Interactions of Cardiac Proteins with Plasma-Synthesized Polypyrrole (PSPy) to Improve Adult Cardiomyocytes Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture of Adult Rat Cardiomyocytes
Cell Culture
2.2. Plasma-Synthesized Polypyrrole Particles
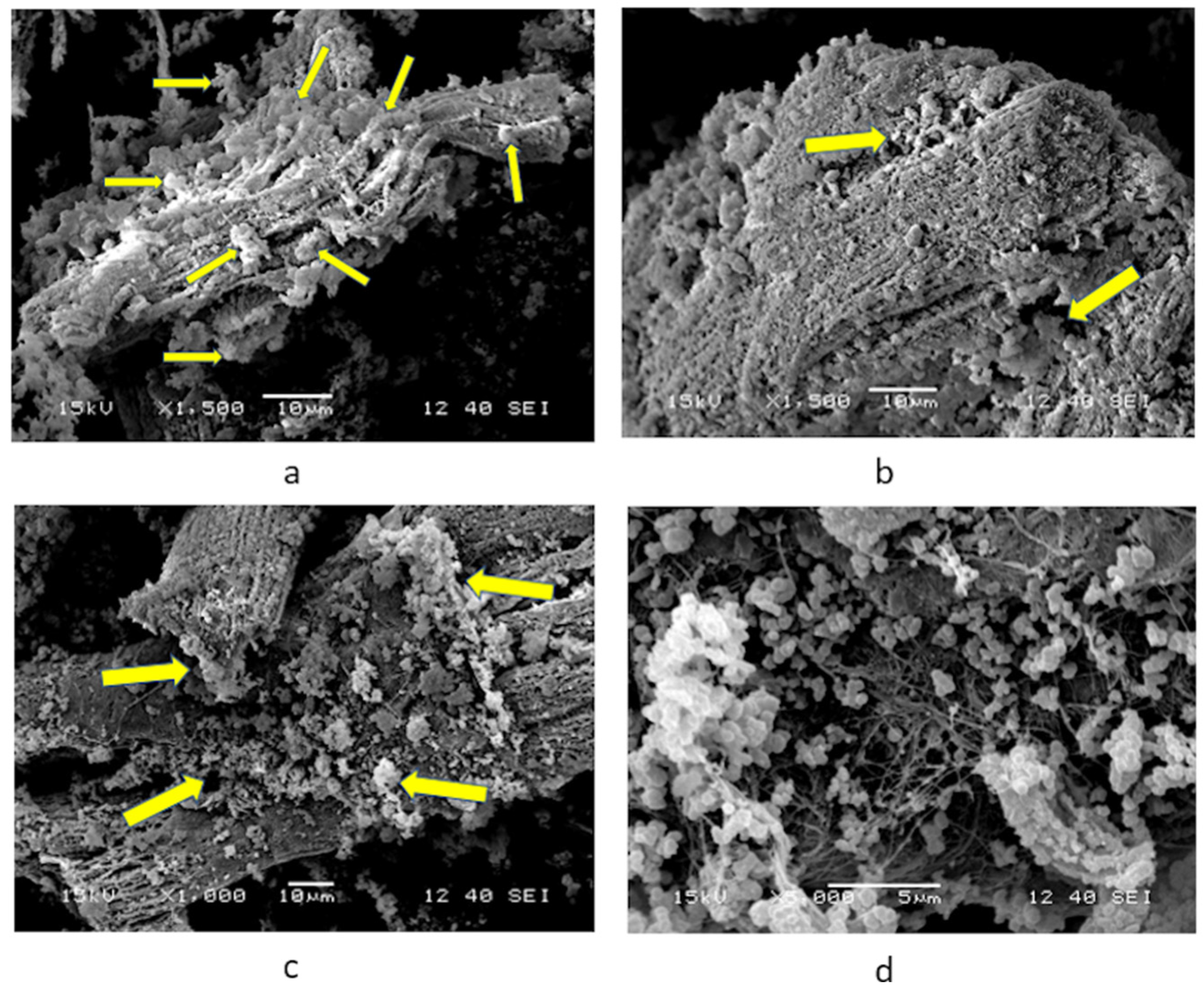
2.3. SEM Micrographs of Cardiomyocytes Cultured with nPSPy
2.4. Preparation of Structures
2.4.1. Receptors
2.4.2. Ligands
- Model 1 has a -NH2 group in R1, a -C≡N group in R2, and R3 with a primary amine -OH.
- Model 2 has in R1 a -OH group, in R2 it has a -C≡N group, and in R3 -NH2.
- Model 3 has a -OH group in R1, a -C≡N group in R2, and R3 with a primary amine -NH2 accompanied by a 5-carbon aliphatic chain (-CH2- CH2-CH2-CH2-CH2-R3). The original structure proposed by Kumar et al. [30] only has a 2-carbon chain (-CH2-CH2-R3).
2.4.3. Molecular Docking Studies
2.4.4. Binding Free Energy Calculations for All Systems
3. Results
3.1. Primary Culture of Cardiomyocytes with PSPy
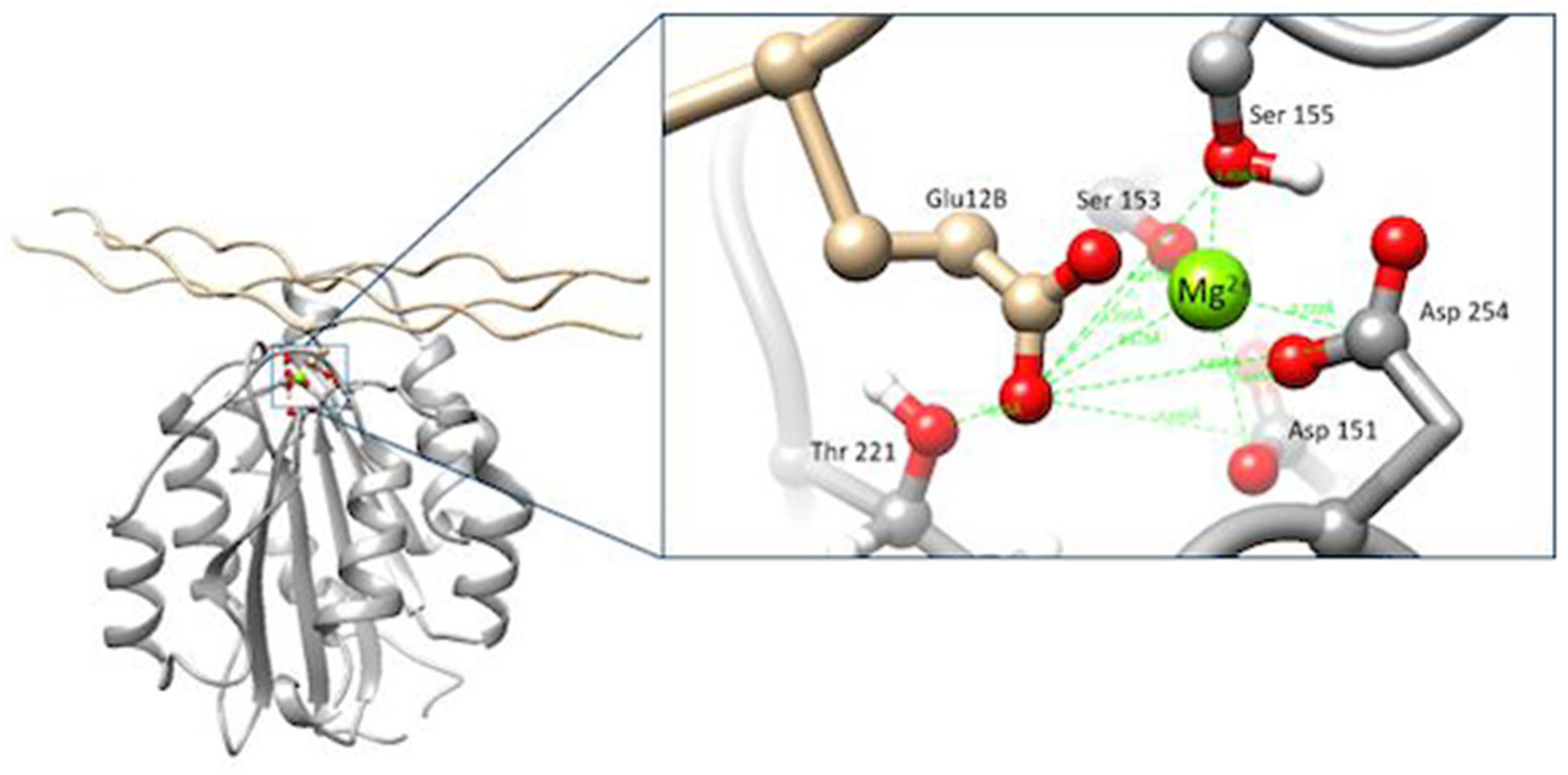
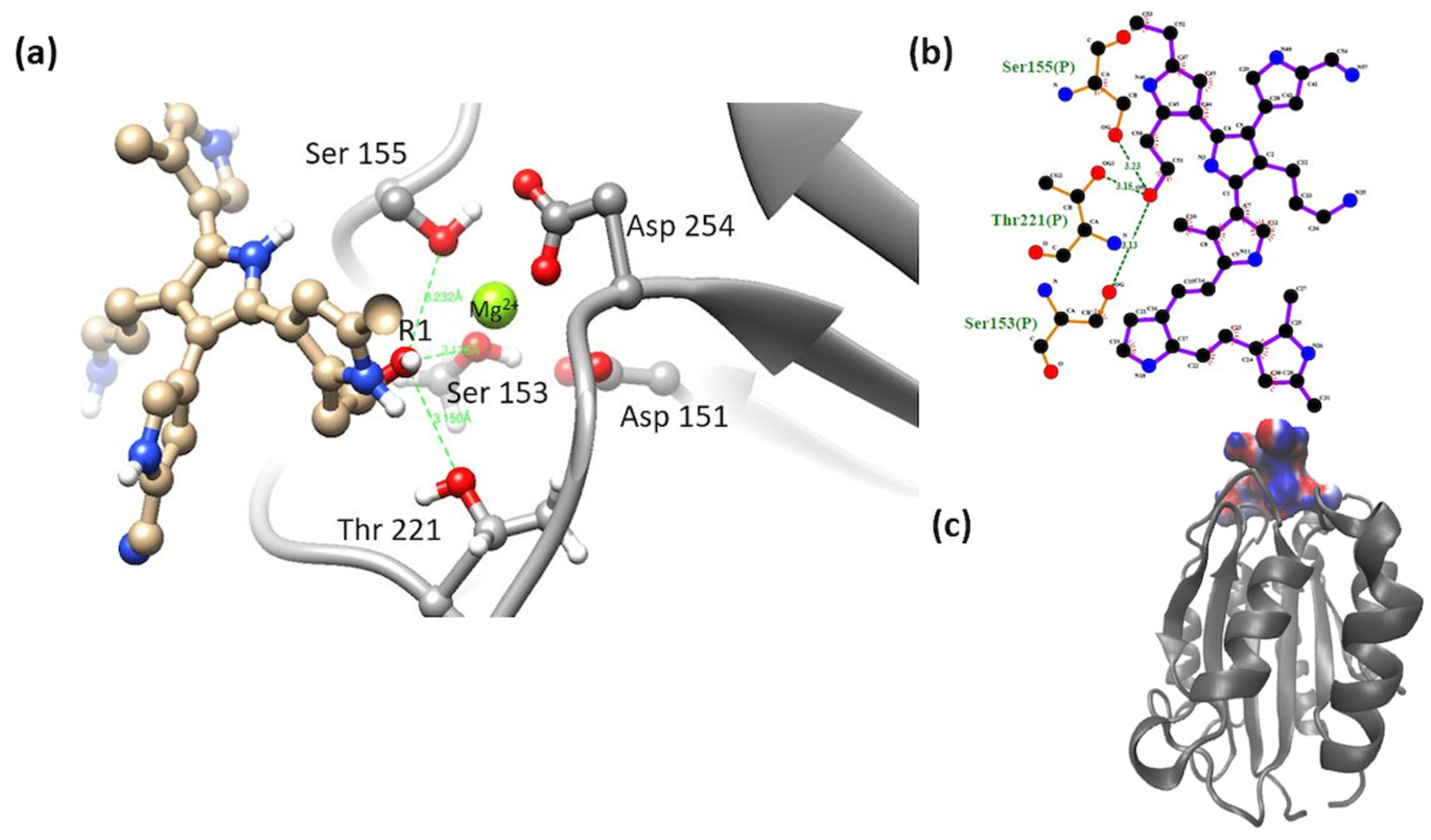
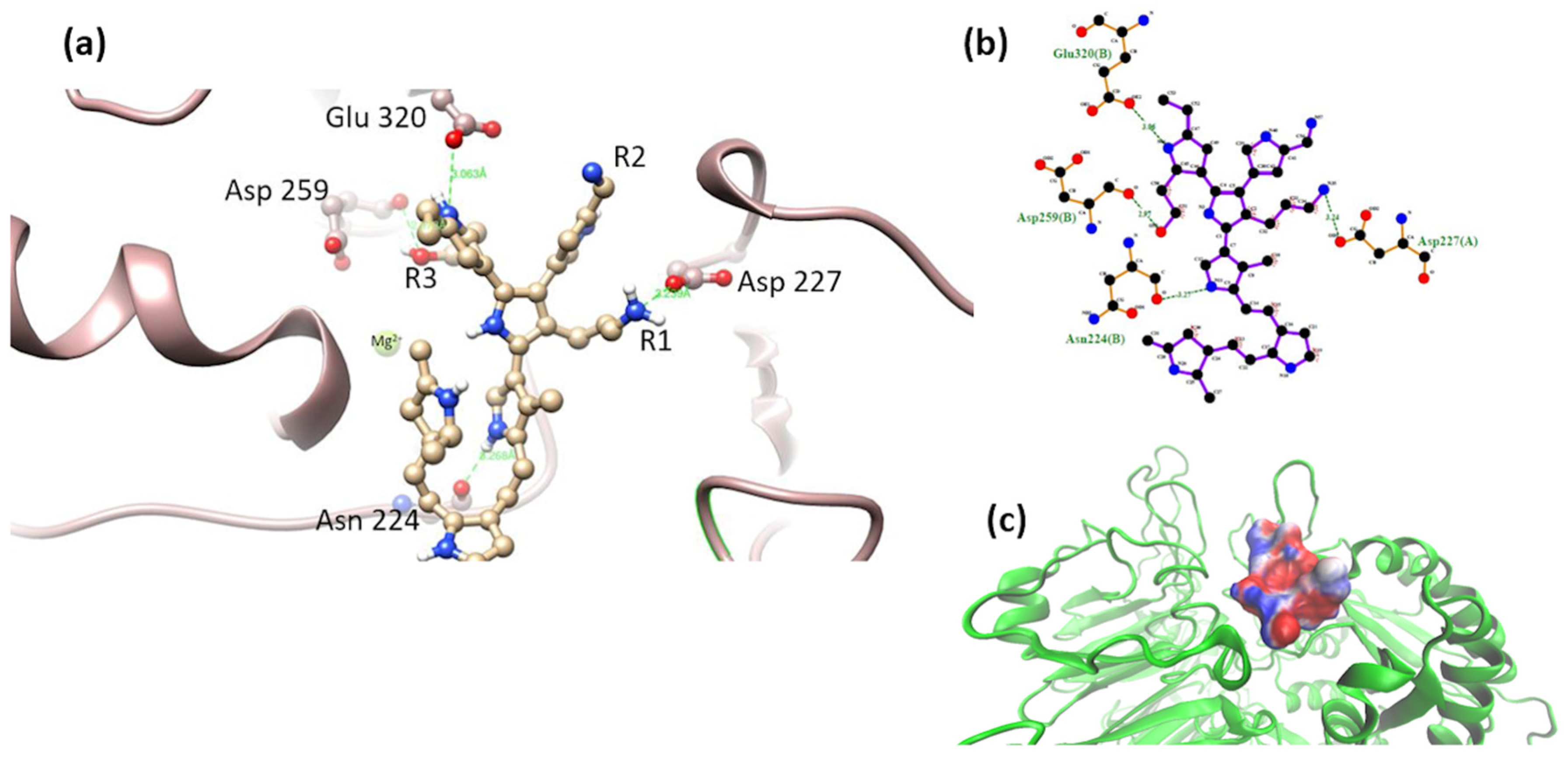
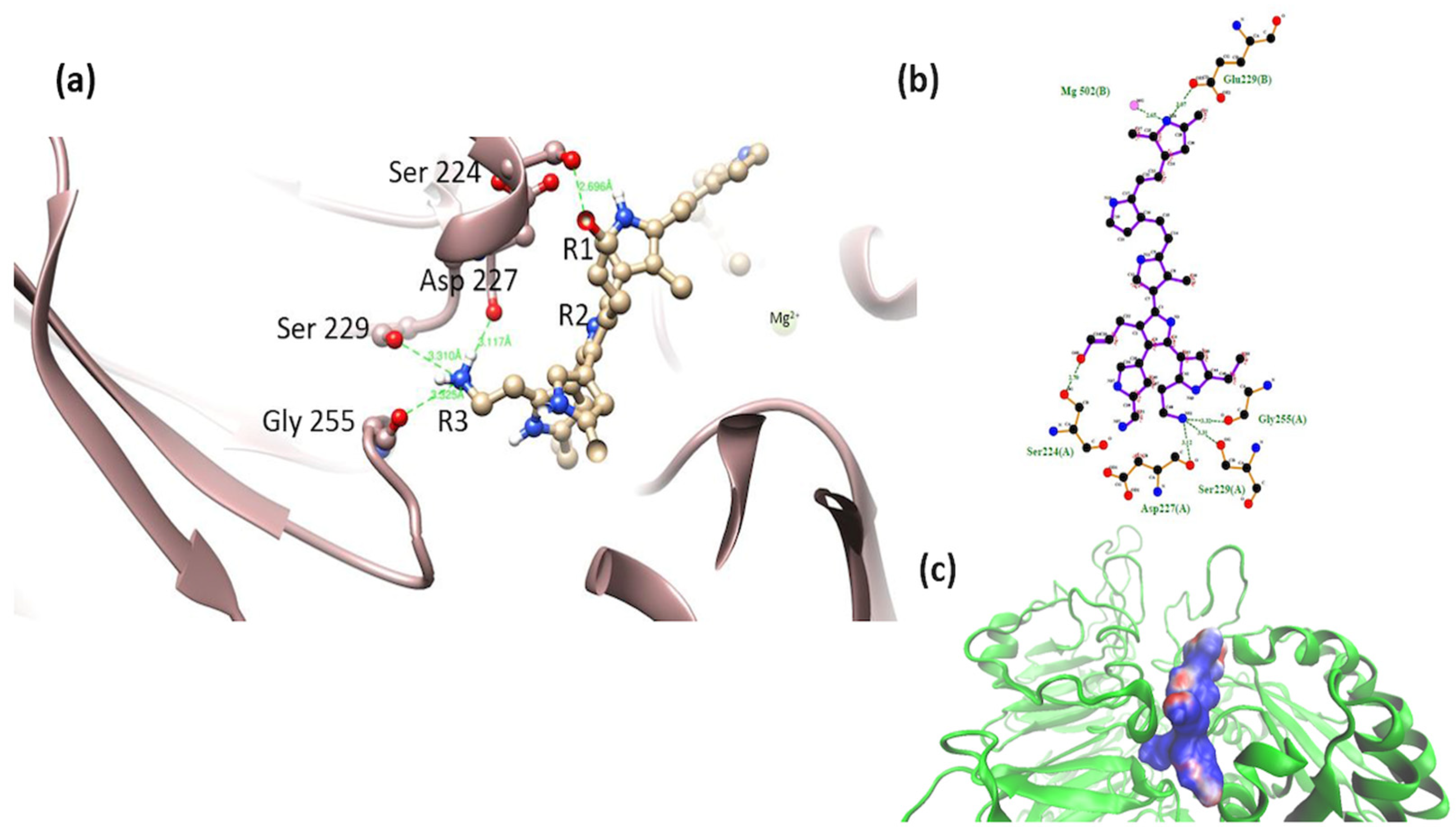
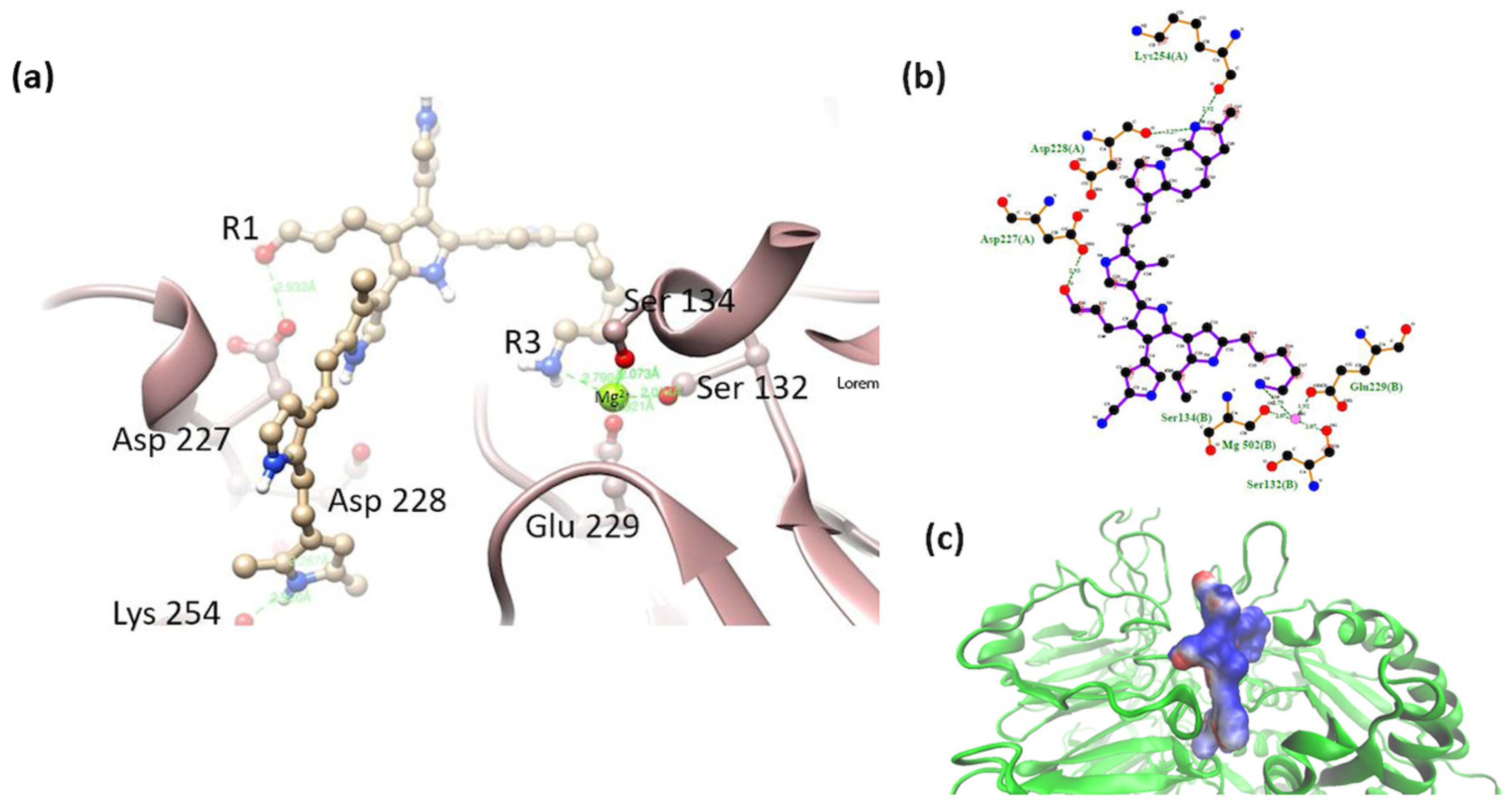
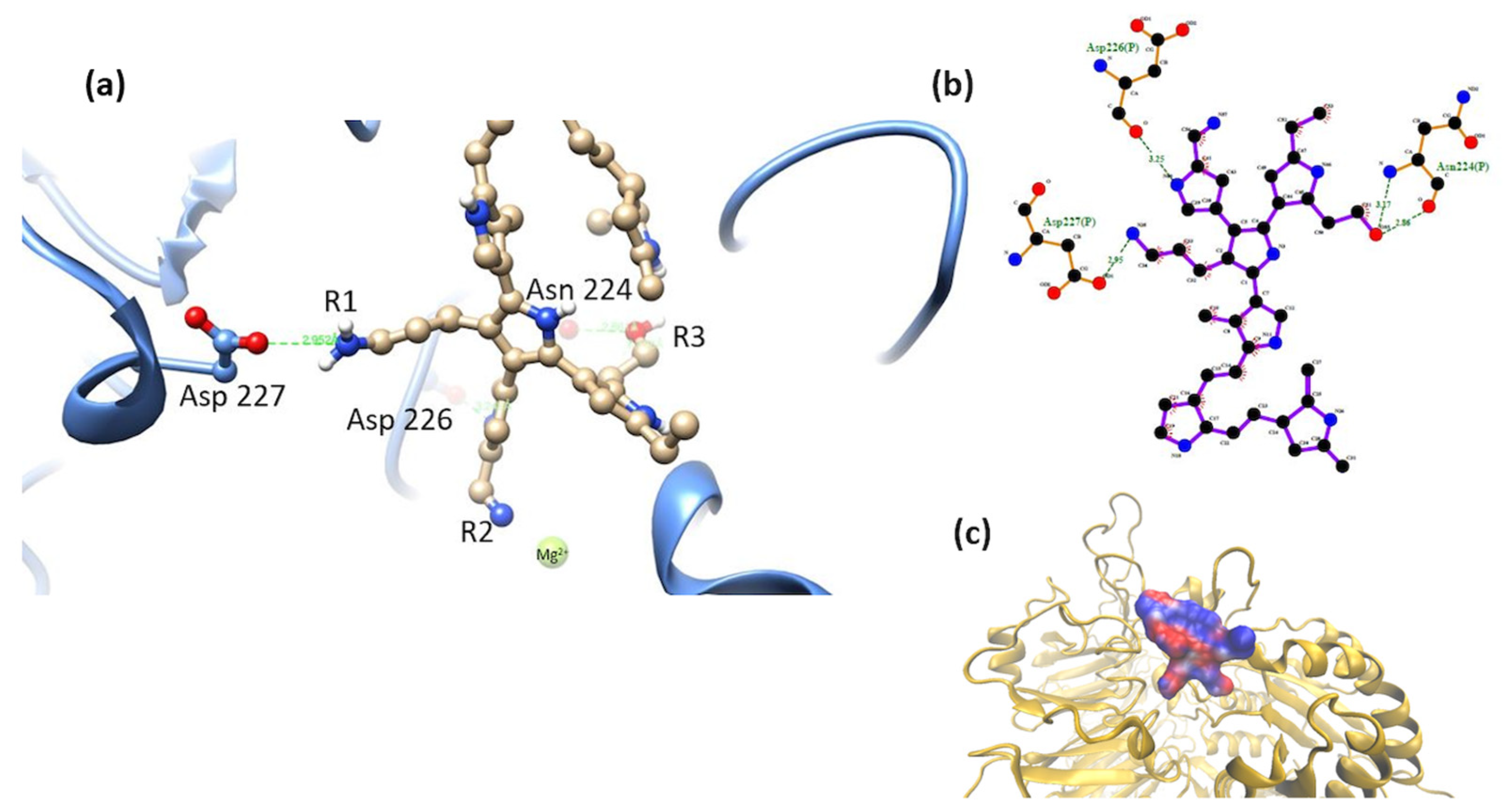
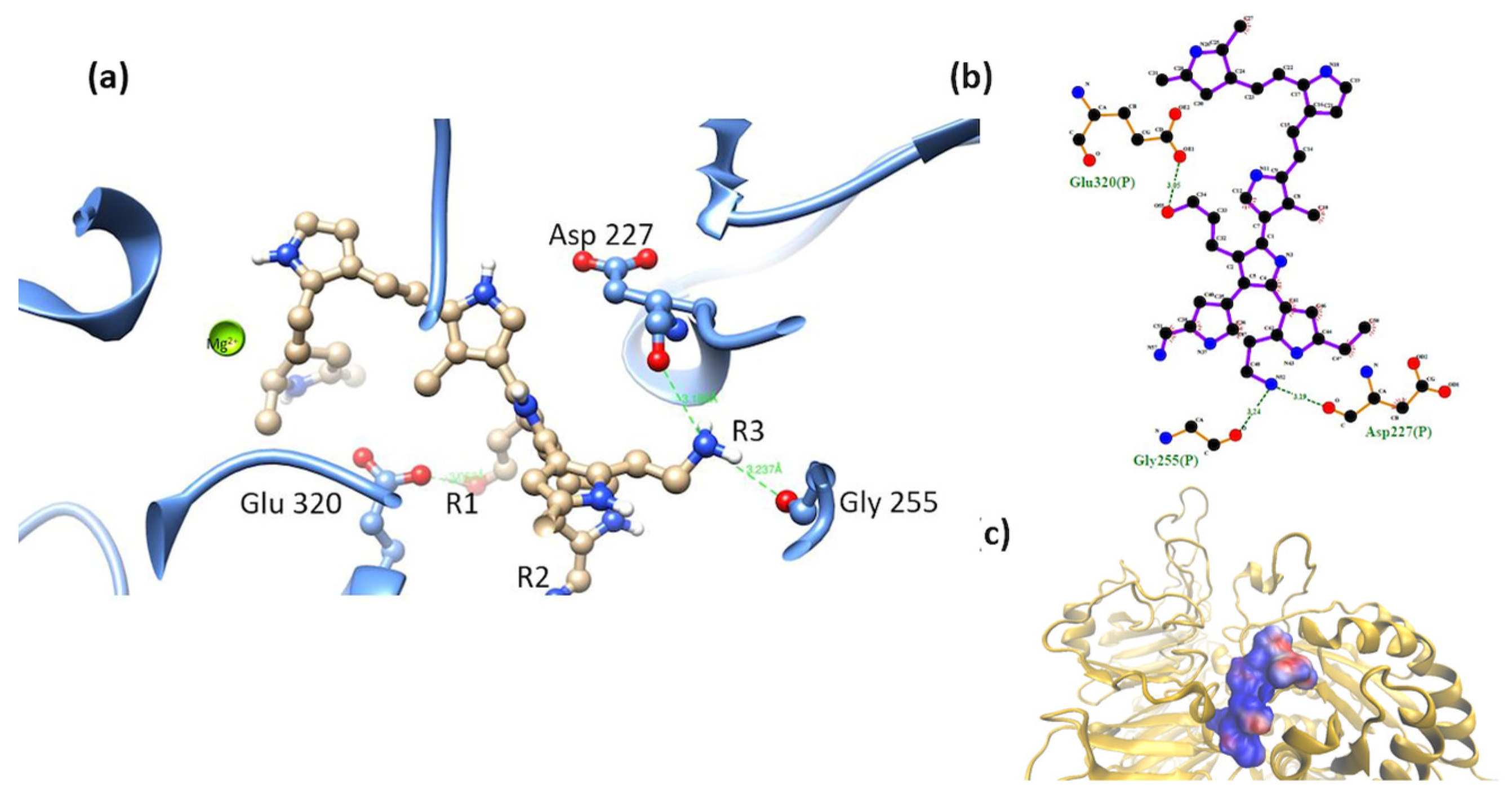
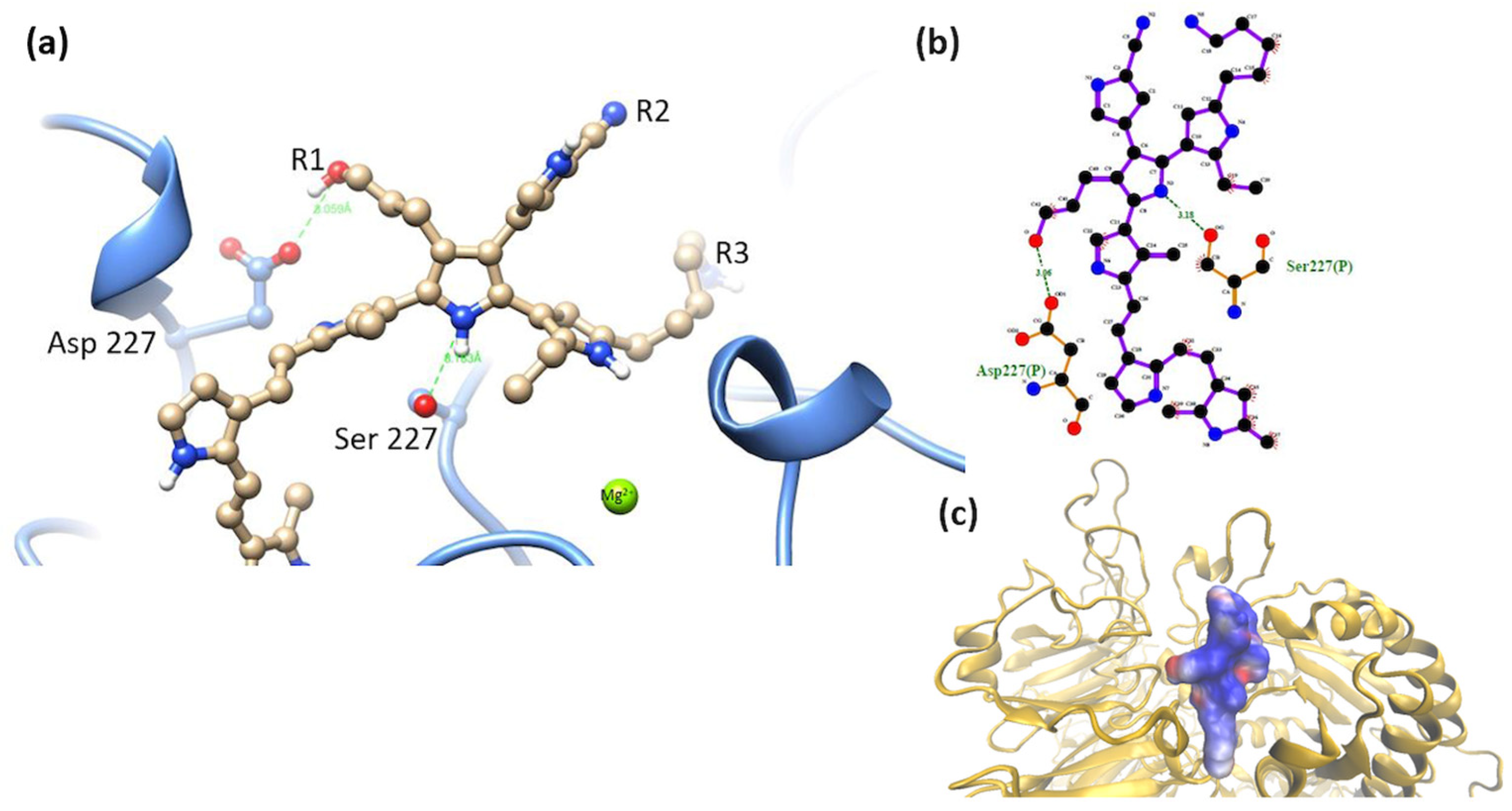
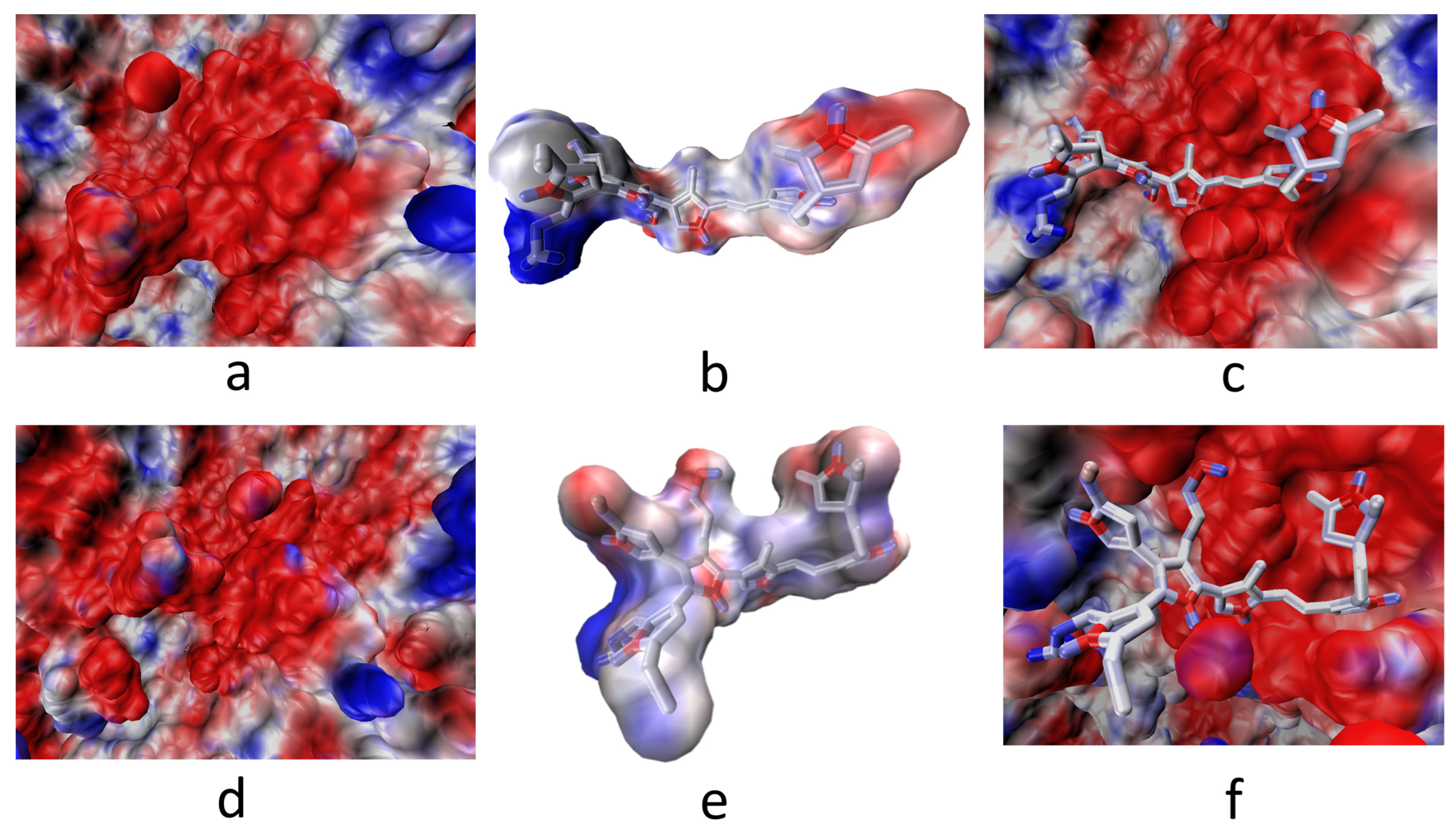
3.2. Interaction of the Integrin Domain with PSPy Ligand
4. Discussion
4.1. Primary Culture of Cardiomyocytes with PSPy
4.2. Molecular Dynamics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gutierrez, J.; Alloubani, A.; Mari, M.; Alzaatreh, M. Cardiovascular Disease Risk Factors: Hypertension, Diabetes Mellitus and Obesity among Tabuk Citizens in Saudi Arabia. Open Cardiovasc. Med. J. 2018, 12, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Rosengren, A. Obesity and cardiovascular health: The size of the problem. Eur. Hear. J. 2021, 42, 3404–3406. [Google Scholar] [CrossRef]
- Amini, M.; Zayefi, F. Trend analysis of cardiovascular disease mortality, incidence, and mortality-to-incidence ratio: Results from global burden of disease study 2017. BMC Public Health 2021, 21, 401. [Google Scholar] [CrossRef]
- Tadevosyan, K.; Iglesias-García, O.; Mazo, M.M.; Prósper, F.; Raya, A. Engineering and Assessing Cardiac Tissue Complexity. Int. J. Mol. Sci. 2021, 22, 1479. [Google Scholar] [CrossRef]
- Kim, I.-C.; Youn, J.-C.; Kobashigawa, J.A. The Past, Present and Future of Heart Transplantation. KCJ 2018, 48, 565–590. [Google Scholar] [CrossRef]
- Hasimoto, H.; Olson, E.N. Therapeutic approaches for cardiac regeneration and repair. Nat. Rev. Cardiol. 2018, 15, 585–600. [Google Scholar]
- Eghbali, A.; Dukes, A.; Toischer, K.; Hasenfuss, G.; Field, L.J. Cell Cycle–Mediated Cardiac Regeneration in the Mouse Heart. Curr. Cardiol. Rep. 2019, 21, 131. [Google Scholar] [CrossRef] [PubMed]
- Pomeroy, J.E.; Helfer, A.; Bursac, N. Biomaterialization the Promise of Cardiac Tissue Engineering. Biotechnol. Adv. 2020, 42, 107353. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhu, X.; Zhang, X.; Chen, G.; Bian, F.; Wang, J.; Zhou, Q.; Wang, D.; Zhao, Y. Induced cardiomyocytes-integrated conductive microneedle patch for treating myocardial infarction. Chem. Eng. J. 2021, 414, 128723. [Google Scholar] [CrossRef]
- LaBarge, W.; Mattappally, S. Maturation of three-dimensional, hiPSC-derived cardiomyocyte spheroids utilizing cyclic, uniaxial stretch and electrical stimulation. PLoS ONE 2019, 14, e0219442. [Google Scholar]
- Li, M.; Ma, J.; Gao, Y.; Yang, L. Cell sheet technology: A promising strategy in regenerative medicine. Cytotherapy 2019, 21, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tejada, E.; Morales-Corona, J.; Gómez-Quiroz, L.; Gutiérrez-Ruiz, M.; Olayo, R. Effect of synthesis variables of plasma synthesized polymers on growth of HepG2 cells. Biocell 2017, 41, 41–43. [Google Scholar] [CrossRef]
- Islas-Arteaga, N.C.; Raya, A. Electrospun scaffolds with surfaces modified by plasma for regeneration of articular cartilage tissue: A pilot study in rabbit. Int. J. Polym. Mater. 2019, 68, 1089–1098. [Google Scholar] [CrossRef]
- Alvarez-Mejía, L.; Morales, J.; Cruz, G.J.; Olayo, M.-G.; Olayo, R.; Díaz-Ruíz, A.; Ríos, C.; Mondragón-Lozano, R.; Sánchez-Torres, S.; Morales-Guadarrama, A.; et al. Functional recovery in spinal cord injured rats using polypyrrole/iodine implants and treadmill training. J. Mater. Sci. Mater. Med. 2015, 26, 209. [Google Scholar] [CrossRef] [PubMed]
- Flores-Sánchez, M.G.; Islas-Arteaga, N.C.; Raya-Rivera, A.M.; Esquiliano-Rendon, D.R.; Morales-Corona, J.; Uribe-Juarez, O.E.; Vivar-Velázquez, F.I.; Ortiz-Vázquez, G.P.; Olayo, R. Effect of a plasma synthesized polypyrrole coverage on polylactic acid/hydroxyapatite scaffolds for bone tissue engineering. J. Biomed. Mater. Res. Part A 2021, 109, 2199–2211. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Juárez, O.; Godínez, R.; Morales-Corona, J.; Velasco, M.; Olayo-Valles, R.; Acosta-García, M.C.; Alvarado, E.J.; Miguel-Alavez, L.; Carrillo-González, O.-J.; Flores-Sánchez, M.G.; et al. Application of plasma polymerized pyrrole nanoparticles to prevent or reduce de-differentiation of adult rat ventricular cardiomyocytes. J. Mater. Sci. Mater. Med. 2021, 32, 121. [Google Scholar] [CrossRef] [PubMed]
- Meagher, P.B.; Lee, X.A.; Lee, J.; Visram, A.; Friedberg, M.K.; Connelly, K.A. Cardiac Fibrosis: Key Role of Integrins in Cardiac Homeostasis and Remodeling. Cells 2021, 10, 770. [Google Scholar] [CrossRef] [PubMed]
- Chastney, M.R.; Conway, J.R.; Ivaska, J. Integrin adhesion complexes. Curr. Biol. 2021, 31, R536–R542. [Google Scholar] [CrossRef] [PubMed]
- Emsley, J.; Knight, C.G. Structural Basis of Collagen Recognition by Integrin a2b1. Cell 2000, 101, 47–56. [Google Scholar] [CrossRef]
- Brown, K.L.; Banerjee, S.; Feigley, A.; Abe, H.; Blackwell, T.S.; Pozzi, A.; Hudson, B.; Zent, R. Salt-bridge modulates differential calcium-mediated ligand binding to integrin α1- and α2-I domains. Sci. Rep. 2018, 8, 2916. [Google Scholar] [CrossRef]
- Qui, Z.; Park, A. The RGD (Arg-Gly-Asp) is a potential cell-binding motif of UNC-52/PERLECAN. Biochem. Biophys. Res. Commun. 2022, 586, 143–149. [Google Scholar]
- Madamanchi, A.; Santoro, S.A. Advances in Experimental Medicine and Biology: I Domain Integrins, 819; Gullberg; SpringerLink: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Emsley, J.; King, S.L. Crystal Structure of the I Domain from Integrin a2b1*. JBC 1997, 272, 28512–28517. [Google Scholar] [CrossRef] [PubMed]
- Carafoli, F.; Hamaia, S.W.; Bihan, D.; Hohenester, E.; Farndale, R.W. An Activating Mutation Reveals a Second Binding Mode of the Integrin a2 I Domain to the GFOGER Motif in Collagens. PLoS ONE 2013, 8, e69833. [Google Scholar] [CrossRef] [PubMed]
- Emsley, J.; Kinght, C.G. Structure of the Integrin alpha2beta1-binding Collagen Peptide. J. Mol. Biol. 2004, 335, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddart, T.D. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Mezu-Ndubuisi, O.; Maheshwar, A. The role of integrins in inflammation and angiogenesis. Pediatr. Res. 2021, 89, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Nagae, M.; Re, S. Crystal structure of a5b1 integrin ectodomain: Atomic details of the fibronectin receptor. JCB 2012, 197, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Serratos, I.N.; Olayo, R.; Millán-Pacheco, C.; Morales-Corona, J.; Vicente-Escobar, J.O.; Soto-Estrada, A.M.; Córdoba-Herrera, J.G.; Uribe, O.; Gómez-Quintero, T.; Arroyo-Ornelas, M.; et al. Modeling integrin and plasma-polymerized pyrrole interactions: Chemical diversity relevance for cell regeneration. Sci. Rep. 2019, 9, 7009. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.D.; Nakamura, K. Optical and electrical characterization of plasma polymerized pyrrole films. J. Appl. Phys. 2003, 93, 2705–2711. [Google Scholar] [CrossRef]
- Serratos, I.N.; Luviano, A.S.; Millan-Pacheco, C.; Morales-Corona, J.; Munoz, E.J.A.; Campos-Teran, J.; Olayo, R. Quartz crystal microbalance application and in silico studies to characterize the interaction of bovine serum albumin with plasma polymerized pyrrole surfaces: Implications for the development of biomaterials. Langmuir 2023, 39, 11213–11223. [Google Scholar] [CrossRef]
- PyMOL Molecular Graphics System, Version 1.3; Schrödinger, LLC: New York, NY, USA, 2010.
- Gómez-Quintero, T.; Serratos-Alvarez, I.; Godínez, R.; Olayo, R. Collagen/Plasma-Polymerized Pyrrole Interaction: Molecular Docking and Binding Energy Calculations. XLV Mex. Conf. Biomed. Eng. 2022, 86, 153–161. [Google Scholar]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Avogadro: An Open-Source Molecular Builder and Visualization Tool. Available online: http://avogadro.cc/ (accessed on 2 February 2024).
- Ramírez, E.R.; Serratos, I. Modeling of the Interaction of Plasma-Polymerized Pyrrole with Immunoglobulin M (IgM) by Bio-Computational Tools; Springer International Publishing: Puerto Vallarta, Mexico, 2022. [Google Scholar]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- LigPlot+. LIGPLOT and DIMPLOT Schematics Diagrams, version 2.2.4. Available online: https://www.ebi.ac.uk/thornton-srv/software/LigPlus/download.html (accessed on 30 April 2024).
- Dolinsky, T.J.; Czodrowki, P. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, 522–525. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Baker, N.A. Poisson–Boltzmann Methods for Biomolecular Electrostatics. Methods Enzym. 2004, 383, 94–118. [Google Scholar] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Genheden, S.; Mikulskis, P.; Hu, L.; Kongsted, J.; Söderhjelm, P.; Ryde, U. Accurate predictions of nonpolar solvation free energies require explicit consideration of binding-site hydration. J. Am. Chem. Soc. 2011, 133, 13081–13092. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvatation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Millán-Pacheco, C.; Arreola, R.; Villalobos-Osnaya, A.; Garza-Ramos, G.; Serratos, I.N.; Díaz-Vilchis, A.; Rudiño-Piñera, E.; Alvarez-Sanchez, M.E. A Putative New Role of Tv-PSP1 Recognizes IRE and ERE Hairpin Structures from Trichomonas vaginalis. Pathogens 2023, 12, 79. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, Q.; Li, Z.; Zhang, J.; Liu, H. Revealing the Interaction Mechanism between Mycobacterium tuberculosis GyrB and Novobiocin, SPR719 through Binding Thermodynamics and Dissociation Kinetics Analysis. Int. J. Mol. Sci. 2024, 25, 3764. [Google Scholar] [CrossRef]
- Santos Nascimento, J.d.I.; Aquino, M.d.T.; Silva-Júnior, F.d.E. Repurposing FDA-approved Drugs Targeting SARS-CoV2 3CLpro: A Study by Applying Virtual Screening, Molecular Dynamics, MM-PBSA Calculations and Covalent Docking. Lett. Drug Des. Discov. 2022, 19, 637–653. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, J.; Zhu, Y.; Tan, S.; Liu, H. Binding Thermodynamics and Dissociation Kinetics Analysis Uncover the Key Structural Motifs of Phenoxyphenol Derivatives as the Direct InhA Inhibitors and the Hotspot Residues of InhA. Int. J. Mol. Sci. 2022, 23, 10102. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Zhang, Q.; Wang, J.; Gao, P.; Xie, G.; Liu, H.; Yao, X. Molecular Modeling Study on the Interaction Mechanism between the LRRK2 G2019S Mutant and Type I Inhibitors by Integrating Molecular Dynamics Simulation, Binding Free Energy Calculations, and Pharmacophore Modeling. ACS Chem. Neurosci. 2022, 13, 599–612. [Google Scholar] [CrossRef]
- Viniegra, M.; Serratos, I.N. Insights into protein-ligand interactions: Thermodynamic signatures. In An eBook on Thermodynamics; Rutherford, K., Ed.; Open Access eBooks: Las Vegas, NV, USA, 2020; pp. 1–13. ISBN 978-93-87500-78-5. Available online: https://openaccessebooks.com/thermodynamics.html (accessed on 9 May 2024).
- Adhav, V.A.; Saikrishnan, K. The Realm of Unconventional Noncovalent Interactions in Proteins: Their Significance in Structure and Function. ACS Omega 2023, 8, 22268–22284. [Google Scholar] [CrossRef]
- NOM-062-ZOO-1999; Especificaciones Técnicas Para la Producción, Cuidado y Uso de Los Animales de Laboratorio. Available online: https://www.gob.mx/cms/uploads/attachment/file/203498/NOM-062-ZOO-1999_220801.pdf (accessed on 19 April 2024).







| Systems | PSPy Ligands | |||
|---|---|---|---|---|
| Model 1 | Model 2 | Model 3 | ||
| Receptors | α2β1 | α2β1-model 1 | α2β1-model 2 | α2β1-model 3 |
| α5β1 | α5β1-model 1 | α5β1-model 2 | α5β1-model 3 | |
| Ligand | Integrin | System | ΔGsol (kJ/mol) | ΔGcoul (kJ/mol) | ΔGnon-polar (kJ/mol) | ΔGb (kJ/mol) |
|---|---|---|---|---|---|---|
| Model 1 | α2β1 | C1 | 85 | −18 | −21 | >0 |
| C2 | 91 | −14 | −20 | >0 | ||
| α5β1 | C1 | 87 | 1 | −21 | >0 | |
| C2 | 98 | −1 | −24 | >0 | ||
| Model 2 | α2β1 | C1 | 81 | −136 | −19 | −74 |
| C2 | 82 | −138 | −18 | −74 | ||
| α5β1 | C1 | 115 | −421 | −25 | −331 | |
| C2 | 125 | −425 | −25 | −325 | ||
| Model 3 | α2β1 | C1 | 92 | −128 | −22 | −58 |
| C2 | 98 | −130 | −21 | −53 | ||
| α5β1 | C1 | 149 | −370 | −26 | −247 | |
| C2 | 188 | −382 | −25 | −219 |
| Ligand | Integrin | System | ΔGsol (kJ/mol) | ΔGcoul (kJ/mol) | ΔGnon-polar (kJ/mol) | ΔGb (kJ/mol) |
|---|---|---|---|---|---|---|
| Model 1 | α5β1 | C1 | 87 | 1 | −21 | >0 |
| C2 | 98 | −1 | −24 | >0 | ||
| α5β1 cluster | C1 | 119 | 2 | −21 | >0 | |
| C2 | 166 | −18 | −26 | >0 | ||
| Model 2 | α5β1 | C1 | 115 | −421 | −25 | −331 |
| C2 | 125 | −425 | −25 | −325 | ||
| α5β1 cluster | C1 | 184 | −482 | −23 | −321 | |
| C2 | 135 | −313 | −21 | −199 | ||
| Model 3 | α5β1 | C1 | 149 | −370 | −26 | −247 |
| C2 | 188 | −382 | −25 | −219 | ||
| α5β1 cluster | C1 | 236 | −299 | −26 | −89 | |
| C2 | 253 | −312 | −27 | −86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Quintero, T.; Olayo, R.; Morales-Corona, J.; Uribe-Juárez, O.E.; Millán-Pacheco, C.; Godínez-Fernández, R.; Serratos, I.N. Interactions of Cardiac Proteins with Plasma-Synthesized Polypyrrole (PSPy) to Improve Adult Cardiomyocytes Culture. Polymers 2024, 16, 1470. https://doi.org/10.3390/polym16111470
Gómez-Quintero T, Olayo R, Morales-Corona J, Uribe-Juárez OE, Millán-Pacheco C, Godínez-Fernández R, Serratos IN. Interactions of Cardiac Proteins with Plasma-Synthesized Polypyrrole (PSPy) to Improve Adult Cardiomyocytes Culture. Polymers. 2024; 16(11):1470. https://doi.org/10.3390/polym16111470
Chicago/Turabian StyleGómez-Quintero, Teresa, Roberto Olayo, Juan Morales-Corona, Omar E. Uribe-Juárez, César Millán-Pacheco, Rafael Godínez-Fernández, and Iris N. Serratos. 2024. "Interactions of Cardiac Proteins with Plasma-Synthesized Polypyrrole (PSPy) to Improve Adult Cardiomyocytes Culture" Polymers 16, no. 11: 1470. https://doi.org/10.3390/polym16111470
APA StyleGómez-Quintero, T., Olayo, R., Morales-Corona, J., Uribe-Juárez, O. E., Millán-Pacheco, C., Godínez-Fernández, R., & Serratos, I. N. (2024). Interactions of Cardiac Proteins with Plasma-Synthesized Polypyrrole (PSPy) to Improve Adult Cardiomyocytes Culture. Polymers, 16(11), 1470. https://doi.org/10.3390/polym16111470








