Bacterial Population Changes during the Degradation Process of a Lactate (LA)-Enriched Biodegradable Polymer in River Water: LA-Cluster Preferable Bacterial Consortium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Several Polymers
2.2. HPLC Analysis of Polymer
2.3. Growth Conditions of Bacteria from the River Water Sample
2.4. Measurement Weight and LA Content of LAHB after Immersion in the River Water Samples
2.5. Measurement of Cell Density Using Flow Cytometry
2.6. Scanning Electron Microscope
2.7. Preparation of Samples for Metagenome Analysis and Metagenome Analysis Using NGS
2.8. Microbial Community Analysis
3. Results and Discussion
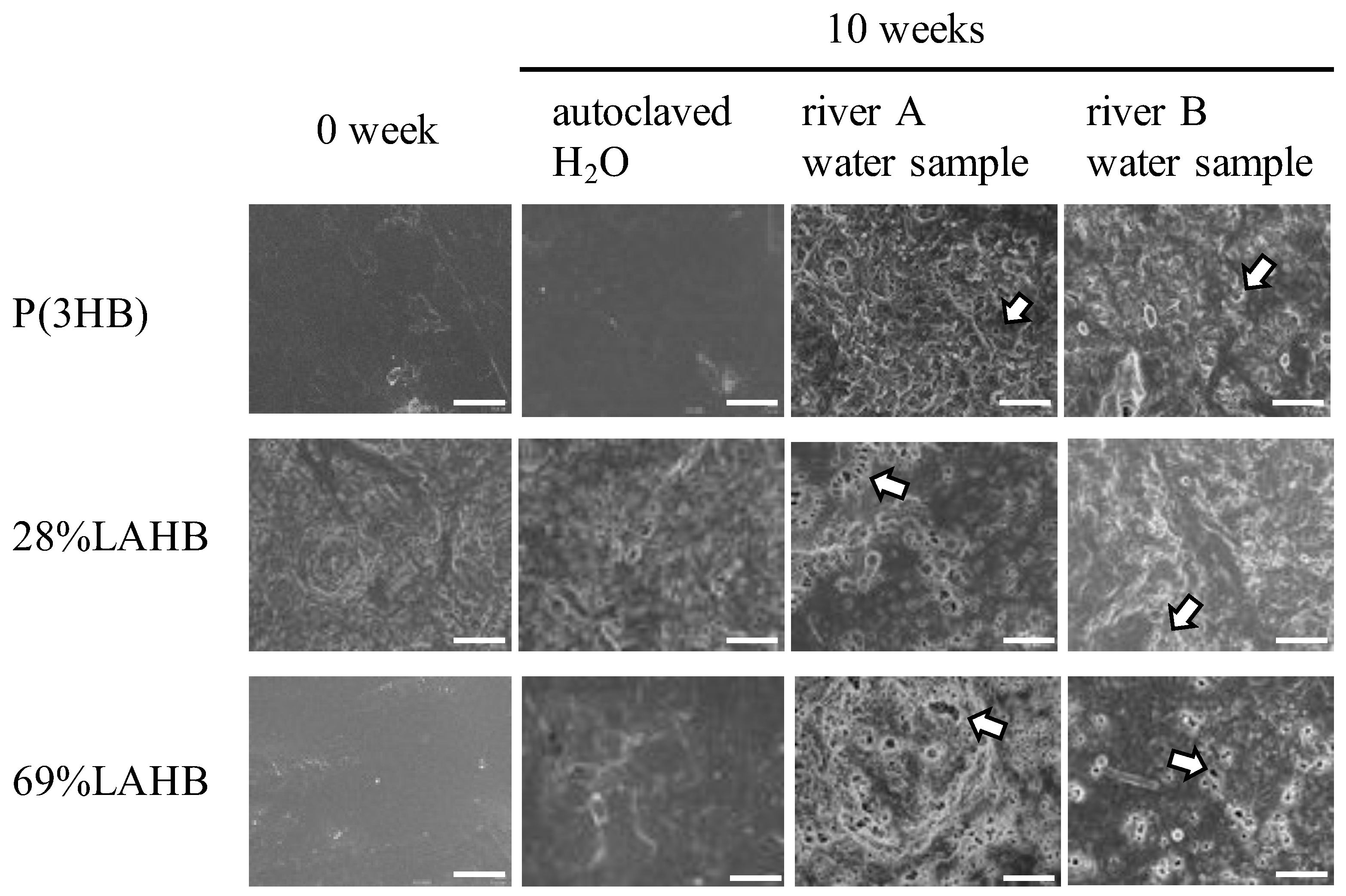
3.1. Microbial Degradation of Polymers Containing LA in River Waters
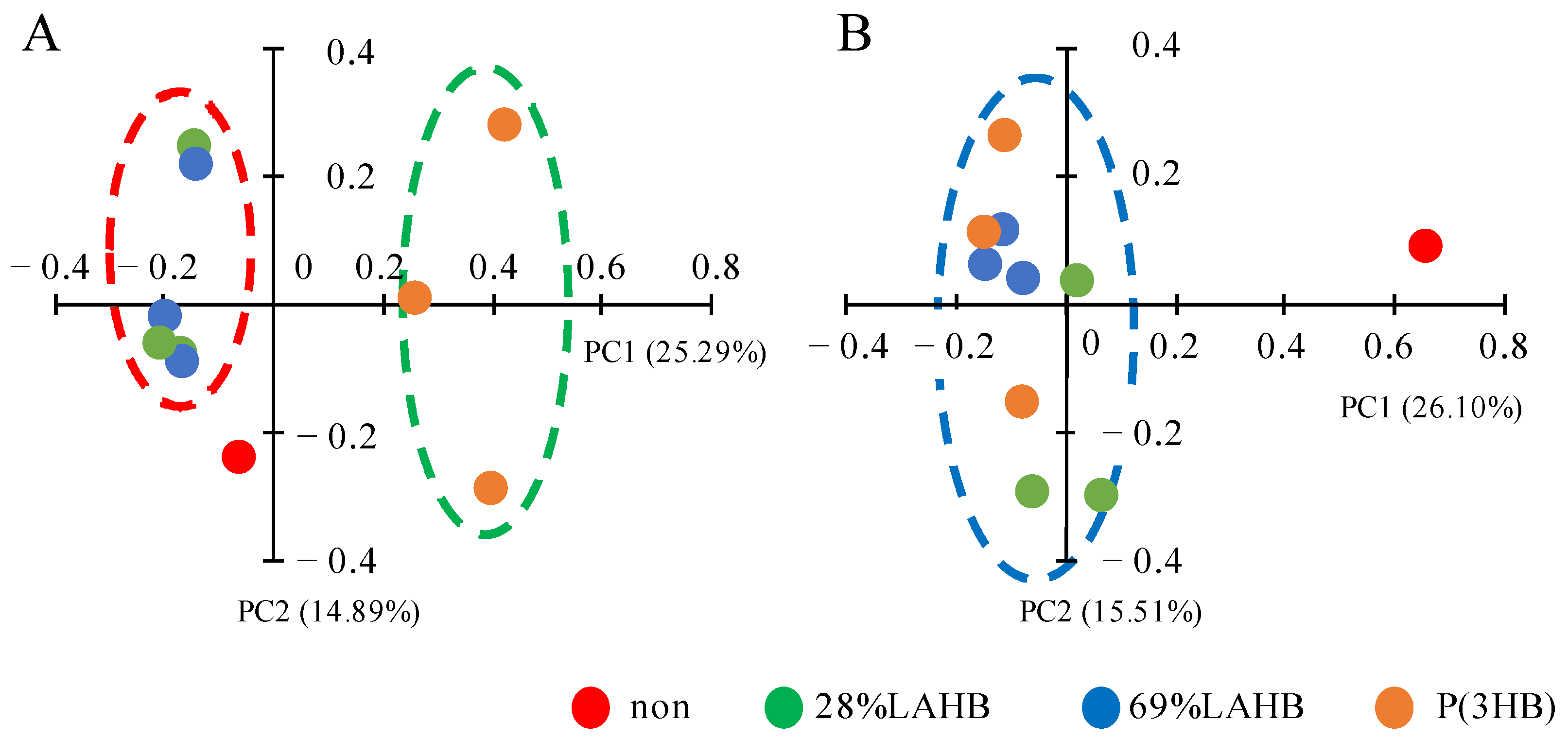
3.2. Statistical Evaluation of Bacterial Diversity in the River Water Samples under LAHB Degradation
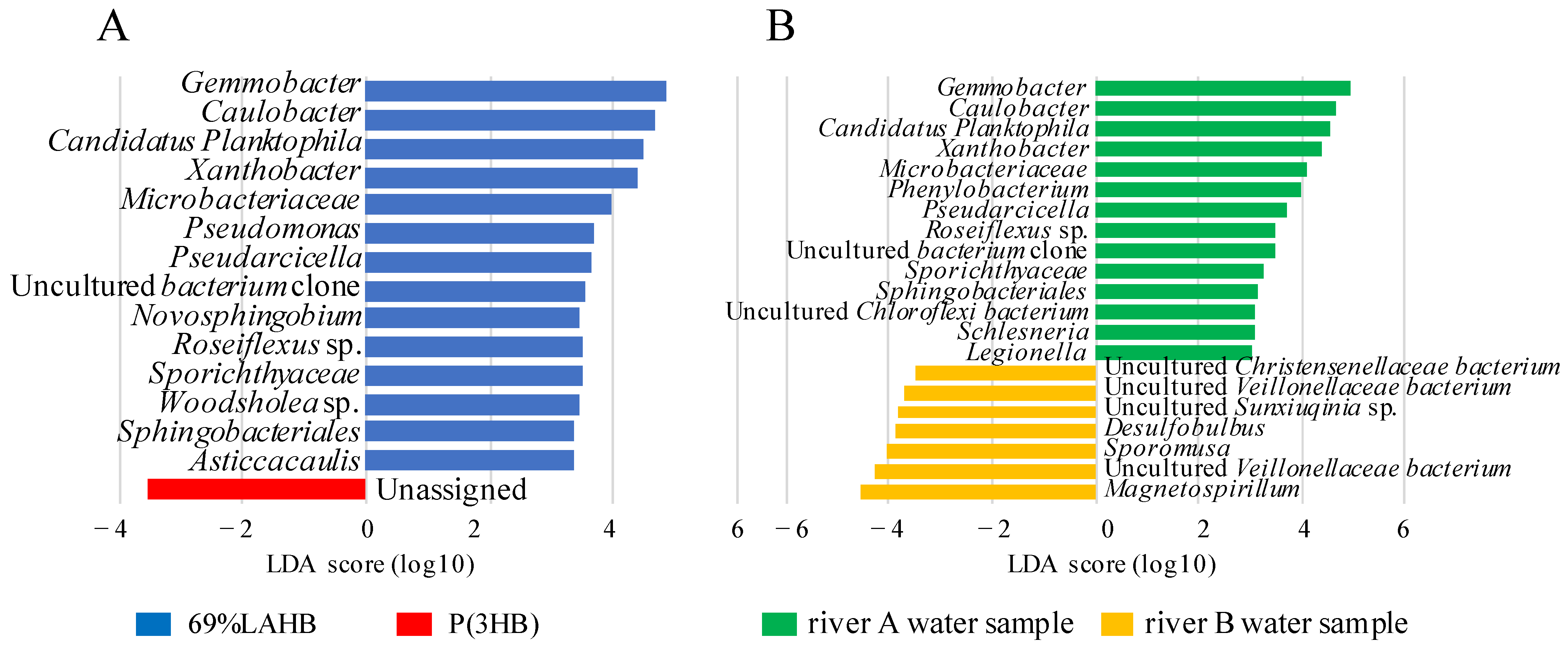
3.3. Characterization of the Microbial Community in the River Water Samples under LAHB Degradation Conditions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jambeck, J.R.; Geyer, R.; Wilcox, C.; Siegler, T.R.; Perryman, M.; Andrady, A.; Narayan, R.; Law, K.L. Marine pollution. Plastic waste inputs from land into the ocean. Science 2015, 347, 768–771. [Google Scholar] [CrossRef]
- Roman, L.; Schuyler, Q.; Wilcox, C.; Hardesty, B.D. Plastic pollution is killing marine megafauna, but how do we prioritize policies to reduce mortality? Conserv. Lett. 2020, 14, e12781. [Google Scholar] [CrossRef]
- Kumar, K.; Umapathi, R.; Ghoreishian, S.M.; Tiwari, J.N.; Hwang, S.K.; Huh, Y.S.; Venkatesu, P.; Shetti, N.P.; Aminabhavi, T.M. Microplastics and biobased polymers to combat plastics waste. Chemosphere 2023, 341, 140000. [Google Scholar] [CrossRef]
- Chubarenko, I.; Stepanova, N. Microplastics in sea coastal zone: Lessons learned from the Baltic amber. Environ. Pollut. 2017, 224, 243–254. [Google Scholar] [CrossRef]
- Liu, S.; Huang, W.; Yang, J.; Xiong, Y.; Huang, Z.; Wang, J.; Cai, T.; Dang, Z.; Yang, C. Formation of environmentally persistent free radicals on microplastics under UV irradiations. J. Hazard. Mater. 2023, 453, 131277. [Google Scholar] [CrossRef]
- Prata, J.C.; da Costa, J.P.; Lopes, I.; Duarte, A.C.; Rocha-Santos, T. Environmental exposure to microplastics: An overview on possible human health effects. Sci. Total Environ. 2020, 702, 134455. [Google Scholar] [CrossRef]
- Bai, Z.; Wang, N.; Wang, M. Effects of microplastics on marine copepods. Ecotoxicol. Environ. Saf. 2021, 217, 112243. [Google Scholar] [CrossRef]
- Taguchi, S.; Matsumoto, K. Evolution of polyhydroxyalkanoate synthesizing systems toward a sustainable plastic industry. Polym. J. 2021, 53, 67–79. [Google Scholar] [CrossRef]
- De Gisi, S.; Gadaleta, G.; Gorrasi, G.; La Mantia, F.P.; Notarnicola, M.; Sorrentino, A. The role of (bio)degradability on the management of petrochemical and bio-based plastic waste. J. Environ. Manag. 2022, 310, 114769. [Google Scholar] [CrossRef]
- Singhvi, M.S.; Zinjarde, S.S.; Gokhale, D.V. Polylactic acid: Synthesis and biomedical applications. J. Appl. Microbiol. 2019, 127, 1612–1626. [Google Scholar] [CrossRef]
- Engler, G.L.; Farias, N.C.; Crespo, S.J.; Gately, N.M.; Major, I.; Pezzoli, R.; Devine, D.M. Designing Sustainable Polymer Blends: Tailoring Mechanical Properties and Degradation Behaviour in PHB/PLA/PCL Blends in a Seawater Environment. Polymers 2023, 15, 2874. [Google Scholar] [CrossRef] [PubMed]
- Dalton, B.; Bhagabati, P.; De Micco, J.; Padamati, R.B.; O’Connor, K. A Review on Biological Synthesis of the Biodegradable Polymers Polyhydroxyalkanoates and the Development of Multiple Applications. Catalysts 2022, 12, 319. [Google Scholar] [CrossRef]
- Nagarajan, D.; Aristya, G.R.; Lin, Y.J.; Chang, J.J.; Yen, H.W.; Chang, J.S. Microbial cell factories for the production of polyhydroxyalkanoates. Essays Biochem. 2021, 65, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, S.; Yamada, M.; Matsumoto, K.; Tajima, K.; Satoh, Y.; Munekata, M.; Ohno, K.; Kohda, K.; Shimamura, T.; Kambe, H.; et al. A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proc. Natl. Acad. Sci. USA 2008, 105, 17323–17327. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Suzuki, N.; Matsumoto, K.; Taguchi, S.; Tanaka, K.; Matsusaki, H. Enhancement of lactate fraction in poly(lactate-co-3-hydroxybutyrate) synthesized by Escherichia coli harboring the D-lactate dehydrogenase gene from Lactobacillus acetotolerans HT. J. Gen. Appl. Microbiol. 2019, 65, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Kadoya, R.; Matsumoto, K.; Takisawa, K.; Ooi, T.; Taguchi, S. Enhanced production of lactate-based polyesters in Escherichia coli from a mixture of glucose and xylose by Mlc-mediated catabolite derepression. J. Biosci. Bioeng. 2018, 125, 365–370. [Google Scholar] [CrossRef]
- Nagao, Y.; Koh, S.; Taguchi, S.; Shimada, T. Cell-growth phase-dependent promoter replacement approach for improved poly(lactate-co-3-hydroxybutyrate) production in Escherichia coli. Microb. Cell Fact. 2023, 22, 131. [Google Scholar] [CrossRef]
- Nduko, J.M.; Taguchi, S. Microbial Production of Biodegradable Lactate-Based Polymers and Oligomeric Building Blocks from Renewable and Waste Resources. Front. Bioeng. Biotechnol. 2020, 8, 618077. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 2021, 759, 143536. [Google Scholar] [CrossRef]
- Garcia-Depraect, O.; Lebrero, R.; Rodriguez-Vega, S.; Bordel, S.; Santos-Beneit, F.; Martinez-Mendoza, L.J.; Aragao Borner, R.; Borner, T.; Munoz, R. Biodegradation of bioplastics under aerobic and anaerobic aqueous conditions: Kinetics, carbon fate and particle size effect. Bioresour. Technol. 2022, 344, 126265. [Google Scholar] [CrossRef]
- Sun, J.; Matsumoto, K.; Nduko, J.M.; Ooi, T.; Taguchi, S. Enzymatic characterization of a depolymerase from the isolated bacterium Variovorax sp. C34 that degrades poly(enriched lactate-co-3-hydroxybutyrate). Polym. Degrad. Stab. 2014, 110, 44–49. [Google Scholar] [CrossRef]
- Sun, J.; Matsumoto, K.; Tabata, Y.; Kadoya, R.; Ooi, T.; Abe, H.; Taguchi, S. Molecular weight-dependent degradation of D-lactate-containing polyesters by polyhydroxyalkanoate depolymerases from Variovorax sp. C34 and Alcaligenes faecalis T1. Appl. Microbiol. Biotechnol. 2015, 99, 9555–9563. [Google Scholar] [CrossRef] [PubMed]
- Hori, C.; Sugiyama, T.; Watanabe, K.; Sun, J.; Kamada, Y.; Ooi, T.; Isono, T.; Satoh, T.; Sato, S.; Taguchi, S.; et al. Isolation of poly[D-lactate (LA)-co-3-hydroxybutyrate)]-degrading bacteria from soil and characterization of D-LA homo-oligomer degradation by the isolated strains. Polym. Degrad. Stab. 2022, 179, 109231. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Wani, S.J.; Alyousef, A.A.; Alqasim, A.; Syed, A.; El-Enshasy, H.A. Purification and kinetics of the PHB depolymerase of Microbacterium paraoxydans RZS6 isolated from a dumping yard. PLoS ONE 2019, 14, e0212324. [Google Scholar] [CrossRef]
- Amir, M.; Bano, N.; Baker, A.; Zia, Q.; Banawas, S.; Zaheer, M.R.; Shariq, M.; Nawaz, M.S.; Khan, M.F.; Azad, Z.; et al. Isolation and optimization of extracellular PHB depolymerase producer Aeromonas caviae Kuk1-(34) for sustainable solid waste management of biodegradable polymers. PLoS ONE 2022, 17, e0264207. [Google Scholar] [CrossRef]
- Kadoya, R.; Tanaka, N.; Fujita, N.; Shiwa, Y.; Taguchi, S. Changed bacterial community in the river water samples upon introduction of biodegradable poly(3-hydroxybutyrate). Polym. Degrad. Stab. 2020, 176, 109144. [Google Scholar] [CrossRef]
- Vigneswari, S.; Lee, T.S.; Bhubalan, K.; Amirul, A.A. Extracellular Polyhydroxyalkanoate Depolymerase by Acidovorax sp. DP5. Enzyme Res. 2015, 2015, 212159. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Kadouri, D.; Jurkevitch, E.; Okon, Y. Poly beta-hydroxybutyrate depolymerase (PhaZ) in Azospirillum brasilense and characterization of a phaZ mutant. Arch. Microbiol. 2003, 180, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Swiontek Brzezinka, M.; Richert, A.; Kalwasinska, A.; Swiatczak, J.; Deja-Sikora, E.; Walczak, M.; Michalska-Sionkowska, M.; Piekarska, K.; Kaczmarek-Szczepanska, B. Microbial degradation of polyhydroxybutyrate with embedded polyhexamethylene guanidine derivatives. Int. J. Biol. Macromol. 2021, 187, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Sheu, D.S.; Sheu, F.S.; Chen, W.M. Gemmobacter tilapiae sp. nov., a poly-beta-hydroxybutyrate-accumulating bacterium isolated from a freshwater pond. Int. J. Syst. Evol. Microbiol. 2013, 63, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Salinas, A.L.; Osorio, A.; Legorreta-Hissner, T.; Lara-Martinez, R.; Jimenez-Garcia, L.F.; Camarena, L.; Poggio, S. A new type of phasin characterized by the presence of a helix-hairpin-helix domain is required for normal polyhydroxybutyrate accumulation and granule organization in Caulobacter crescentus. Mol. Microbiol. 2023, 120, 307–323. [Google Scholar] [CrossRef]
- Neuenschwander, S.M.; Ghai, R.; Pernthaler, J.; Salcher, M.M. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2018, 12, 185–198. [Google Scholar] [CrossRef]
- Brison, A.; Rossi, P.; Derlon, N. Influent carbon to phosphorus ratio drives the selection of PHA-storing organisms in a single CSTR. Water Res. X 2022, 16, 100150. [Google Scholar] [CrossRef]
- Parada-Pinilla, M.P.; Ferreira, M.A.; Roncallo, J.C.; Santos, S.N.; Melo, I.S.; Assef, A.N.B.; Wilke, D.V.; Silva, L.F.; Garrido, L.M.; Araujo, W.L.; et al. Biopolymer production by halotolerant bacteria isolated from Caatinga biome. Braz. J. Microbiol. 2021, 52, 547–559. [Google Scholar] [CrossRef]
- Mohanan, N.; Wong, M.C.; Budisa, N.; Levin, D.B. Polymer-Degrading Enzymes of Pseudomonas chloroaphis PA23 Display Broad Substrate Preferences. Int. J. Mol. Sci. 2023, 24, 4501. [Google Scholar] [CrossRef]
- Teeka, J.; Imai, T.; Reungsang, A.; Cheng, X.; Yuliani, E.; Thiantanankul, J.; Poomipuk, N.; Yamaguchi, J.; Jeenanong, A.; Higuchi, T.; et al. Characterization of polyhydroxyalkanoates (PHAs) biosynthesis by isolated Novosphingobium sp. THA_AIK7 using crude glycerol. J. Ind. Microbiol. Biotechnol. 2012, 39, 749–758. [Google Scholar] [CrossRef]
- Brown, P.J.; Kysela, D.T.; Buechlein, A.; Hemmerich, C.; Brun, Y.V. Genome sequences of eight morphologically diverse Alphaproteobacteria. J. Bacteriol. 2011, 193, 4567–4568. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadoya, R.; Soga, H.; Matsuda, M.; Sato, M.; Taguchi, S. Bacterial Population Changes during the Degradation Process of a Lactate (LA)-Enriched Biodegradable Polymer in River Water: LA-Cluster Preferable Bacterial Consortium. Polymers 2023, 15, 4111. https://doi.org/10.3390/polym15204111
Kadoya R, Soga H, Matsuda M, Sato M, Taguchi S. Bacterial Population Changes during the Degradation Process of a Lactate (LA)-Enriched Biodegradable Polymer in River Water: LA-Cluster Preferable Bacterial Consortium. Polymers. 2023; 15(20):4111. https://doi.org/10.3390/polym15204111
Chicago/Turabian StyleKadoya, Ryosuke, Hitomi Soga, Miki Matsuda, Michio Sato, and Seiichi Taguchi. 2023. "Bacterial Population Changes during the Degradation Process of a Lactate (LA)-Enriched Biodegradable Polymer in River Water: LA-Cluster Preferable Bacterial Consortium" Polymers 15, no. 20: 4111. https://doi.org/10.3390/polym15204111
APA StyleKadoya, R., Soga, H., Matsuda, M., Sato, M., & Taguchi, S. (2023). Bacterial Population Changes during the Degradation Process of a Lactate (LA)-Enriched Biodegradable Polymer in River Water: LA-Cluster Preferable Bacterial Consortium. Polymers, 15(20), 4111. https://doi.org/10.3390/polym15204111





