Glucosinolate Biosynthesis and the Glucosinolate–Myrosinase System in Plant Defense
Abstract
1. Introduction
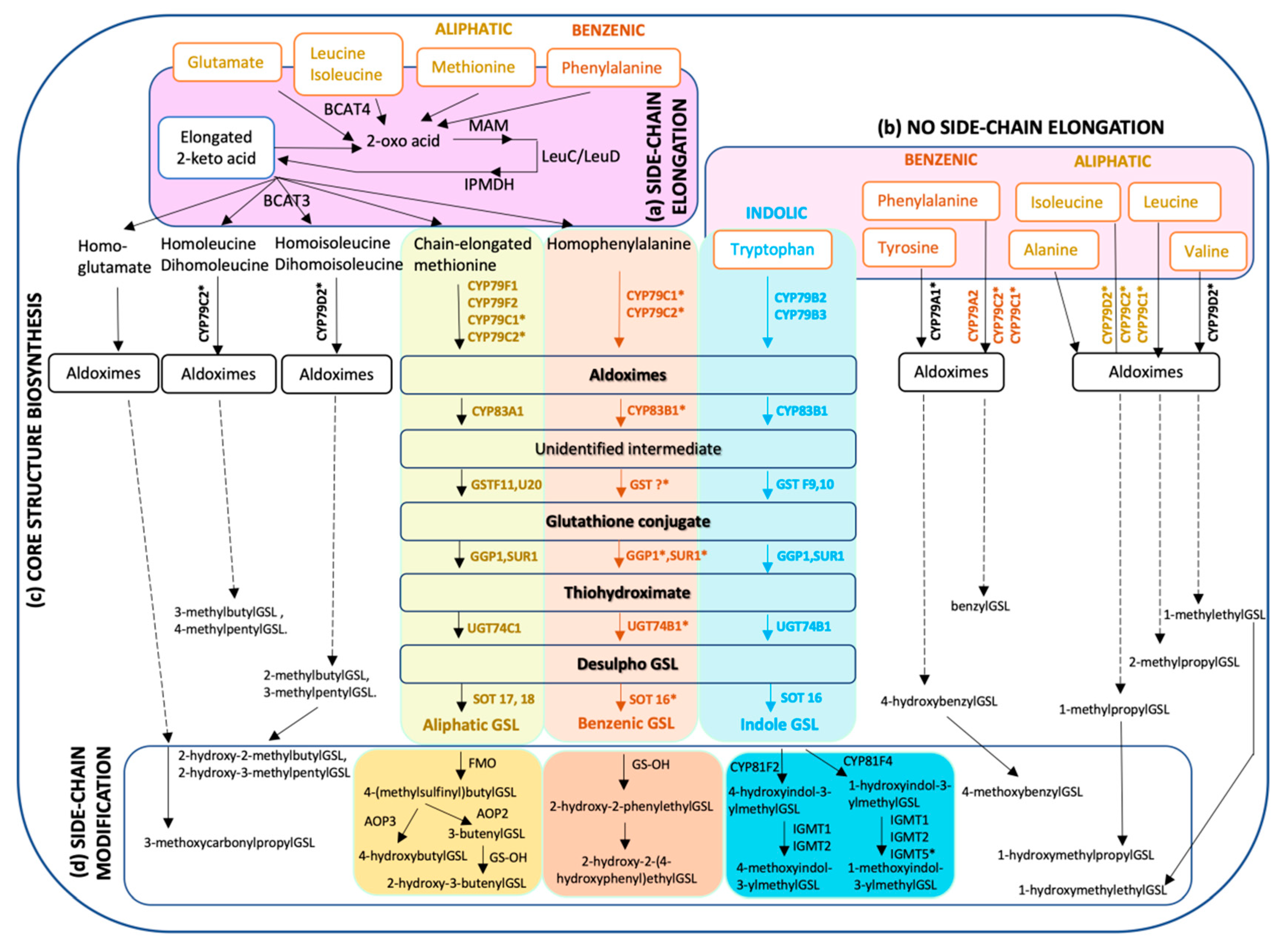
2. Diversity of Glucosinolate Structure and Biosynthesis
2.1. Biosynthesis of Aliphatic Glucosinolates
2.2. Biosynthesis of Indole Glucosinolates
2.3. Knowledge Gaps in the Biosynthesis of Glucosinolates
3. Regulation of Glucosinolate Biosynthesis
3.1. Transcriptional Regulators Controlling Glucosinolate Biosynthesis
3.2. Signaling Networks in the Control of Glucosinolate Biosynthesis
3.3. Crosstalk of Glucosinolate Biosynthesis with Other Metabolic Pathways
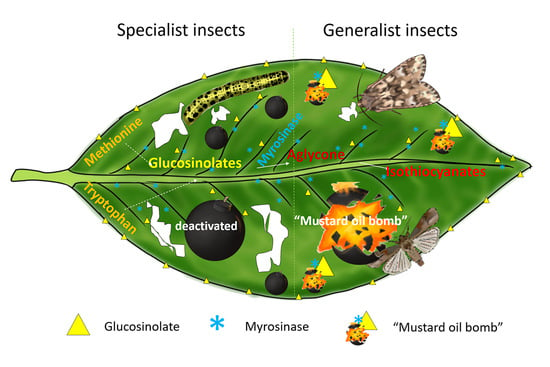
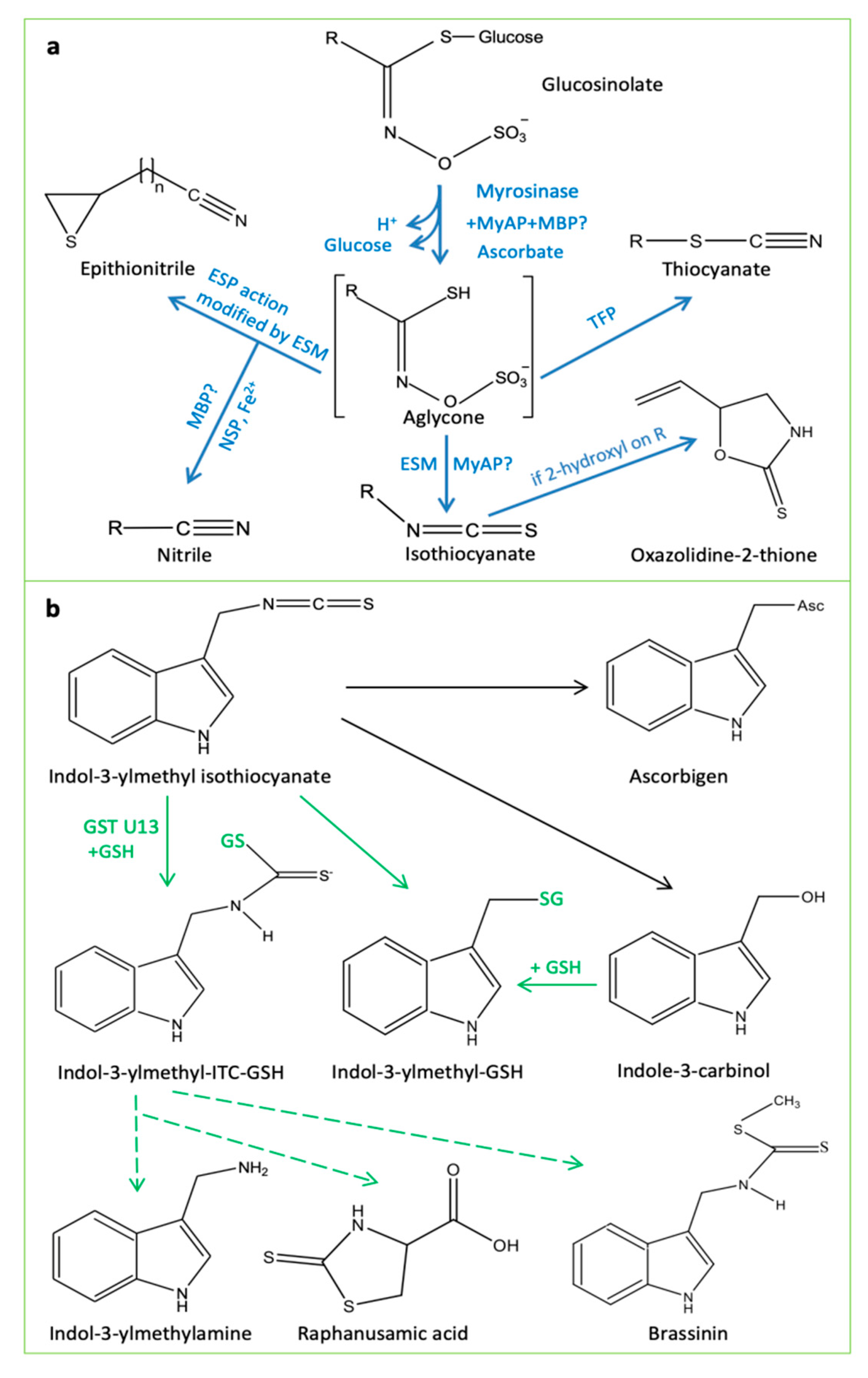
4. Glucosinolate–Myrosinase System: The “Mustard Oil Bomb”
4.1. Myrosinases and Myrosinase-Interacting Proteins
4.2. “Mustard Oil Bomb” in Bacterial and Fungal Pathogen Defense
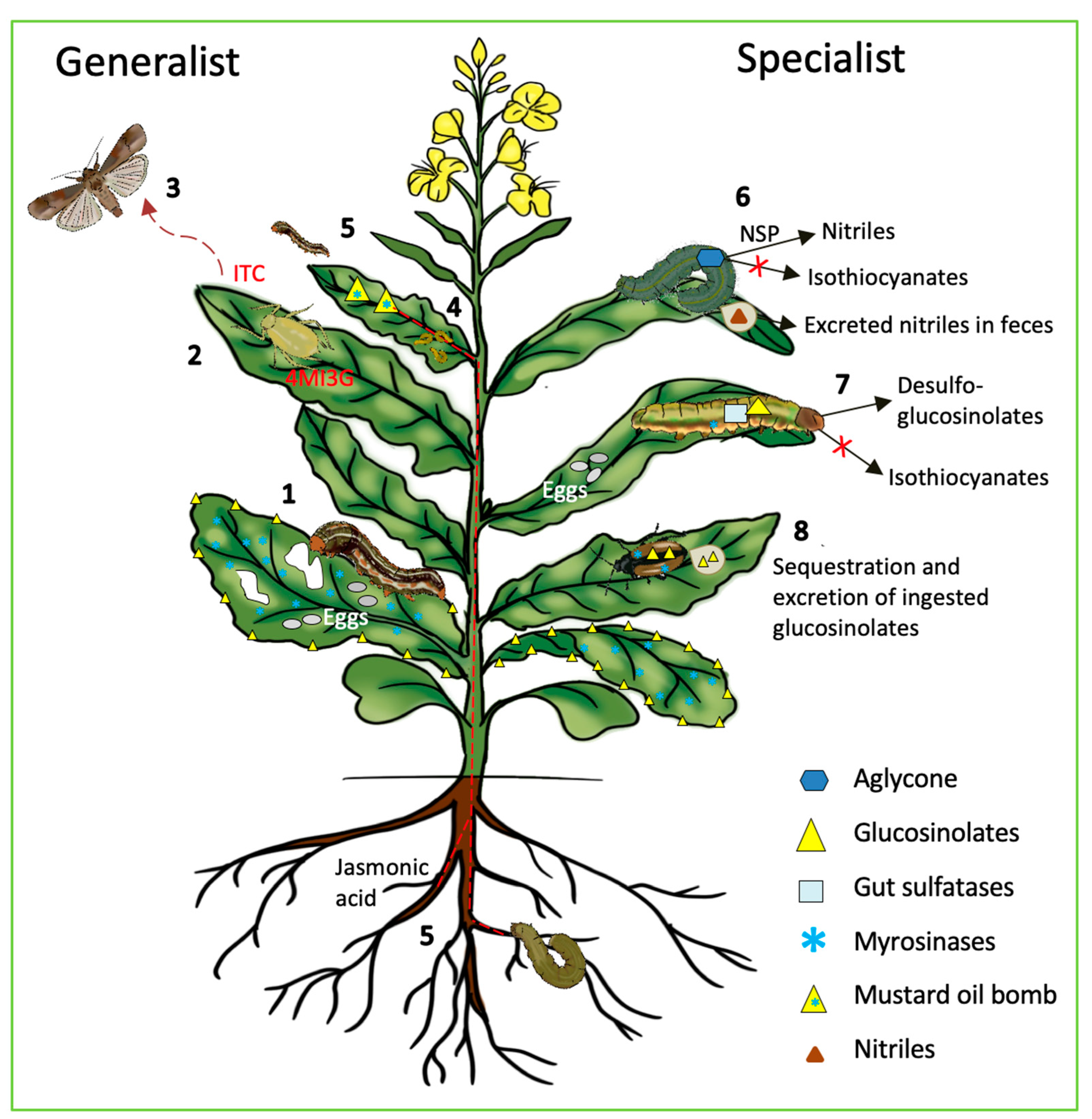
5. Role of the “Mustard Oil Bomb” in Plant Insect Defense
5.1. Disarming the “Mustard Oil Bomb”
5.2. “Mustard Oil Bomb” in Tug of War between Plants and Insects
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Kliebenstein:, D.J.; Kroymann, J.; Mitchell-Olds, T. The glucosinolate–myrosinase system in an ecological and evolutionary context. Curr. Opin. Plant Biol. 2005, 8, 264–271. [Google Scholar] [CrossRef]
- Yan, X.; Chen, S. Regulation of plant glucosinolate metabolism. Planta 2007, 226, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed]
- Horbowicz, M. The occurrence, role and contents of glucosinolates in Brassica vegetables. Veg. Crops Res. Bull. 2003, 58, 23–40. [Google Scholar]
- Possenti, M.; Baima, S.; Raffo, A.; Durazzo, A.; Giusti, A.M.; Natella, F. Glucosinolates in food. In Glucosinolates, Reference Series in Phytochemistry; Merillon, J.M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 87–132. [Google Scholar]
- Edger, P.P.; Hall, J.C.; Harkess, A.; Tang, M.; Coombs, J.; Mohammadin, S.; Schranz, M.E.; Xiong, Z.; Leebens-Mack, J.; Meyers, B.C.; et al. Brassicales phylogeny inferred from 72 plastid genes: A reanalysis of the phylogenetic localization of two paleopolyploid events and origin of novel chemical defenses. Am. J. Bot. 2018, 105, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S. Inhibition of carcinogenesis by isothiocyanates. Drug Metab. Rev. 2000, 32, 395–411. [Google Scholar] [CrossRef]
- Johnson, T.; Dinkova-Kostova, A.; Fahey, J.W. Glucosinolates from the Brassica vegetables and their health effects. In Encyclopedia of Food and Health; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 248–255. [Google Scholar]
- Kaiser, S.J.; Mutters, N.T.; Blessing, B.; Gunther, F. Natural isothiocyanates express antimicrobial activity against developing and mature biofilms of Pseudomonas aeruginosa. Fitoterapia 2017, 119, 57–63. [Google Scholar] [CrossRef]
- Bednarek, P.; Piślewska-Bednarek, M.; Svatoš, A.; Schneider, B.; Doubský, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef]
- Fei, M.; Harvey, J.A.; Weldegergis, B.T.; Huang, T.; Reijngoudt, K.; Vet, L.M.; Gols, R. Integrating insect life history and food plant phenology: Flexible maternal choice is adaptive. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Humphrey, P.T.; Gloss, A.D.; Alexandre, N.M.; Villalobos, M.M.; Fremgen, M.R.; Groen, S.C.; Meihls, L.N.; Jander, G.; Whiteman, N.K. Aversion and attraction to harmful plant secondary compounds jointly shape the foraging ecology of a specialist herbivore. Ecol. Evol. 2016, 6, 3256–3268. [Google Scholar] [CrossRef]
- Kim, J.H.; Jander, G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 2007, 49, 1008–1019. [Google Scholar] [CrossRef]
- Moyers, B.T. Symphony of the Regulators: How do plants control complex responses to environmental signals? Plant Cell 2018, 30, 258–259. [Google Scholar] [CrossRef] [PubMed]
- Nintemann, S.J.; Vik, D.; Svozil, J.; Bak, M.; Baerenfaller, K.; Burow, M.; Halkier, B.A. Unravelling protein-protein interaction networks linked to aliphatic and indole glucosinolate biosynthetic pathways in arabidopsis. Front. Plant Sci. 2017, 8, 2028. [Google Scholar] [CrossRef] [PubMed]
- Pilarska, M.; Wiciarz, M.; Jajic, I.; Kozieradzka-Kiszkurno, M.; Dobrev, P.; Vankova, R.; Niewiadomska, E. A Different pattern of production and scavenging of reactive oxygen species in halophytic Eutrema salsugineum (Thellungiella salsuginea) plants in comparison to Arabidopsis thaliana and its relation to salt stress signaling. Front. Plant Sci. 2016, 7, 1179. [Google Scholar] [CrossRef] [PubMed]
- Reichelt, M.; Brown, P.D.; Schneider, B.; Oldham, N.J.; Stauber, E.; Tokuhisa, J.; Kliebenstein, D.J.; Mitchell-Olds, T.; Gershenzon, J. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 2002, 59, 663–671. [Google Scholar] [CrossRef]
- Brown, P.D.; Tokushisa, J.; Reichelt, M.; Gershenzon, J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 2003, 62, 471–481. [Google Scholar] [CrossRef]
- Jensen, L.M.; Jepsen, H.K.; Halkier, B.A.; Kliebenstein, D.J.; Burow, M. Natural variation in cross-talk between glucosinolates and onset of flowering in Arabidopsis. Front. Plant Sci. 2015, 8, 697. [Google Scholar] [CrossRef]
- Kim, J.I.; Zhang, X.; Pascuzzi, P.E.; Liu, C.J.; Chapple, C. Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol. 2020, 225, 154–168. [Google Scholar] [CrossRef]
- Frisch, T.; Motawia, M.S.; Olsen, C.E.; Agerbirk, N.; Moller, B.L.; Bjarnholt, N. Diversified glucosinolate metabolism: Biosynthesis of hydrogen cyanide and of the hydroxynitrile glucoside alliarinoside in relation to sinigrin metabolism in Alliaria petiolata. Front. Plant Sci. 2015, 6, 926. [Google Scholar] [CrossRef]
- Angelino, D.; Dosz, E.B.; Sun, J.; Hoeflinger, J.L.; Van Tassell, M.L.; Chen, P.; Harnly, J.M.; Miller, M.J.; Jeffery, E.H. Myrosinase-dependent and–independent formation and control of isothiocyanate products of glucosinolate hydrolysis. Front. Plant Sci. 2015, 6, 831. [Google Scholar] [CrossRef] [PubMed]
- Møldrup, M.E.; Geu-Flores, F.; de Vos, M.; Olsen, C.E.; Sun, J.; Jander, G.; Halkier, B. Engineering of benzylglucosinolate in tobacco provides proof-of-concept for dead-end trap crops genetically modified to attract Plutella xylostella (diamondback moth). Plant Biotechnol. J. 2012, 10, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Pislewska-Bednarek, M.; Nakano, R.T.; Hiruma, K.; Pastorczyk, M.; Sanchez-Vallet, A.; Singkaravanit-Ogawa, S.; Ciesiolka, D.; Takano, Y.; Molina, A.; Schulze-Lefert, P.; et al. Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol. 2018, 176, 538–551. [Google Scholar] [CrossRef]
- Agerbirk, N.; De Vos, M.; Kim, J.H.; Jander, G. Indole glucosinolate breakdown and its biological effects. Phytochem. Rev. 2009, 8, 101. [Google Scholar] [CrossRef]
- Bell, L. The biosynthesis of glucosinolates: Insights, inconsistencies, and unknowns. Annu. Plant Rev. Online 2018, 969–1000. [Google Scholar]
- Blažević, I.; Montaut, S.; Burčul, F.; Olsen, C.E.; Burow, M.; Rollin, P.; Agerbirk, N. Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 2020, 169, 112100. [Google Scholar] [CrossRef]
- Agerbirk, N.; Olsen, C.E. Glucosinolate structures in evolution. Phytochemistry 2012, 77, 16–45. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Textor, S.; De Kraker, J.W.; Hause, B.; Gershenzon, J.; Tokuhisa, J.G. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 2007, 144, 60–71. [Google Scholar] [CrossRef]
- Kroymann, J.; Textor, S.; Tokuhisa, J.G.; Falk, K.L.; Bartram, S.; Gershenzon, J.; Mitchell-Olds, T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 2001, 127, 1077–1088. [Google Scholar] [CrossRef]
- Kumar, R.; Lee, S.G.; Augustine, R.; Reichelt, M.; Vassão, D.G.; Palavalli, M.H.; Allen, A.; Gershenzon, J.; Jez, J.M.; Bisht, N.C. Molecular basis of the evolution of methylthioalkylmalate synthase and the diversity of methionine-derived glucosinolates. Plant Cell 2019, 31, 1633–1647. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Galant, A.; Pang, Q.; Strul, J.M.; Balogun, S.F.; Jez, J.M.; Chen, S. Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 28794–28801. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Mawhinney, T.P.; Preuss, M.L.; Schroeder, A.C.; Chen, B.; Abraham, L.; Jez, J.M.; Chen, S. A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 2009, 60, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Selmar, D. Biosynthesis of cyanogenic glycosides, glucosinolates and non-protein amino acids. Annu. Plant Revi. Online 2018, 15, 92–181. [Google Scholar]
- Reed, D.W.; Davin, L.; Jain, J.C.; Deluca, V.; Nelson, L.; Underhill, E.W. Purification and properties of UDP-glucose: Thiohydroximate glucosyltransferase from Brassica napus L. seedlings. Arch. Biochem. Biophys. 1993, 305, 526–532. [Google Scholar] [CrossRef]
- Hirschmann, F.; Krause, F.; Baruch, P.; Chizhov, I.; Mueller, J.W.; Manstein, D.J.; Papenbrock, J.; Fedorov, R. Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism. Sci. Rep. 2017, 7, 4160. [Google Scholar] [CrossRef]
- Kakizaki, T.; Kitashiba, H.; Zou, Z.; Li, F.; Fukino, N.; Ohara, T.; Nishio, T.; Ishida, M. A 2-Oxoglutarate-dependent dioxygenase mediates the biosynthesis of glucoraphasatin in radish. Plant Physiol. 2017, 173, 1583–1593. [Google Scholar] [CrossRef]
- Zuluaga, D.L.; Graham, N.S.; Klinder, A.; van Ommen Kloeke, A.E.E.; Marcotrigiano, A.R.; Wagstaff, C.; Verkerk, R.; Sonnante, G.; Aarts, M.G.M. Overexpression of the MYB29 transcription factor affects aliphatic glucosinolate synthesis in Brassica oleracea. Plant Mol. Biol. 2019, 101, 65–79. [Google Scholar] [CrossRef]
- Hansen, B.G.; Kerwin, R.E.; Ober, J.A.; Lambrix, V.M.; Mitchell-Olds, T.; Gershenzon, J.; Halkier, B.A.; Kliebenstein, D.J. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008, 148, 2096–2108. [Google Scholar] [CrossRef]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2–oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J.; Lambrix, V.M.; Reichelt, M.; Gershenzon, J.; Mitchell-Olds, T. Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 2001, 13, 681–693. [Google Scholar] [PubMed]
- Jensen, L.M.; Kliebenstein, D.J.; Burow, M. Investigation of the multifunctional gene AOP3 expands the regulatory network fine-tuning glucosinolate production in Arabidopsis. Front. Plant Sci. 2015, 6, 762. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.Y.; Sugiyama, K.; Sawada, Y.; Tohge, T.; Obayashi, T.; Suzuki, A.; Araki, R.; Sakurai, N.; Suzuki, H.; Aoki, K.; et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 6478–6483. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.M.; Halkier, B.A.; Burow, M. How to discover a metabolic pathway? An update on gene identification in aliphatic glucosinolate biosynthesis, regulation and transport. Biol. Chem. 2014, 395, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Böttcher, C.; Baatout, D.; Gigolashvili, T. Glucosinolates are produced in trichomes of Arabidopsis thaliana. Front. Plant Sci. 2012, 3, 242. [Google Scholar] [CrossRef]
- Yan, H.; Yoo, M.J.; Koh, J.; Liu, L.; Chen, Y.; Acikgoz, D.; Wang, Q.; Chen, S. Molecular reprogramming of Arabidopsis in response to perturbation of jasmonate signaling. J. Proteom. Res. 2014, 13, 5751–5766. [Google Scholar] [CrossRef]
- Pfalz, M.; Mikkelsen, M.D.; Bednarek, P.; Olsen, C.E.; Halkier, B.A.; Kroymann, J. Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 2011, 23, 716–729. [Google Scholar] [CrossRef]
- Mostafa, I.; Yoo, M.J.; Zhu, N.; Geng, S.; Dufresne, C.; Abou-Hashem, M.; El-Domiaty, M.; Chen, S. Membrane proteomics of arabidopsis glucosinolate mutants cyp79b2/b3 and myb28/29. Front. Plant Sci. 2017, 8, 534. [Google Scholar] [CrossRef]
- Rahikainen, M.; Trotta, A.; Alegre, S.; Pascual, J.; Vuorinen, K.; Overmyer, K.; Moffatt, B.; Ravanel, S.; Glawischnig, E.; Kangasjarvi, S. PP2A-B’gamma modulates foliar trans-methylation capacity and the formation of 4-methoxy-indol-3-yl-methyl glucosinolate in Arabidopsis leaves. Plant J. 2017, 89, 112–127. [Google Scholar] [CrossRef]
- Pfalz, M.; Mukhaimar, M.; Perreau, F.; Kirk, J.; Hansen, C.I.; Olsen, C.E.; Agerbirk, N.; Kroymann, J. Methyl transfer in glucosinolate biosynthesis mediated by indole glucosinolate o-methyltransferase 5. Plant Physiol. 2016, 172, 2190–2203. [Google Scholar] [CrossRef] [PubMed]
- Durian, G.; Rahikainen, M.; Alegre, S.; Brosché, M.; Kangasjärvi, S. Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front. Plant Sci. 2016, 7, 812. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Halkier, B.A. Metabolic engineering of valine-and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from cassava. Plant Physiol. 2003, 131, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dissing, M.M.; Agerbirk, N.; Crocoll, C.; Halkier, B.A. Characterization of Arabidopsis CYP79C1 and CYP79C2 by glucosinolate pathway engineering in Nicotiana benthamiana shows substrate specificity toward a range of aliphatic and aromatic amino acids. Front. Plant Sci. 2020, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Olsen, C.E.; Petersen, B.L.; Møller, B.L.; Halkier, B.A. Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J. 1999, 20, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dissing, M.M.; Agerbirk, N.; Crocoll, C.; Halkier, B.A. Engineering and optimization of the 2-phenylethylglucosinolate production in Nicotiana benthamiana by combining biosynthetic genes from Barbarea vulgari and Arabidopsis thaliana. Biorxiv 2020. [Google Scholar]
- Liu, T.; Zhang, X.; Yang, H.; Agerbirk, N.; Qiu, Y.; Wang, H.; Shen, D.; Song, J.; Li, X. Aromatic glucosinolate biosynthesis pathway in Barbarea vulgaris and its response to Plutella xylostella infestation. Front. Plant Sci. 2016, 7, 83. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, T.; Duan, M.; Song, J.; Li, X. De novo transcriptome analysis of Sinapis alba in revealing the glucosinolate and phytochelatin pathways. Front. Plant Sci. 2016, 7, 259. [Google Scholar] [CrossRef]
- Petersen, A.; Hansen, L.G.; Mirza, N.; Crocoll, C.; Mirza, O.; Halkier, B.A. Changing substrate specificity and iteration of amino acid chain elongation in glucosinolate biosynthesis through targeted mutagenesis of Arabidopsis methylthioalkylmalate synthase 1. Biosci. Rep. 2019, 39, BSR20190446. [Google Scholar] [CrossRef]
- Kittipol, V.; He, Z.; Wang, L.; Doheny-Adams, T.; Langer, S.; Bancroft, I. Genetic architecture of glucosinolate variation in Brassica napus. J. Plant Physiol. 2019, 240, 152988. [Google Scholar] [CrossRef]
- Wittstock, U.; Halkier, B.A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002, 7, 263–270. [Google Scholar] [CrossRef]
- Mewis, I.; Appel, H.M.; Hom, A.; Raina, R.; Schultz, J.C. Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 2005, 138, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A.; Inoue, E.; Watanabe-Takahashi, A.; Yamaya, T.; Takahashi, H. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 2003, 132, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Wang, Q.; Kaspi, R.; Parrella, M.P.; Abel, S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005, 43, 79–96. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Nakamura, Y.; Tohge, T.; Saito, K.; Takahashi, H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 2006, 18, 3235–3251. [Google Scholar] [CrossRef]
- Seo, M.S.; Kim, J.S. Understanding of MYB Transcription Factors Involved in Glucosinolate Biosynthesis in Brassicaceae. Molecules 2017, 22. [Google Scholar]
- Sønderby, I.E.; Hansen, B.G.; Bjarnholt, N.; Ticconi, C.; Halkier, B.A.; Kliebenstein, D.J. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2007, 2, e1322. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Engqvist, M.; Yatusevich, R.; Müller, C.; Flügge, U.I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008, 177, 627–642. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Müller, C.; Flügge, U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 51, 247–261. [Google Scholar] [CrossRef]
- Celenza, J.L.; Quiel, J.A.; Smolen, G.A.; Merrikh, H.; Silvestro, A.R.; Normanly, J.; Bender, J. The Arabidopsis ATR1 Myb transcription factor controls indole glucosinolate homeostasis. Plant Physiol. 2005, 137, 253–262. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Berger, B.; Gigolashvili, T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014, 166, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Gigolashvili, T.; Berger, B.; Mock, H.P.; Müller, C.; Weisshaar, B.; Flügge, U.I. The transcription factor HIG1/MYB51 regulates indole glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 50, 886–901. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate indole glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar] [CrossRef]
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef]
- Xu, J.; Meng, J.; Meng, X.; Zhao, Y.; Liu, J.; Sun, T.; Liu, Y.; Wang, Q.; Zhang, S. Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in arabidopsis immunity. Plant Cell 2016, 28, 1144–1162. [Google Scholar] [CrossRef]
- Fröschel, C.; Iven, T.; Walper, E.; Bachmann, V.; Weiste, C.; Dröge-Laser, W. A Gain-of-function screen reveals redundant erf transcription factors providing opportunities for resistance breeding toward the vascular fungal pathogen Verticillium longisporum. Mol. Plant Microbe Interact. 2019, 32, 1095–1109. [Google Scholar] [CrossRef]
- Skirycz, A.; Reichelt, M.; Burow, M.; Birkemeyer, C.; Rolcik, J.; Kopka, J.; Zanor, M.I.; Gershenzon, J.; Strnad, M.; Szopa, J. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006, 47, 10–24. [Google Scholar] [CrossRef]
- Miao, H.Y.; Wang, M.Y.; Chang, J.Q.; Tao, H.; Sun, B.; Wang, Q.M. Effects of glucose and gibberellic acid on glucosinolate content and antioxidant properties of Chinese kale sprouts. J. Zhejiang Univ. Sci. B 2017, 18, 1093–1100. [Google Scholar] [CrossRef]
- Miao, H.; Cai, C.; Wei, J.; Huang, J.; Chang, J.; Qian, H.; Zhang, X.; Zhao, Y.; Sun, B.; Wang, B.; et al. Glucose enhances indolic glucosinolate biosynthesis without reducing primary sulfur assimilation. Sci. Rep. 2016, 6, 31854. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kong, W.; Zhao, H.; Li, R.; Yang, Y.; Li, J. Transcriptome-wide identification of indole glucosinolate dependent flg22-response genes in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 520, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Khoshfarman-Borji, H.; Yali, M.P.; Bozorg-Amirkalaee, M. Induction of resistance against Brevicoryne brassicae by Pseudomonas putida and salicylic acid in canola. Bull. Entomolog. Res. 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Piślewska-Bednarek, M.; Sánchez-Vallet, A.; Molina, A.; Glawischnig, E.; Gigolashvili, T.; Bednarek, P. Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 2016, 9, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Frederickson Matika, D.E.; Loake, G.J. Redox regulation in plant immune function. Antioxid. Redox Signal. 2014, 21, 1373–1388. [Google Scholar] [CrossRef] [PubMed]
- Czerniawski, P.; Bednarek, P. Glutathione S-transferases in the biosynthesis of sulfur-containing secondary metabolites in brassicaceae plants. Front. Plant Sci. 2018, 9, 1639. [Google Scholar] [CrossRef]
- Fukushima, A.; Iwasa, M.; Nakabayashi, R.; Kobayashi, M.; Nishizawa, T.; Okazaki, Y.; Saito, K.; Kusano, M. Effects of combined low glutathione with mild oxidative and low phosphorus stress on the metabolism of Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1464. [Google Scholar] [CrossRef]
- Rasool, B.; Karpinska, B.; Pascual, J.; Kangasjarvi, S.; Foyer, C.H. Catalase, glutathione, and protein phosphatase 2A-dependent organellar redox signalling regulate aphid fecundity under moderate and high irradiance. Plant Cell Environ. 2020, 43, 209–222. [Google Scholar] [CrossRef]
- Katsarou, D.; Omirou, M.; Liadaki, K.; Tsikou, D.; Delis, C.; Garagounis, C.; Krokida, A.; Zambounis, A.; Papadopoulou, K.K. Glucosinolate biosynthesis in Eruca sativa. Plant Physiol. Biochem. 2016, 109, 452–466. [Google Scholar] [CrossRef]
- Jeschke, V.; Weber, K.; Moore, S.S.; Burow, M. Coordination of glucosinolate biosynthesis and turnover under different nutrient conditions. Front. Plant Sci. 2019, 10, 1560. [Google Scholar] [CrossRef]
- Baek, S.A.; Im, K.H.; Park, S.U.; Oh, S.D.; Choi, J.; Kim, J.K. Dynamics of short-term metabolic profiling in radish sprouts (Raphanus sativus L.) in response to nitrogen deficiency. Plants 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Aarabi, F.; Kusajima, M.; Tohge, T.; Konishi, T.; Gigolashvili, T.; Takamune, M.; Sasazaki, Y.; Watanabe, M.; Nakashita, H.; Fernie, A.R.; et al. Sulfur deficiency–induced repressor proteins optimize glucosinolate biosynthesis in plants. Sci. Adv. 2016, 2, e1601087. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, H.M.; Shin, M.; Arasu, M.V.; Chung, D.Y.; Al-Dhabi, N.A.; Kim, S.J. Effect of different proportion of sulphur treatments on the contents of glucosinolate in kale (Brassica oleracea var. acephala) commonly consumed in Republic of Korea. Saudi J. Biol. Sci. 2018, 25, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Aghajanzadeh, T.; Kopriva, S.; Hawkesford, M.J.; Koprivova, A.; De Kok, L.J. Atmospheric H2S and SO2 as sulfur source for Brassica juncea and Brassica rapa: Impact on the glucosinolate composition. Front. Plant Sci. 2015, 6, 924. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A. Metabolic changes sustain the plant life in low-sulfur environments. Curr. Opin. Plant Biol. 2017, 39, 144–151. [Google Scholar] [CrossRef]
- Marino, D.; Ariz, I.; Lasa, B.; Santamaria, E.; Fernandez-Irigoyen, J.; Gonzalez-Murua, C.; Aparicio Tejo, P.M. Quantitative proteomics reveals the importance of nitrogen source to control glucosinolate metabolism in Arabidopsis thaliana and Brassica oleracea. J. Exp. Bot. 2016, 67, 3313–3323. [Google Scholar] [CrossRef]
- Hiruma, K. Roles of plant-derived secondary metabolites during interactions with pathogenic and beneficial microbes under conditions of environmental stress. Microorganisms 2019, 7. [Google Scholar] [CrossRef]
- Almuziny, M.; Decker, C.; Wang, D.; Gerard, P.; Tharayil, N. Nutrient supply and simulated herbivory differentially alter the metabolite pools and the efficacy of the glucosinolate-based defense system in Brassica species. J. Chem. Ecol. 2017, 43, 129–142. [Google Scholar] [CrossRef]
- Rodriguez-Celma, J.; Tsai, Y.H.; Wen, T.N.; Wu, Y.C.; Curie, C.; Schmidt, W. Systems-wide analysis of manganese deficiency-induced changes in gene activity of Arabidopsis roots. Sci. Rep. 2016, 6, 35846. [Google Scholar] [CrossRef]
- McKenzie, M.; Matich, A.; Hunter, D.; Esfandiari, A.; Trolove, S.; Chen, R.; Lill, R. Selenium application during Radish (Raphanus sativus) plant development alters glucosinolate metabolic gene expression and results in the production of 4-(methylseleno)but-3-enyl glucosinolate. Plants 2019, 8. [Google Scholar] [CrossRef]
- Tian, M.; Yang, Y.; Ávila, F.W.; Fish, T.; Yuan, H.; Hui, M.; Pan, S.; Thannhauser, T.W.; Li, L. Effects of selenium supplementation on glucosinolate biosynthesis in broccoli. J. Agric. Food Chem. 2018, 66, 8036–8044. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Xu, X.; Liu, F.; Fan, X.; Pan, S. Untargeted metabolomics reveals predominant alterations in primary metabolites of broccoli sprouts in response to pre-harvest selenium treatment. Food Res. Int. 2018, 111, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Liu, R.; Ruan, Y.; Liu, C. Sodium chloride primes JA-independent defense against Spodoptera litura (Fabricius) larvae in Arabidopsis thaliana. Plant Signal. Behav. 2019, 14, 1607466. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Bisht, N.C. Biotic elicitors and mechanical damage modulate glucosinolate accumulation by co-ordinated interplay of glucosinolate biosynthesis regulators in polyploid Brassica juncea. Phytochemistry 2015, 117, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.M.; Samara, R.; Renaud, J.; Sumarah, M. Plant growth regulator-mediated anti-herbivore responses of cabbage (Brassica oleracea) against cabbage looper Trichoplusia ni Hübner (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 2017, 141, 9–17. [Google Scholar] [CrossRef]
- Brader, G.; Tas, E.; Palva, E.T. Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 2001, 126, 849–860. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Petersen, B.L.; Glawischnig, E.; Jensen, A.B.; Andreasson, E.; Halkier, B.A. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 2003, 131, 298–308. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Thilmony, R.; Underwood, W.; He, S.Y. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006, 46, 34–53. [Google Scholar] [CrossRef]
- Jarad, M.; Mariappan, K.; Almeida-Trapp, M.; Mette, M.F.; Mithofer, A.; Rayapuram, N.; Hirt, H. The lamin-like little nuclei 1 (LINC1) regulates pattern-triggered immunity and jasmonic acid signaling. Front. Plant Sci. 2019, 10, 1639. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites—Pathways, transcription factors and applied aspects—A brief review. New Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of growth-defense balance by the Jasmonate zim-domain (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari, M.; Glauser, G.; Rasmann, S. Root JA induction modifies glucosinolate profiles and increases subsequent aboveground resistance to herbivore attack in Cardamine hirsuta. Front. Plant Sci. 2018, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Ku, K.M.; Becker, T.M.; Juvik, J.A. Transcriptome and metabolome analyses of glucosinolates in two broccoli cultivars following jasmonate treatment for the induction of glucosinolate defense to Trichoplusia ni (Hubner). Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.C.; Juvik, J.A.; Ku, K.M. Targeted metabolomic and transcriptomic analyses of "Red Russian" Kale (Brassicae napus var. pabularia) following methyl jasmonate treatment and larval infestation by the cabbage looper (Trichoplusia ni Hubner). Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Rechner, O.; Neugart, S.; Schreiner, M.; Wu, S.; Poehling, H.-M. Different narrow-band light ranges alter plant secondary metabolism and plant defense response to aphids. J. Chem. Ecol. 2016, 42, 989–1003. [Google Scholar] [CrossRef]
- Rechner, O.; Neugart, S.; Schreiner, M.; Wu, S.; Poehling, H.M. Can narrow-bandwidth light from UV-A to green alter secondary plant metabolism and increase Brassica plant defenses against aphids? PLoS ONE 2017, 12, e0188522. [Google Scholar] [CrossRef]
- Qi, J.; Zhang, M.; Lu, C.; Hettenhausen, C.; Tan, Q.; Cao, G.; Zhu, X.; Wu, G.; Wu, J. Ultraviolet-B enhances the resistance of multiple plant species to lepidopteran insect herbivory through the jasmonic acid pathway. Sci. Rep. 2018, 8, 277. [Google Scholar] [CrossRef]
- Ishiga, Y.; Watanabe, M.; Ishiga, T.; Tohge, T.; Matsuura, T.; Ikeda, Y.; Hoefgen, R.; Fernie, A.R.; Mysore, K.S. The SAL-PAP chloroplast retrograde pathway contributes to plant immunity by regulating glucosinolate pathway and phytohormone signaling. Mol. Plant Microbe Interact. 2017, 30, 829–841. [Google Scholar] [CrossRef]
- Liao, K.; Peng, Y.-J.; Yuan, L.-B.; Dai, Y.-S.; Chen, Q.-F.; Yu, L.-J.; Bai, M.-Y.; Zhang, W.-Q.; Xie, L.-J.; Xiao, S. Brassinosteroids antagonize jasmonate-activated plant defense responses through bri1-ems-suppressor1 (BES1). Plant Physiol. 2020, 182, 1066–1082. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Qian, H.; Shen, W.; Liu, L.; Zhang, M.; Cai, C.; Zhao, Y.; Qiao, J.; Wang, Q. BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 2013, 64, 2401–2412. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, J.; Kim, H.; Chae, W.B.; Kim, S.J.; Lim, Y.P.; Oh, M.H. Brassinosteroids regulate glucosinolate biosynthesis in Arabidopsis thaliana. Physiolog. Plantarum 2018, 163, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Hillwig, M.S.; Chiozza, M.; Casteel, C.L.; Lau, S.T.; Hohenstein, J.; Hernandez, E.; Jander, G.; MacIntosh, G.C. Abscisic acid deficiency increases defence responses against Myzus persicae in Arabidopsis. Mol. Plant Pathol. 2016, 17, 225–235. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y.; Guan, R.; Li, S.; Xu, X.; Zhang, S.; Xu, J. Co-regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. I. Integr. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Malka, S.K.; Cheng, Y. Possible interactions between the biosynthetic pathways of indole glucosinolate and auxin. Front. Plant Sci. 2017, 8, 2131. [Google Scholar] [CrossRef]
- Hemm, M.R.; Ruegger, M.O.; Chapple, C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 2003, 15, 179–194. [Google Scholar] [CrossRef]
- Kim, J.I.; Dolan, W.L.; Anderson, N.A.; Chapple, C. Indole glucosinolate biosynthesis limits phenylpropanoid accumulation in Arabidopsis thaliana. Plant Cell 2015, 27, 1529–1546. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Garbowicz, K.; Brotman, Y.; Tohge, T.; Fernie, A.R. The acetate pathway supports flavonoid and lipid biosynthesis in Arabidopsis. Plant Physiol. 2020, 182, 857–869. [Google Scholar] [CrossRef]
- Muzac, I.; Wang, J.; Anzellotti, D.; Zhang, H.; Ibrahim, R.K. Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch. Biochem. Biophys. 2000, 375, 385–388. [Google Scholar] [CrossRef]
- Mostafa, I.; Zhu, N.; Yoo, M.J.; Balmant, K.M.; Misra, B.B.; Dufresne, C.; Abou-Hashem, M.; Chen, S.; El-Domiaty, M. New nodes and edges in the glucosinolate molecular network revealed by proteomics and metabolomics of Arabidopsis myb28/29 and cyp79B2/B3 glucosinolate mutants. J. Proteom. 2016, 138, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kloth, K.J.; Abreu, I.N.; Delhomme, N.; Petrik, I.; Villard, C.; Strom, C.; Amini, F.; Novak, O.; Moritz, T.; Albrectsen, B.R. Pectin acetylesterase9 affects the transcriptome and metabolome and delays aphid feeding. Plant Physiol. 2019, 181, 1704–1720. [Google Scholar] [CrossRef]
- Hopkins, R.J.; van Dam, N.M.; van Loon, J.J. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 2009, 54. [Google Scholar] [CrossRef] [PubMed]
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: Gene family evolution and herbivory defense in Brassicaceae. Plant Mol. Biol. 2000, 42, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Vyas, D. Myrosinase: Insights on structural, catalytic, regulatory, and environmental interactions. Crit. Rev. Biotechnol. 2019, 39, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Ratzka, A.; Vogel, H.; Kliebenstein, D.J.; Mitchell-Olds, T.; Kroymann, J. Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 2002, 99, 11223–11228. [Google Scholar] [CrossRef] [PubMed]
- Chhajed, S.; Misra, B.B.; Tello, N.; Chen, S. Chemodiversity of the Glucosinolate-Myrosinase System at the Single Cell Type Resolution. Front. Plant Sci. 2019, 10, 618. [Google Scholar] [CrossRef]
- Wittstock, U.; Kurzbach, E.; Herfurth, A.M.; Stauber, E.J. Glucosinolate breakdown. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2016; Volume 80, pp. 125–169. [Google Scholar]
- Wittstock, U.; Burow, M. Tipping the scales-specifier proteins in glucosinolate hydrolysis. IUBMB Life 2007, 59, 744–751. [Google Scholar] [CrossRef]
- Backenkohler, A.; Eisenschmidt, D.; Schneegans, N.; Strieker, M.; Brandt, W.; Wittstock, U. Iron is a centrally bound cofactor of specifier proteins involved in glucosinolate breakdown. PLoS ONE 2018, 13, e0205755. [Google Scholar] [CrossRef]
- Shirakawa, M.; Hara-Nishimura, I. Specialized vacuoles of myrosin cells: Chemical defense strategy in brassicales plants. Plant Cell Physiol. 2018, 59, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, R.; Hirai, M.Y. Atypical myrosinase as a mediator of glucosinolate functions in plants. Front. Plant Sci. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, E.; Provart, N.J. The botany array resource: E-northerns, expression angling and promoter analysis. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef]
- Zhang, J.; Pontoppidan, B.; Xue, J.; Rask, L.; Meijer, J. The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiologia Plantarum. 2002, 115, 25–34. [Google Scholar] [CrossRef]
- Wang, M.; Sun, X.; Tan, D.; Gong, S.; Meijer, J.; Zhang, J. The two non-functional myrosinase genes TGG3 and TGG6 in Arabidopsis are expressed predominantly in pollen. Plant Sci. 2009, 177, 371–375. [Google Scholar] [CrossRef]
- Nakano, R.T.; Pislewska-Bednarek, M.; Yamada, K.; Edger, P.P.; Miyahara, M.; Kondo, M.; Bottcher, C.; Mori, M.; Nishimura, M.; Schulze-Lefert, P.; et al. PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana. Plant J. 2017, 89, 204–220. [Google Scholar] [CrossRef]
- Crane, R.A.; Cardenas Valdez, M.; Castaneda, N.; Jackson, C.L.; Riley, C.J.; Mostafa, I.; Kong, W.; Chhajed, S.; Chen, S.; Brusslan, J.A. Negative regulation of age-related developmental leaf senescence by the IAOx pathway, PEN1, and PEN3. Front. Plant Sci. 2019, 10, 1202. [Google Scholar] [CrossRef]
- Morikawa-Ichinose, T.; Miura, D.; Zhang, L.; Kim, S.-J.; Maruyama-Nakashita, A. Involvement of BGLU30 in glucosinolate catabolism in the Arabidopsis leaf under dark conditions. Plant Cell Physiol. 2020. [Google Scholar] [CrossRef]
- Fujiki, Y.; Yoshikawa, Y.; Sato, T.; Inada, N.; Ito, M.; Nishida, I.; Watanabe, A. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiolog. Plantarum 2001, 111, 345–352. [Google Scholar] [CrossRef]
- Zhang, L.; Kawaguchi, R.; Morikawa-Ichinose, T.; Allahham, A.; Kim, S.-J.; Maruyama-Nakashita, A. Sulfur deficiency-induced glucosinolate catabolism attributed to two β-Glucosidases, BGLU28 and BGLU30, is required for plant growth maintenance under sulfur deficiency. Plant Cell Physiol. 2020, 61, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Agerbirk, N.; Matthes, A.; Erthmann, P.Ø.; Ugolini, L.; Cinti, S.; Lazaridi, E.; Nuzillard, J.-M.; Müller, C.; Bak, S.; Rollin, P. Glucosinolate turnover in Brassicales species to an oxazolidin-2-one, formed via the 2-thione and without formation of thioamide. Phytochemistry 2018, 153, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ober, J.A.; Kliebenstein, D.J. The gene controlling the quantitative trait locus epithiospecifier modifier1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 2006, 18, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Textor, S.; Gershenzon, J. Herbivore induction of the glucosinolate–myrosinase defense system: Major trends, biochemical bases and ecological significance. Phytochem. Rev. 2009, 8, 149–170. [Google Scholar] [CrossRef]
- Ahuja, I.; de Vos, R.C.; Rohloff, J.; Stoopen, G.M.; Halle, K.K.; Ahmad, S.J.; Hoang, L.; Hall, R.D.; Bones, A.M. Arabidopsis myrosinases link the glucosinolate-myrosinase system and the cuticle. Sci. Rep. 2016, 6, 38990. [Google Scholar] [CrossRef] [PubMed]
- Dubey, O.; Dubey, S.; Schnee, S.; Glauser, G.; Nawrath, C.; Gindro, K.; Farmer, E.E. Plant surface metabolites as potent antifungal agents. Plant Physiol. Biochem. 2020. [Google Scholar] [CrossRef]
- Zhang, K.; Su, H.; Zhou, J.; Liang, W.; Liu, D.; Li, J. Overexpressing the myrosinase gene TGG1 enhances stomatal defense against Pseudomonas syringae and delays flowering in Arabidopsis. Front. Plant Sci. 2019, 10, 1230. [Google Scholar] [CrossRef]
- Calmes, B.; N’Guyen, G.; Dumur, J.; Brisach, C.A.; Campion, C.; Iacomi, B.; Pigné, S.; Dias, E.; Macherel, D.; Guillemette, T. Glucosinolate-derived isothiocyanates impact mitochondrial function in fungal cells and elicit an oxidative stress response necessary for growth recovery. Front. Plant Sci. 2015, 6, 414. [Google Scholar] [CrossRef]
- Kuhn, H.; Lorek, J.; Kwaaitaal, M.; Consonni, C.; Becker, K.; Micali, C.; Ver Loren van Themaat, E.; Bednarek, P.; Raaymakers, T.M.; Appiano, M.; et al. Key Components of different plant defense pathways are dispensable for powdery mildew resistance of the Arabidopsis mlo2 mlo6 mlo12 triple mutant. Front. Plant Sci. 2017, 8, 1006. [Google Scholar] [CrossRef]
- Fukunaga, S.; Sogame, M.; Hata, M.; Singkaravanit-Ogawa, S.; Pislewska-Bednarek, M.; Onozawa-Komori, M.; Nishiuchi, T.; Hiruma, K.; Saitoh, H.; Terauchi, R.; et al. Dysfunction of Arabidopsis MACPF domain protein activates programmed cell death via tryptophan metabolism in MAMP-triggered immunity. Plant J. 2017, 89, 381–393. [Google Scholar] [CrossRef]
- Barrameda-Medina, Y.; Blasco, B.; Lentini, M.; Esposito, S.; Baenas, N.; Moreno, D.A.; Ruiz, J.M. Zinc biofortification improves phytochemicals and amino-acidic profile in Brassica oleracea cv. Bronco. Plant Sci. 2017, 258, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Gallego, B.; Martos, S.; Cabot, C.; Barcelo, J.; Poschenrieder, C. Zinc hyperaccumulation substitutes for defense failures beyond salicylate and jasmonate signaling pathways of Alternaria brassicicola attack in Noccaea caerulescens. Physiolog. Plantarum 2017, 159, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Rouphael, Y.; Colla, G.; Bernardo, L.; Kane, D.; Trevisan, M.; Lucini, L. Zinc excess triggered polyamines accumulation in lettuce root metabolome, as compared to osmotic stress under high salinity. Front. Plant Sci. 2016, 7, 842. [Google Scholar] [CrossRef] [PubMed]
- Samira, R.; Li, B.; Kliebenstein, D.; Li, C.; Davis, E.; Gillikin, J.W.; Long, T.A. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol. Biol. 2018, 97, 297–309. [Google Scholar] [CrossRef]
- Dahlin, P.; Hallmann, J. New Insights on the Role of Allyl Isothiocyanate in Controlling the Root Knot Nematode Meloidogyne hapla. Plants 2020, 9. [Google Scholar] [CrossRef]
- Larkin, R.P.; Lynch, R.P. Use and effects of different Brassica and other rotation crops on soilborne diseases and yield of potato. Horticulturae 2018, 4, 37. [Google Scholar] [CrossRef]
- Park, W.; Lee, Y.-H.; Kim, K.-S.; Cha, Y.-L.; Moon, Y.-H.; Song, Y.-S.; Kwon, D.-E.; Lee, J.-E. The optimal mixing ratio of Brassica napus and Brassica juncea meal improve nematode Meloidogyne hapla effects. Plant Signal. Behav. 2019, 14, 1678369. [Google Scholar] [CrossRef]
- Buxdorf, K.; Yaffe, H.; Barda, O.; Levy, M. The effects of glucosinolates and their breakdown products on necrotrophic fungi. PLoS ONE 2013, 8, e70771. [Google Scholar] [CrossRef]
- Mitchell, C.; Brennan, R.M.; Graham, J.; Karley, A.J. Plant defense against herbivorous pests: Exploiting resistance and tolerance traits for sustainable crop protection. Front. Plant Sci. 2016, 7, 1132. [Google Scholar] [CrossRef]
- Agnihotri, A.R.; Hulagabali, C.V.; Adhav, A.S.; Joshi, R.S. Mechanistic insight in potential dual role of sinigrin against Helicoverpa armigera. Phytochemistry 2018, 145, 121–127. [Google Scholar] [CrossRef]
- Ma, X.L.; He, W.Y.; Chen, W.; Xu, X.J.; Qi, W.P.; Zou, M.M.; You, Y.C.; Baxter, S.W.; Wang, P.; You, M.S. Structure and expression of sulfatase and sulfatase modifying factor genes in the diamondback moth, Plutella xylostella. Insect Sci. 2018, 25, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, V.; Kearney, E.E.; Schramm, K.; Kunert, G.; Shekhov, A.; Gershenzon, J.; Vassão, D.G. How glucosinolates affect generalist lepidopteran larvae: Growth, development and glucosinolate metabolism. Front. Plant Sci. 2017, 8, 1995. [Google Scholar] [CrossRef] [PubMed]
- Wittstock, U.; Agerbirk, N.; Stauber, E.J.; Olsen, C.E.; Hippler, M.; Mitchell-Olds, T.; Gershenzon, J.; Vogel, H. Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc. Natl. Acad. Sci. USA 2004, 101, 4859–4864. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.L.; Kunert, G.; Sporer, T.; Kornig, J.; Beran, F. Glucosinolate abundance and composition in brassicaceae influence sequestration in a specialist flea beetle. J. Chem. Ecol. 2020, 46, 186–197. [Google Scholar] [CrossRef]
- Khakimov, B.; Kuzina, V.; Erthmann, P.Ø.; Fukushima, E.O.; Augustin, J.M.; Olsen, C.E.; Scholtalbers, J.; Volpin, H.; Andersen, S.B.; Hauser, T.P.; et al. Identification and genome organization of saponin pathway genes from a wild crucifer, and their use for transient production of saponins in Nicotiana benthamiana. Plant J. 2015, 84, 478–490. [Google Scholar] [CrossRef]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Bak, S. Plant defense against insect herbivores. Int. J. Mol. Sci. 2013, 14, 10242–10297. [Google Scholar] [CrossRef]
- Eakteiman, G.; Moses-Koch, R.; Moshitzky, P.; Mestre-Rincon, N.; Vassão, D.G.; Luck, K.; Sertchook, R.; Malka, O.; Morin, S. Targeting detoxification genes by phloem-mediated RNAi: A new approach for controlling phloem-feeding insect pests. Insect Biochem. Mol. Biol. 2018, 100, 10–21. [Google Scholar] [CrossRef]
- Shroff, R.; Vergara, F.; Muck, A.; Svatoš, A.; Gershenzon, J. Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 2008, 105, 6196–6201. [Google Scholar] [CrossRef]
- Lei, J.; Jayaprakasha, G.K.; Singh, J.; Uckoo, R.; Borrego, E.J.; Finlayson, S.; Kolomiets, M.; Patil, B.S.; Braam, J.; Zhu-Salzman, K. Circadian clock-associated1 controls resistance to aphids by altering indole glucosinolate production. Plant Physiol. 2019, 181, 1344–1359. [Google Scholar] [CrossRef]
- van Hulten, M.; Pelser, M.; Van Loon, L.; Pieterse, C.M.; Ton, J. Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 5602–5607. [Google Scholar] [CrossRef]
- Ton, J.; D’Alessandro, M.; Jourdie, V.; Jakab, G.; Karlen, D.; Held, M.; Mauch-Mani, B.; Turlings, T.C. Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 2007, 49, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Poelman, E.; Van Wees, S.; Dicke, M. Induced plant responses to microbes and insects. Front. Plant Sci. 2013, 4, 475. [Google Scholar] [CrossRef] [PubMed]
- Pierre, S.P.; Dugravot, S.; Hervé, M.R.; Hasan, H.; Van Dam, N.M.; Cortesero, A.M. Belowground induction by Delia radicum or phytohormones affect aboveground herbivore communities on field-grown broccoli. Front. Plant Sci. 2013, 4, 305. [Google Scholar] [CrossRef] [PubMed]
- Geem, M.V.; Raaijmakers, C.E.; Harvey, J.A.; Gols, R. Interactions between a belowground herbivore and primary and secondary root metabolites in wild cabbage. J. Chem. Ecol. 2015, 41, 696–707. [Google Scholar] [CrossRef]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef]
- Papadopoulou, G.V.; van Dam, N.M. Mechanisms and ecological implications of plant-mediated interactions between belowground and aboveground insect herbivores. Ecol. Res. 2017, 32, 13–26. [Google Scholar] [CrossRef]
- De Vos, M.; Van Oosten, V.R.; Van Poecke, R.M.; Van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Van Loon, L.; Dicke, M. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef]
- Ali, J.G.; Agrawal, A.A. Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci. 2012, 17, 293–302. [Google Scholar] [CrossRef]
- Cao, H.H.; Liu, H.R.; Zhang, Z.F.; Liu, T.X. The green peach aphid Myzus persicae perform better on pre-infested Chinese cabbage Brassica pekinensis by enhancing host plant nutritional quality. Sci. Rep. 2016, 6, 21954. [Google Scholar] [CrossRef]
- Cao, H.H.; Zhang, Z.F.; Wang, X.F.; Liu, T.X. Nutrition versus defense: Why Myzus persicae (green peach aphid) prefers and performs better on young leaves of cabbage. PLoS ONE 2018, 13, e0196219. [Google Scholar] [CrossRef]
- Hubbard, C.J.; Li, B.; McMinn, R.; Brock, M.T.; Maignien, L.; Ewers, B.E.; Kliebenstein, D.; Weinig, C. The effect of rhizosphere microbes outweighs host plant genetics in reducing insect herbivory. Mol. Ecol. 2019, 28, 1801–1811. [Google Scholar] [CrossRef] [PubMed]
- Broekgaarden, C.; Snoeren, T.A.; Dicke, M.; Vosman, B. Exploiting natural variation to identify insect-resistance genes. Plant Biotechnol. J. 2011, 9, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Malka, O.; Shekhov, A.; Reichelt, M.; Gershenzon, J.; Vassão, D.G.; Morin, S. Glucosinolate desulfation by the phloem-feeding insect Bemisia tabaci. J. Chem. Ecol. 2016, 42, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Pineda, A.; Pangesti, N.; Soler, R.; van Dam, N.M.; van Loon, J.J.; Dicke, M. Negative impact of drought stress on a generalist leaf chewer and a phloem feeder is associated with, but not explained by an increase in herbivore-induced indole glucosinolates. Environ. Exp. Bot. 2016, 123, 88–97. [Google Scholar] [CrossRef]
- Khan, M.A.M.; Ulrichs, C.; Mewis, I. Drought stress-impact on glucosinolate profile and performance of phloem feeding cruciferous insects. In Proceedings of the XXVIII International Horticultural Congress on Science and Horticulture for People (IHC2010): International Symposium on Plant, Lisbon, Portugal, 22–27 August 2010; Volume 917, pp. 111–117. [Google Scholar]
- Travers-Martin, N.; Müller, C. Specificity of induction responses in Sinapis alba L. and their effects on a specialist herbivore. J. Chem. Ecol. 2007, 33, 1582–1597. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhu, N.; Song, W.y.; Harmon, A.C.; Assmann, S.M.; Chen, S. Thiol-based redox proteins in abscisic acid and methyl jasmonate signaling in Brassica napus guard cells. Plant J. 2014, 78, 491–515. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chhajed, S.; Mostafa, I.; He, Y.; Abou-Hashem, M.; El-Domiaty, M.; Chen, S. Glucosinolate Biosynthesis and the Glucosinolate–Myrosinase System in Plant Defense. Agronomy 2020, 10, 1786. https://doi.org/10.3390/agronomy10111786
Chhajed S, Mostafa I, He Y, Abou-Hashem M, El-Domiaty M, Chen S. Glucosinolate Biosynthesis and the Glucosinolate–Myrosinase System in Plant Defense. Agronomy. 2020; 10(11):1786. https://doi.org/10.3390/agronomy10111786
Chicago/Turabian StyleChhajed, Shweta, Islam Mostafa, Yan He, Maged Abou-Hashem, Maher El-Domiaty, and Sixue Chen. 2020. "Glucosinolate Biosynthesis and the Glucosinolate–Myrosinase System in Plant Defense" Agronomy 10, no. 11: 1786. https://doi.org/10.3390/agronomy10111786
APA StyleChhajed, S., Mostafa, I., He, Y., Abou-Hashem, M., El-Domiaty, M., & Chen, S. (2020). Glucosinolate Biosynthesis and the Glucosinolate–Myrosinase System in Plant Defense. Agronomy, 10(11), 1786. https://doi.org/10.3390/agronomy10111786