Development of a PVY Resistant Flue-Cured Tobacco Line via EMS Mutagenesis of eIF4E
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and EMS (Ethyl Methane Sulfonate) Treatment
2.2. DNA Extraction and Sample Pooling
2.3. PCR Amplification
2.4. Digestion with CEL1 endonuclease and Capillary Electrophoresis
2.5. Validation of eIF4E1-S Mutants Selected in the M3 Population
2.6. Virus Inoculation and Detection
2.7. Genotyping by dCAPS Markers
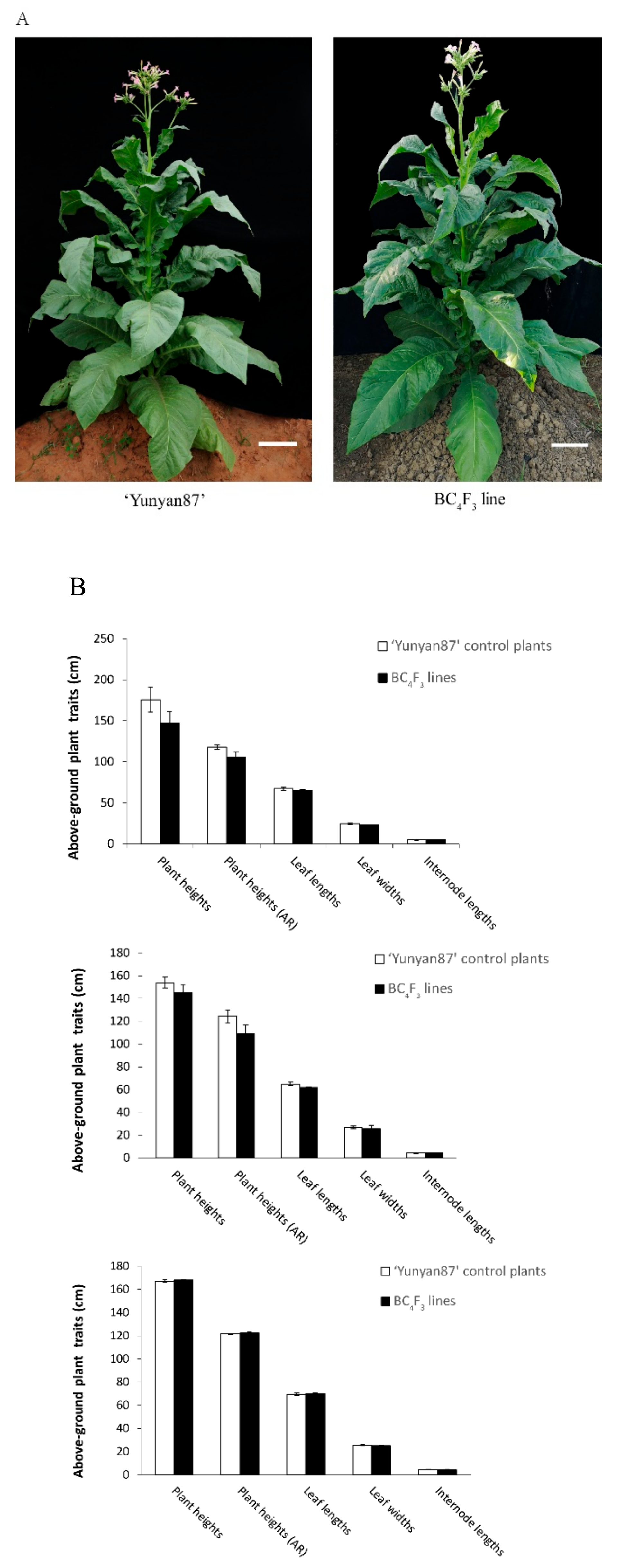
2.8. Pilot Experiments for Agronomic Trait Analysis
3. Results
3.1. Characterization of the eIF4E1-S Mutants Selected in the M2 Generation
3.2. M3 Progeny of Line 918P Exhibit PVY Resistance
3.3. Selection of Backcross Progenies using dCAPS Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; Lopez-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Bastet, A.; Robaglia, C.; Gallois, J.L. eIF4E Resistance: Natural variation should guide gene editing. Trends Plant Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Crosslin, J.M.; Hamm, P.B.; Shiel, P.J.; Hane, D.C.; Brown, C.R.; Berger, P.H. Serological and molecular detection of tobacco veinal necrosis isolates of Potato virus Y (PVYN) from potatoes grown in the western United States. Am. J. Potato Res. 2005, 82, 263–269. [Google Scholar] [CrossRef]
- Nie, X.; Singh, R.P. A new approach for the simultaneous differentiation of biological and geographical strains of Potato virus Y by uniplex and multiplex RT-PCR. J. Virol. Methods 2002, 104, 41–54. [Google Scholar] [CrossRef]
- Navarrete, I.; Panchi, N.; Kromann, P.; Forbes, G.; Andrade-Piedra, J. Health quality of seed potato and yield losses in Ecuador. bioRxiv Artic. 2017, 108712. [Google Scholar] [CrossRef]
- Faurez, F.; Baldwin, T.; Tribodet, M.; Jacquot, E. Identification of new Potato virus Y (PVY) molecular determinants for the induction of vein necrosis in tobacco. Mol. Plant Pathol. 2012, 13, 948–959. [Google Scholar] [CrossRef]
- Miller, R.D. TN 86: A Burley Tobacco Resistant to TVMV, TEV, and PVY; University of Tennessee, Agricultural Experiment Station: Knoxville, TN, USA, 1987. [Google Scholar]
- Dluge, K.L.; Song, Z.; Wang, B.; Tyler Steede, W.; Xiao, B.; Liu, Y.; Dewey, R.E. Characterization of Nicotiana tabacum genotypes possessing deletion mutations that affect potyvirus resistance and the production of trichome exudates. BMC Genom. 2018, 19, 484. [Google Scholar] [CrossRef]
- Acosta-Leal, R.; Xiong, Z. Complementary functions of two recessive R-genes determine resistance durability of tobacco ‘Virgin A Mutant’ (VAM) to Potato virus Y. Virology 2008, 379, 275–283. [Google Scholar] [CrossRef]
- Nielsen, M.T.; Akers, C.P.; Jarlford, V.E.; Wagner, G.J.; Berger, S. Comparative ultrastructural features of secreting and non-secreting glandular trichomes of two genotypes of Nicotiana tabacum L. Bot. Gaz. 1991, 152, 13–22. [Google Scholar] [CrossRef]
- Nielsen, M.T.; Jones, G.A.; Collins, G.B. Inheritance pattern for secreting and nonsecreting glandular trichomes in tobacco. Crop Sci. 1982, 22, 1051–1053. [Google Scholar] [CrossRef]
- Diaz-Pendon, J.A.; Truniger, V.; Nieto, C.; Garcia-Mas, J.; Bendahmane, A.; Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004, 5, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009, 75, 119–159. [Google Scholar] [PubMed]
- Robaglia, C.; Caranta, C. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 2006, 11, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef]
- Decroocq, V.; Sicard, O.; Alamillo, J.M.; Lansac, M.; Eyquard, J.P.; Garcia, J.A.; Candresse, T.; Le Gall, O.; Revers, F. Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana. Mol. Plant Microbe Interact 2006, 19, 541–549. [Google Scholar] [CrossRef]
- Sato, M.; Nakahara, K.; Yoshii, M.; Ishikawa, M.; Uyeda, I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 2005, 579, 1167–1171. [Google Scholar] [CrossRef]
- Ruffel, S.; Dussault, M.H.; Palloix, A.; Moury, B.; Bendahmane, A.; Robaglia, C.; Caranta, C. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 2002, 32, 1067–1075. [Google Scholar] [CrossRef]
- Ruffel, S.; Gallois, J.L.; Lesage, M.L.; Caranta, C. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Gen. Genom. 2005, 274, 346–353. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Julio, E.; Cotucheau, J.; Decorps, C. A eukaryotic translation initiation factor 4E (eIF4E) is responsible for the “va” tobacco recessive resistance to potyviruses. Plant Mol. Biol. Rep. 2015, 33, 609–623. [Google Scholar] [CrossRef]
- Gao, Y.L.; Yao, X.F.; Li, W.Z.; Song, Z.B.; Wang, B.W.; Wu, Y.P.; Shi, J.L.; Liu, G.S.; Li, Y.P.; Liu, C.M. An efficient TILLING platform for cultivated tobacco. J. Integr. Plant Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Athow, K.L.; Laviolette, F.A.; Wilcox, J.R.; Abney, T.S. Registration of ‘TN 86’ burley tobacco. Crop Sci. 1987, 27, 365–366. [Google Scholar] [CrossRef]
- Mazier, M.; Flamain, F.; Nicolai, M.; Sarnette, V.; Caranta, C. Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS ONE 2011, 6, e29595. [Google Scholar] [CrossRef]
- Rodriguez-Hernandez, A.M.; Gosalvez, B.; Sempere, R.N.; Burgos, L.; Aranda, M.A.; Truniger, V. Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 2012, 13, 755–763. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef]
- Wang, X.; Kohalmi, S.E.; Svircev, A.; Wang, A.; Sanfacon, H.; Tian, L. Silencing of the host factor eIF(iso)4E gene confers Plum pox virus resistance in plum. PLoS ONE 2013, 8, e50627. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Li, H.; Perez, K.W.; Jahn, M.M. Ectopic expression of a recessive resistance gene generates dominant potyvirus resistance in plants. Plant Biotechnol. J. 2007, 5, 526–536. [Google Scholar] [CrossRef]
- Arcibal, E.; Gold, M.K.; Flaherty, S. A mutant eIF4E confers resistance to Potato virus Y strains and is inherited in a dominant manner in the potato varieties Atlantic and Russet Norkotah. Am. J. Potato Res. 2015, 93, 64–71. [Google Scholar] [CrossRef]
- Jones, H.D. Regulatory uncertainty over genome editing. Nat. Plants 2015, 1, 14011. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Julio, E.; Candresse, T.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Moury, B.; Glais, L.; Jacquot, E.; de Borne, F.D.; et al. A complex eIF4E locus impacts the durability of va resistance to Potato virus Y in tobacco. Mol. Plant Pathol. 2019, 20, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Masuta, C.; Nishimura, M.; Morishita, H.; Hataya, T. A single amino acid change in viral genome-associated protein of Potato virus Y correlates with resistance breaking in ‘Virgin A Mutant’ tobacco. Phytopathology 1999, 89, 118–123. [Google Scholar] [CrossRef]
- Lebaron, C.; Rosado, A.; Sauvage, C.; Gauffier, C.; German-Retana, S.; Moury, B.; Gallois, J.L. A new eIF4E1 allele characterized by RNAseq data mining is associated with resistance to Potato virus Y in tomato albeit with a low durability. J. Gen. Virol. 2016, 97, 3063–3072. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Nicolai, M.; Gallois, J.L.; Robaglia, C.; Moury, B.; Palloix, A.; Caranta, C. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008, 54, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Gallois, J.L.; Charron, C.; Sanchez, F.; Pagny, G.; Houvenaghel, M.C.; Moretti, A.; Ponz, F.; Revers, F.; Caranta, C.; German-Retana, S. Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J. Gen. Virol. 2010, 91, 288–293. [Google Scholar] [CrossRef]
- Takakura, Y.; Udagawa, H.; Shinjo, A.; Koga, K. Mutation of a Nicotiana tabacum L. eukaryotic translation-initiation factor gene reduces susceptibility to a resistance-breaking strain of Potato virus Y. Mol. Plant Pathol. 2018, 19, 2124–2133. [Google Scholar] [CrossRef]



| Mutant Line (M2) | Nucleic Acid Variation 1 | Amino Acid Variation | PROVEAN Score | Prediction | Target Region |
|---|---|---|---|---|---|
| 844 | GAA/AAA | Glu/Lys, E4K | −0.801 | Neutral | exon |
| 1001 | GAA/AAA | Glu/Lys, E4K | −0.801 | Neutral | exon |
| 113 | GAA/AAA | Glu/Lys, E4K | −0.801 | Neutral | exon |
| 139 | intron | ||||
| 815 | AGA/AAA | Arg/Lys, R62K | 0.9 | Neutral | exon |
| 814 | AGA/AAA | Arg/Lys, R62K | 0.9 | Neutral | exon |
| 939 | CGG/TGG | Arg/Trp, R9W | −1.587 | Neutral | exon |
| 962 | intron | ||||
| 911 | GAT/AAT | Asp/Asn, D81N | −3.302 | Deleterious | exon |
| 918 | TGG/TGA | STOP | exon | ||
| 1381 | GAG/AAG | Glu/Lys, E3K | −1.256 | Neutral | exon |
| 278 | TCC/TTC | Ser/Phe, S77F | −3.942 | Deleterious | exon |
| 1500 | GAA/AAA | Glu/Lys, E80K | −3.679 | Deleterious | exon |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Li, W.; Wang, B.; Gao, Y.; Sui, X.; Liu, Y.; Chen, X.; Yao, X.; Jiao, F.; Song, Z. Development of a PVY Resistant Flue-Cured Tobacco Line via EMS Mutagenesis of eIF4E. Agronomy 2020, 10, 36. https://doi.org/10.3390/agronomy10010036
Zhao L, Li W, Wang B, Gao Y, Sui X, Liu Y, Chen X, Yao X, Jiao F, Song Z. Development of a PVY Resistant Flue-Cured Tobacco Line via EMS Mutagenesis of eIF4E. Agronomy. 2020; 10(1):36. https://doi.org/10.3390/agronomy10010036
Chicago/Turabian StyleZhao, Lu, Wenzheng Li, Bingwu Wang, Yulong Gao, Xueyi Sui, Yong Liu, Xuejun Chen, Xuefeng Yao, Fangchan Jiao, and Zhongbang Song. 2020. "Development of a PVY Resistant Flue-Cured Tobacco Line via EMS Mutagenesis of eIF4E" Agronomy 10, no. 1: 36. https://doi.org/10.3390/agronomy10010036
APA StyleZhao, L., Li, W., Wang, B., Gao, Y., Sui, X., Liu, Y., Chen, X., Yao, X., Jiao, F., & Song, Z. (2020). Development of a PVY Resistant Flue-Cured Tobacco Line via EMS Mutagenesis of eIF4E. Agronomy, 10(1), 36. https://doi.org/10.3390/agronomy10010036





