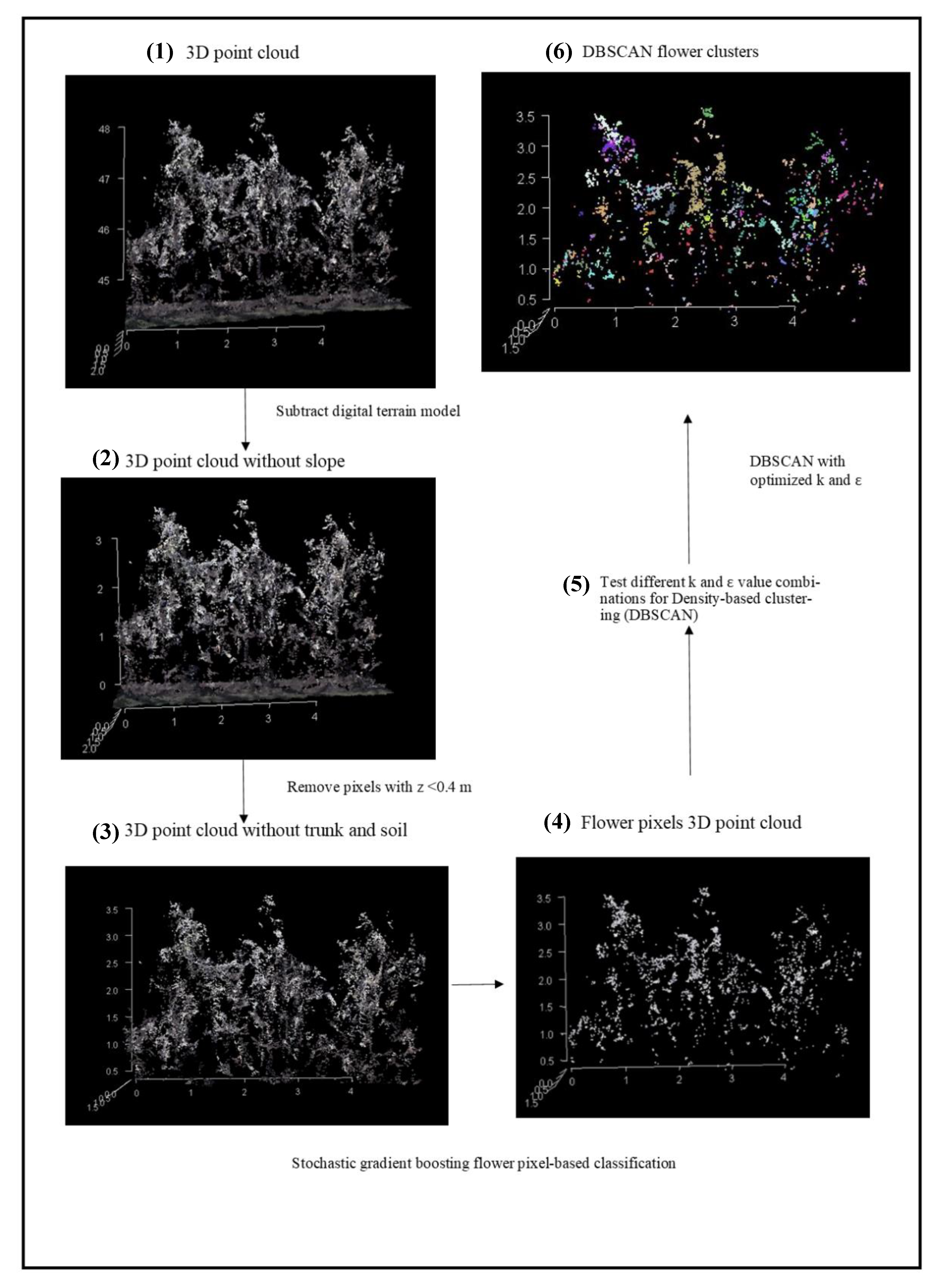
2.3. Data Processing
2.3.1. Colored Dense Point Cloud, Digital Elevation Model and Orthomosaic Generation
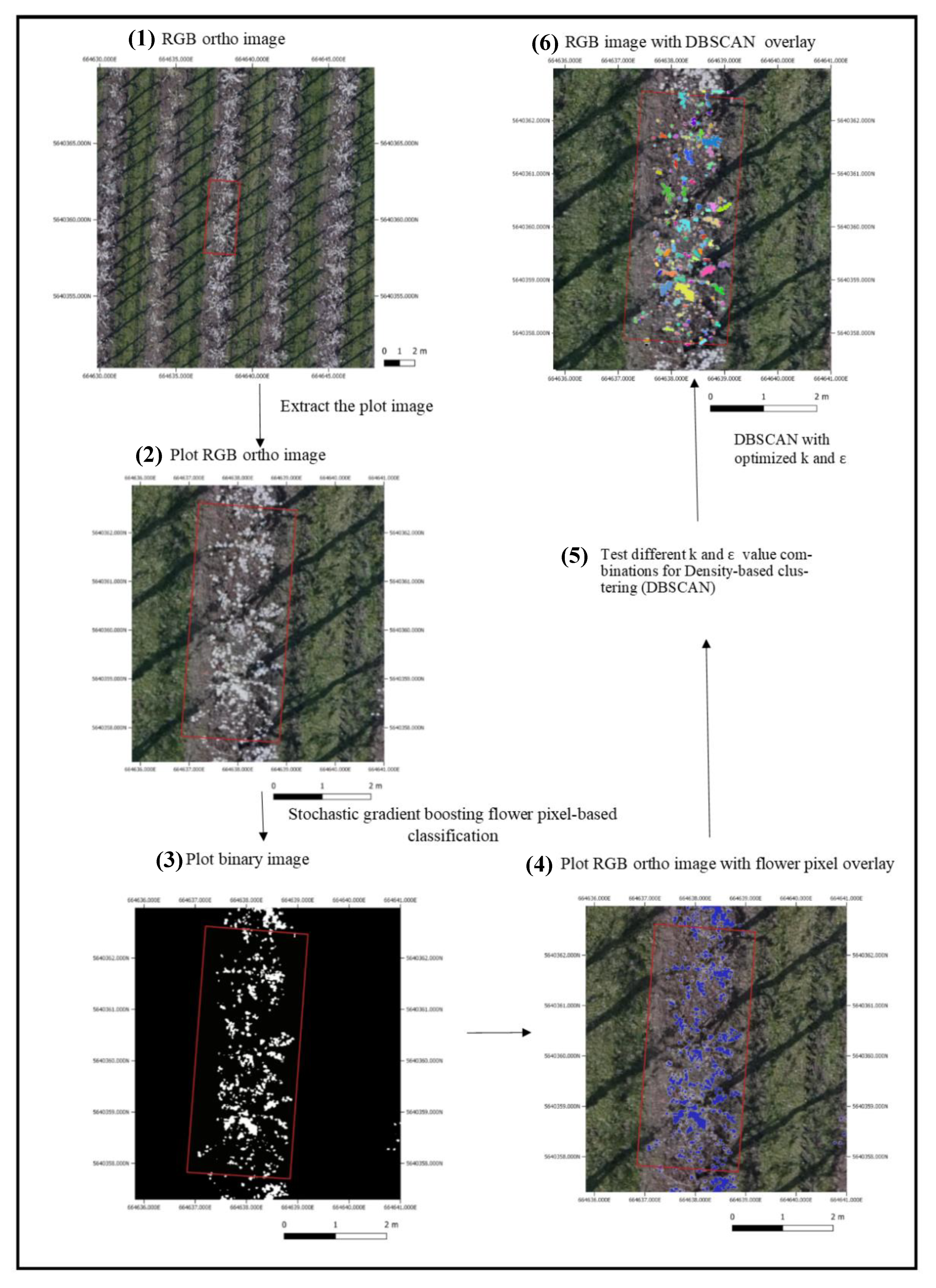
2.3.2. Pixel-Based Flower Classification
2.3.3. Flower Pixel Clustering
2.4. Magnitude of the Flower Occlusion Problem
2.5. Accuracy Assessment
2.5.1. Pixel-Based Classification
2.5.2. Optimization of DBSCAN Parameters
2.5.3. Flower Cluster Estimation Methods
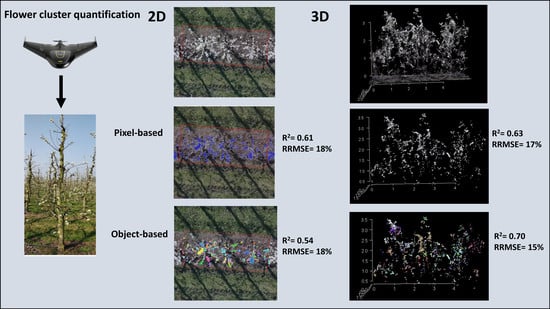
3. Results
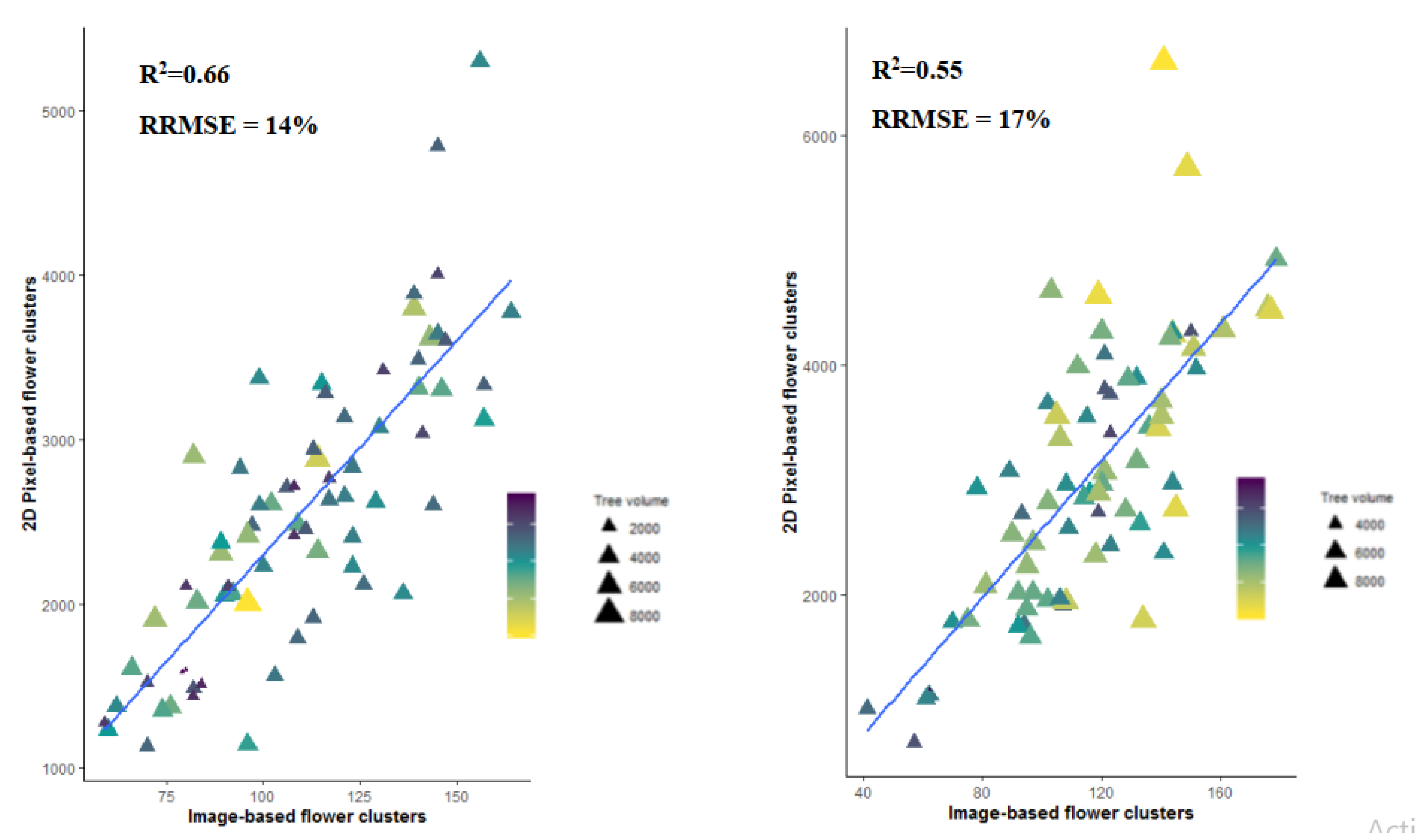
3.1. Magnitude of the Flower Occlusion Problem
3.2. Accuracy of the Pixel-Based Classification
3.3. Optimalisation of DBSCAN Parameters
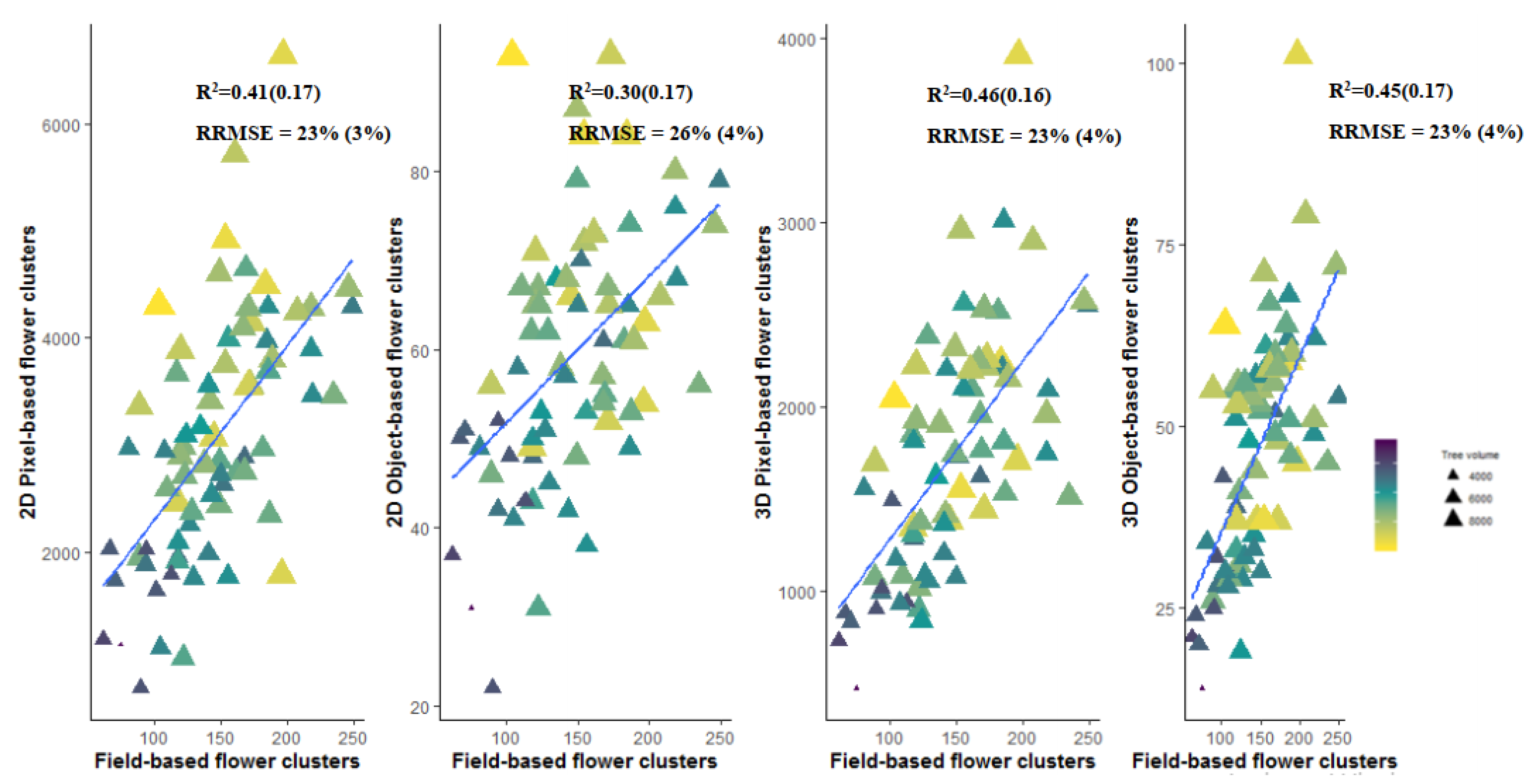
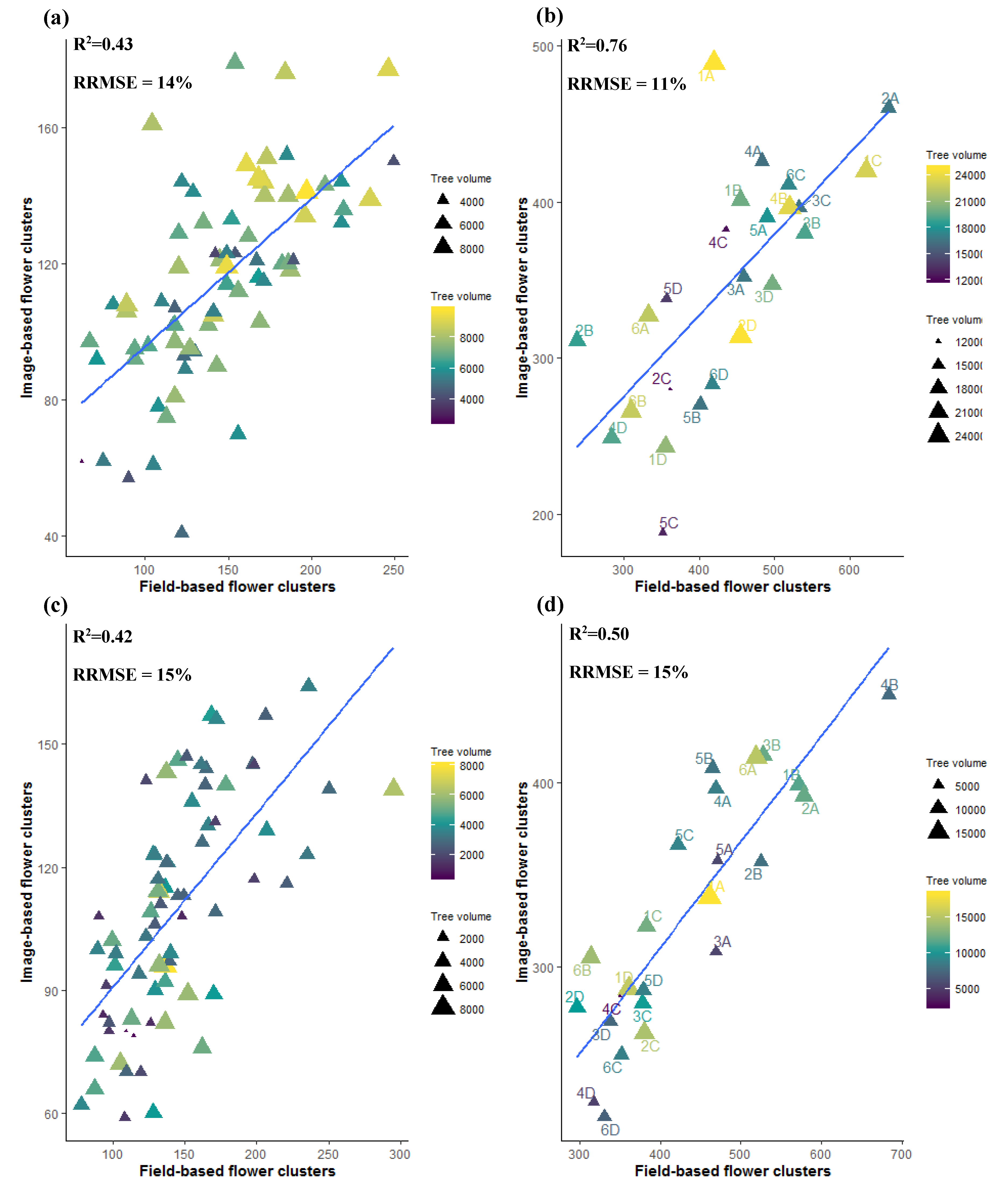
3.4. Flower Cluster Estimation Models
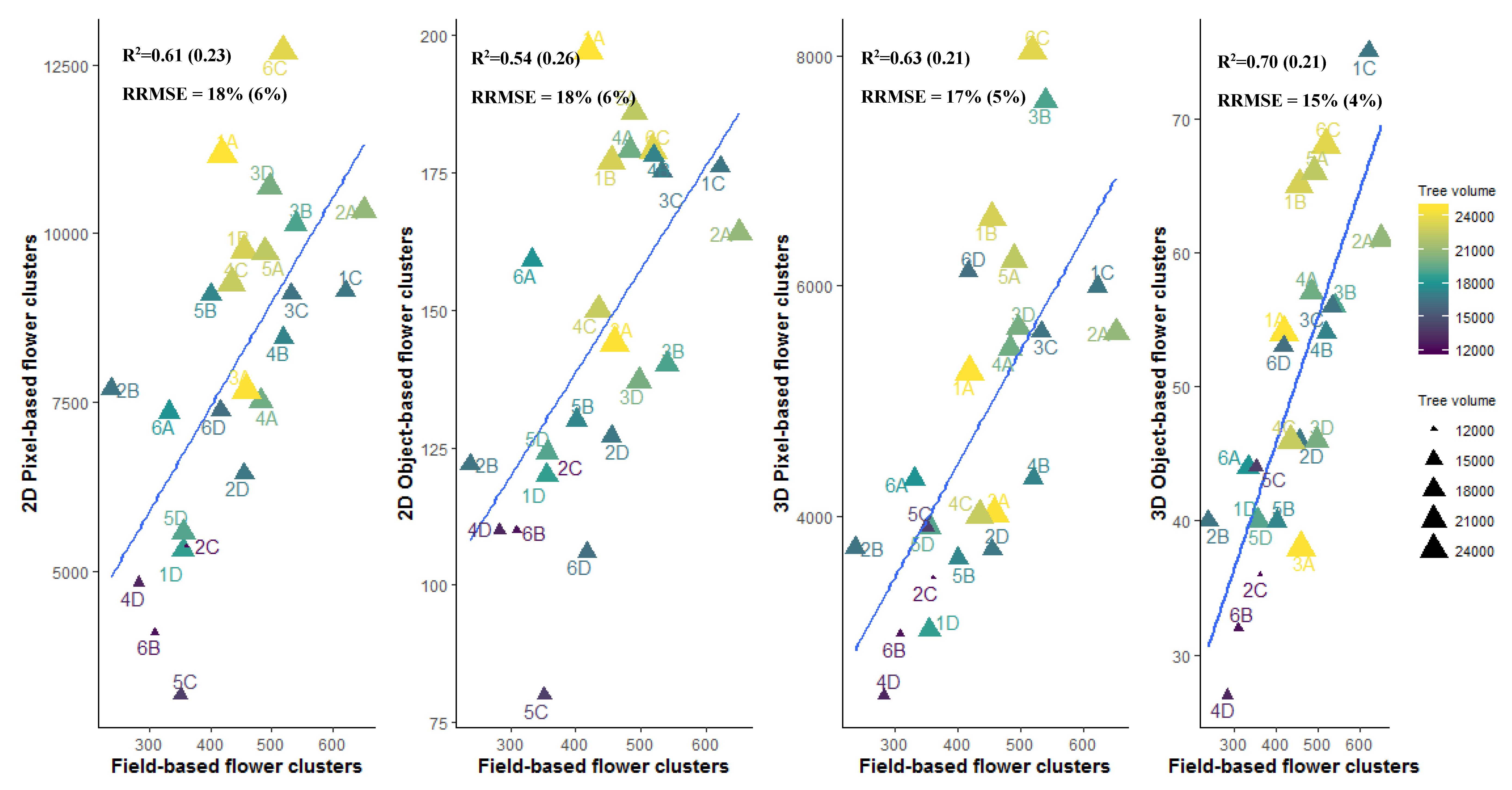
3.4.1. Individual Tree Level
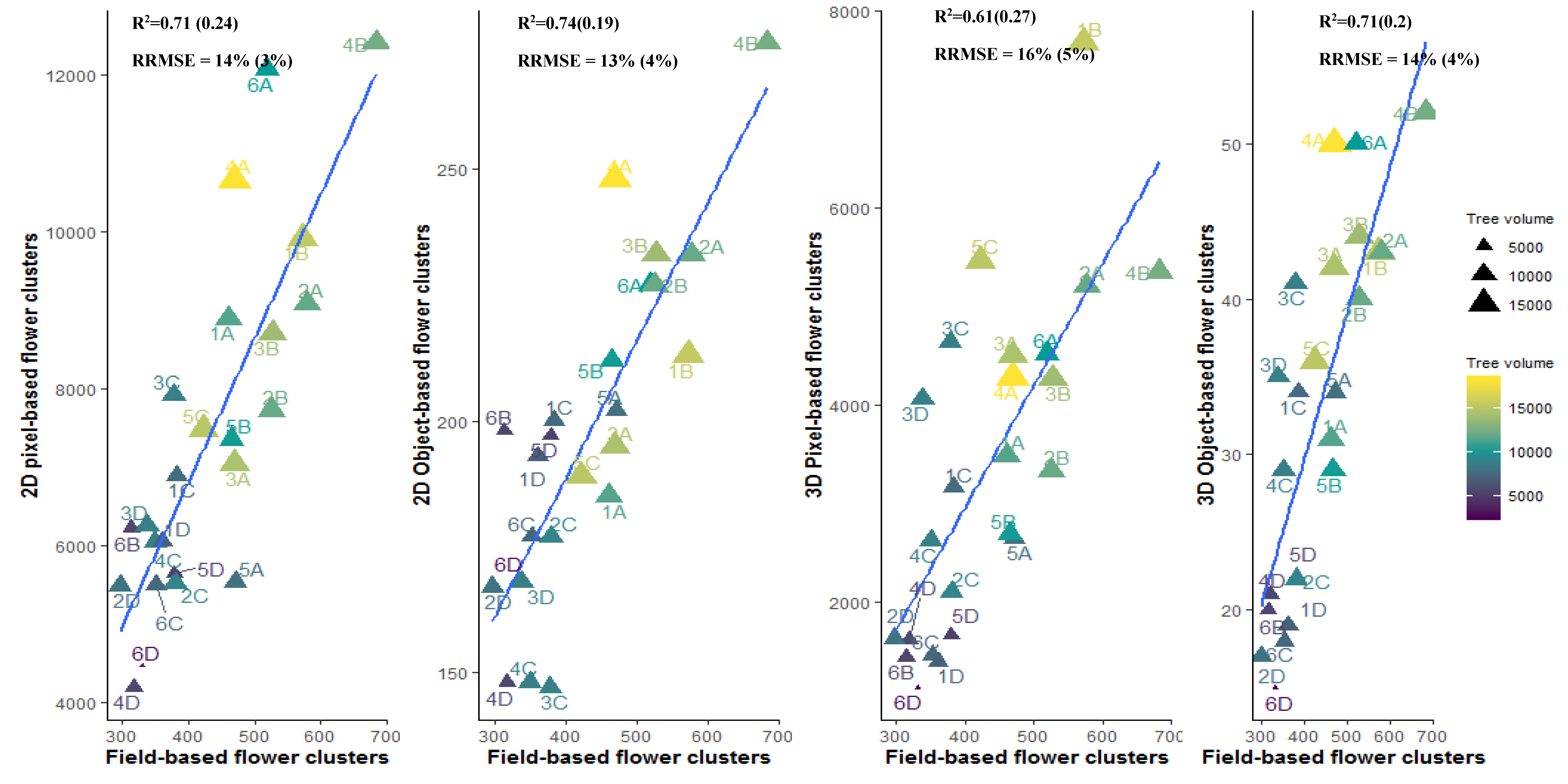
3.4.2. Plot or Multiple Tree Level
4. Discussion
4.1. Viewing Perspective
4.2. Tree Delineation
4.3. Pixel-Based Versus Object-Based Classification
4.4. Limitations and Recommendations for Future Research
5. Conclusions
- (i)
- The top-view perspective for flower cluster estimations can suffice if the trees have an open tree architecture but fail if the canopies have a lot of flower occlusion due to flower overlap.
- (ii)
- The object-based flower cluster estimation model based on colored dense point clouds gives the most accurate results for the flower cluster estimations for both orchards.
- (iii)
- It is better to work on plot level (i.e., multiple tree level) than on tree level for reducing the estimation error caused by errors in delineation of individual trees.
- (iv)
- In future research flower clusters should be counted per running meter instead of per tree to reduce errors in the ground truth and errors due to overlapping branches from neighboring trees.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A



References
- Aggelopoulou, K.; Wulfsohn, D.; Fountas, S.; Gemtos, T.; Nanos, G.; Blackmore, S. Spatial variation in yield and quality in a small apple orchard. Precis. Agric. 2010, 11, 538–556. [Google Scholar] [CrossRef]
- Konopatzki, M.R.; Souza, E.G.; Nóbrega, L.H.; Uribe-Opazo, M.A.; Suszek, G. Spatial variability of yield and other parameters associated with pear trees. Eng. Agrícola 2012, 32, 381–392. [Google Scholar] [CrossRef][Green Version]
- Teodorescu, G.; Moise, V.; Cosac, A.C. Spatial Variation in Blooming and Yield in an Apple Orchard, in Romania. Ann. Valahia Univ. Targoviste Agric. 2016, 10, 1–6. [Google Scholar] [CrossRef][Green Version]
- Karlsen, S.R.; Ramfjord, H.; Høgda, K.A.; Johansen, B.; Danks, F.S.; Brobakk, T.E. A satellite-based map of onset of birch (Betula) flowering in Norway. Aerobiologia 2009, 25, 15. [Google Scholar] [CrossRef]
- Khwarahm, N.R.; Dash, J.; Skjøth, C.; Newnham, R.M.; Adams-Groom, B.; Head, K.; Caulton, E.; Atkinson, P.M. Mapping the birch and grass pollen seasons in the UK using satellite sensor time-series. Sci. Total Environ. 2017, 578, 586–600. [Google Scholar] [CrossRef]
- Landmann, T.; Piiroinen, R.; Makori, D.M.; Abdel-Rahman, E.M.; Makau, S.; Pellikka, P.; Raina, S.K. Application of hyperspectral remote sensing for flower mapping in African savannas. Remote Sens. Environ. 2015, 166, 50–60. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.; Makori, D.; Landmann, T.; Piiroinen, R.; Gasim, S.; Pellikka, P.; Raina, S. The utility of AISA eagle hyperspectral data and random forest classifier for flower mapping. Remote Sens. 2015, 7, 13298–13318. [Google Scholar] [CrossRef]
- Xu, R.; Li, C.; Paterson, A.H.; Jiang, Y.; Sun, S.; Robertson, J.S. Aerial images and convolutional neural network for cotton bloom detection. Front. Plant Sci. 2018, 8, 2235. [Google Scholar] [CrossRef]
- Aggelopoulou, A.; Bochtis, D.; Fountas, S.; Swain, K.C.; Gemtos, T.; Nanos, G. Yield prediction in apple orchards based on image processing. Precis. Agric. 2011, 12, 448–456. [Google Scholar] [CrossRef]
- Hočevar, M.; Širok, B.; Godeša, T.; Stopar, M. Flowering estimation in apple orchards by image analysis. Precis. Agric. 2014, 15, 466–478. [Google Scholar] [CrossRef]
- Liakos, V.; Tagarakis, A.; Aggelopoulou, K.; Fountas, S.; Nanos, G.; Gemtos, T. In-season prediction of yield variability in an apple orchard. Eur. J. Hortic. Sci. 2017, 82, 251–259. [Google Scholar] [CrossRef]
- Dias, P.A.; Tabb, A.; Medeiros, H. Apple flower detection using deep convolutional networks. Comput. Ind. 2018, 99, 17–28. [Google Scholar] [CrossRef]
- Liu, D.; Xia, F. Assessing object-based classification: Advantages and limitations. Remote Sens. Lett. 2010, 1, 187–194. [Google Scholar] [CrossRef]
- Underwood, J.P.; Hung, C.; Whelan, B.; Sukkarieh, S. Mapping almond orchard canopy volume, flowers, fruit and yield using LiDAR and vision sensors. Comput. Electron. Agric. 2016, 130, 83–96. [Google Scholar] [CrossRef]
- Carl, C.; Landgraf, D.; van der Maaten-Theunissen, M.; Biber, P.; Pretzsch, H. Robinia pseudoacacia L. Flower Analyzed by Using An Unmanned Aerial Vehicle (UAV). Remote Sens. 2017, 9, 1091. [Google Scholar] [CrossRef]
- Tubau Comas, A.; Valente, J.; Kooistra, L. Automatic apple tree blossom estimation from uav RGB imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019. [Google Scholar] [CrossRef]
- Pal, N.R.; Pal, S.K. A review on image segmentation techniques. Pattern Recognit. 1993, 26, 1277–1294. [Google Scholar] [CrossRef]
- Xiao, C.; Zheng, L.; Sun, H. Estimation of the Apple Flowers Based on Aerial Multispectral Image. In Proceedings of the 2014 Montreal, Montreal, QC, Canada, 13–16 July 2014; p. 1. [Google Scholar]
- Zawbaa, H.M.; Abbass, M.; Basha, S.H.; Hazman, M.; Hassenian, A.E. An automatic flower classification approach using machine learning algorithms. In Proceedings of the 2014 International Conference on Advances in Computing, Communications and Informatics (ICACCI), New Delhi, India, 24–27 September 2014; pp. 895–901. [Google Scholar]
- Biradar, B.V.; Shrikhande, S.P. Flower detection and counting using morphological and segmentation technique. Int. J. Comput. Sci. Inform. Technol. 2015, 6, 2498–2501. [Google Scholar]
- Dias, P.A.; Tabb, A.; Medeiros, H. Multispecies fruit flower detection using a refined semantic segmentation network. IEEE Robot. Autom. Lett. 2018, 3, 3003–3010. [Google Scholar] [CrossRef]
- Chen, Y.; Lee, W.S.; Gan, H.; Peres, N.; Fraisse, C.; Zhang, Y.; He, Y. Strawberry Yield Prediction Based on a Deep Neural Network Using High-Resolution Aerial Orthoimages. Remote Sens. 2019, 11, 1584. [Google Scholar] [CrossRef]
- Horton, R.; Cano, E.; Bulanon, D.; Fallahi, E. Peach flower monitoring using aerial multispectral imaging. J. Imaging 2017, 3, 2. [Google Scholar] [CrossRef]
- López-Granados, F.; Torres-Sánchez, J.; Jiménez-Brenes, F.M.; Arquero, O.; Lovera, M.; de Castro, A.I. An efficient RGB-UAV-based platform for field almond tree phenotyping: 3-D architecture and flowering traits. Plant Methods 2019, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Wang, X.; Ding, Y.; Lu, W.; Sun, Y.J.A. Remote Measurement of Apple Orchard Canopy Information Using Unmanned Aerial Vehicle Photogrammetry. Agronomy 2019, 9, 774. [Google Scholar] [CrossRef]
- Meier, U. BBCH-Monograph. In Growth Stages of Plants–Entwicklungsstadien von Pflanzen–Estadios de las Plantas–Développement des Plantes; Blackwell: Berlin, Germany; Wien, Austria, 1997. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W.J.N.m. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J.; Van Etten, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Lamigueiro, O.P.; Bevan, A.; Racine, E.B.; Shortridge, A.J.R.p. Package ‘Raster’; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Roussel, J.-R.; Auty, D.; De Boissieu, F.; Meador, A.S. lidR: Airborne LiDAR data manipulation and visualization for forestry applications. 2018. Available online: https://cran.r-project.org/web/packages/lidR/index.html (accessed on 17 March 2020).
- Team, R.C. R: A Language and Environment for Statistical Computing. 2018. Available online: https://www.r-project.org (accessed on 17 March 2020).
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O.; Abdel-Rahman, E.M.; Ismail, R.; Slotow, R. Predicting Eucalyptus spp. stand volume in Zululand, South Africa: An analysis using a stochastic gradient boosting regression ensemble with multi-source data sets. Int. J. Remote Sens. 2015, 36, 3751–3772. [Google Scholar] [CrossRef]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B. Caret: Classification and Regression Training; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Ridgeway, G. gbm: Generalized Boosted Regression Models. 2017. Available online: https://mran.microsoft.com/snapshot/2017-12-11/web/packages/gbm/index.html (accessed on 17 March 2020).
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Kdd, Portland, OR, USA, 2 August 1996; pp. 226–231. [Google Scholar]
- Hahsler, M.; Piekenbrock, M.; Doran, D. dbscan: Fast density-based clustering with r. J. Stat. Softw. 2019, 25, 409–416. [Google Scholar] [CrossRef]
- Schubert, E.; Sander, J.; Ester, M.; Kriegel, H.P.; Xu, X. DBSCAN revisited, revisited: Why and how you should (still) use DBSCAN. ACM Trans. Database Syst. (TODS) 2017, 42, 19. [Google Scholar] [CrossRef]
- Starczewski, A.; Cader, A. Determining the Eps Parameter of the DBSCAN Algorithm. In Proceedings of the International Conference on Artificial Intelligence and Soft Computing, Zakopane, Poland, 16–20 June 2019; pp. 420–430. [Google Scholar]
- Story, M.; Congalton, R.G. Accuracy assessment: A user’s perspective. Photogramm. Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Congalton, R.G.; Mead, R.A. A quantitative method to test for consistency and correctness in photointerpretation. Photogramm. Eng. Remote Sens. 1983, 49, 69–74. [Google Scholar]
- Banko, G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data and of Methods Including Remote Sensing Data in Forest Inventory; International Institution for Applied Systems Analysis: Laxenburg, Austria, 1998. [Google Scholar]
- Baeck, P.; Lewyckyj, N.; Beusen, B.; Horsten, W.; Pauly, K. Drone based near real-time human detection with geographic localization. Int. Arch. Photogramm. Remote Sens. Spat. Inform. Sci. 2019, XLII-3/W8, 49–53. [Google Scholar] [CrossRef]
- Lordan, J.; Reginato, G.H.; Lakso, A.N.; Francescatto, P.; Robinson, T.L. Natural fruitlet abscission as related to apple tree carbon balance estimated with the MaluSim model. Sci. Hortic. 2019, 247, 296–309. [Google Scholar] [CrossRef]
- Johnson, K.; Stockwell, V.O. Management of fire blight: A case study in microbial ecology. Annu. Rev. Phytopathol. 1998, 36, 227–248. [Google Scholar] [CrossRef] [PubMed]
- Quinet, M.; Jacquemart, A.-L. Cultivar placement affects pollination efficiency and fruit production in European pear (Pyrus communis) orchards. Eur. J. Agron. 2017, 91, 84–92. [Google Scholar] [CrossRef]
- Tu, Y.-H.; Phinn, S.; Johansen, K.; Robson, A.; Wu, D. Optimising drone flight planning for measuring horticultural tree crop structure. ISPRS-J. Photogramm. Remote Sens. 2020, 160, 83–96. [Google Scholar] [CrossRef]
| K | ε | R2 | RMSE | RRMSE | |
|---|---|---|---|---|---|
| 2D Object-based tree | 1 | 0.005 | 0.72 (0.10) | 15 (2) | 14% (2%) |
| 2D Object-based plot | 2 | 0.012 | 0.81 (0.14) | 34 (8) | 10% (2%) |
| 3D Object-based tree | 12 | 0.11 | 0.44 (0.15) | 33 (8) | 23% (6%) |
| 3D Object-based plot | 16 | 0.135 | 0.71 (0.2) | 60 (16) | 14% (4 %) |
| K | ε | R2 | RMSE | RRMSE | |
|---|---|---|---|---|---|
| 2D Object-based tree | 9 | 0.019 | 0.60 (0.16) | 19 (4) | 16% (3%) |
| 2D Object-based plot | 9 | 0.022 | 0.87 (0.10) | 32 (9) | 9% (3%) |
| 3D Object-based tree | 1 | 0.06 | 0.45 (0.17) | 33 (6) | 23% (4%) |
| 3D Object-based plot | 13 | 0.12 | 0.70 (0.21) | 64 (18) | 15%(4%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanbrabant, Y.; Delalieux, S.; Tits, L.; Pauly, K.; Vandermaesen, J.; Somers, B. Pear Flower Cluster Quantification Using RGB Drone Imagery. Agronomy 2020, 10, 407. https://doi.org/10.3390/agronomy10030407
Vanbrabant Y, Delalieux S, Tits L, Pauly K, Vandermaesen J, Somers B. Pear Flower Cluster Quantification Using RGB Drone Imagery. Agronomy. 2020; 10(3):407. https://doi.org/10.3390/agronomy10030407
Chicago/Turabian StyleVanbrabant, Yasmin, Stephanie Delalieux, Laurent Tits, Klaas Pauly, Joke Vandermaesen, and Ben Somers. 2020. "Pear Flower Cluster Quantification Using RGB Drone Imagery" Agronomy 10, no. 3: 407. https://doi.org/10.3390/agronomy10030407
APA StyleVanbrabant, Y., Delalieux, S., Tits, L., Pauly, K., Vandermaesen, J., & Somers, B. (2020). Pear Flower Cluster Quantification Using RGB Drone Imagery. Agronomy, 10(3), 407. https://doi.org/10.3390/agronomy10030407