Evaluation of the Binding Relationship of the RdRp Enzyme to Novel Thiazole/Acid Hydrazone Hybrids Obtainable through Green Synthetic Procedure
Abstract
:1. Introduction
2. Results and Discussion
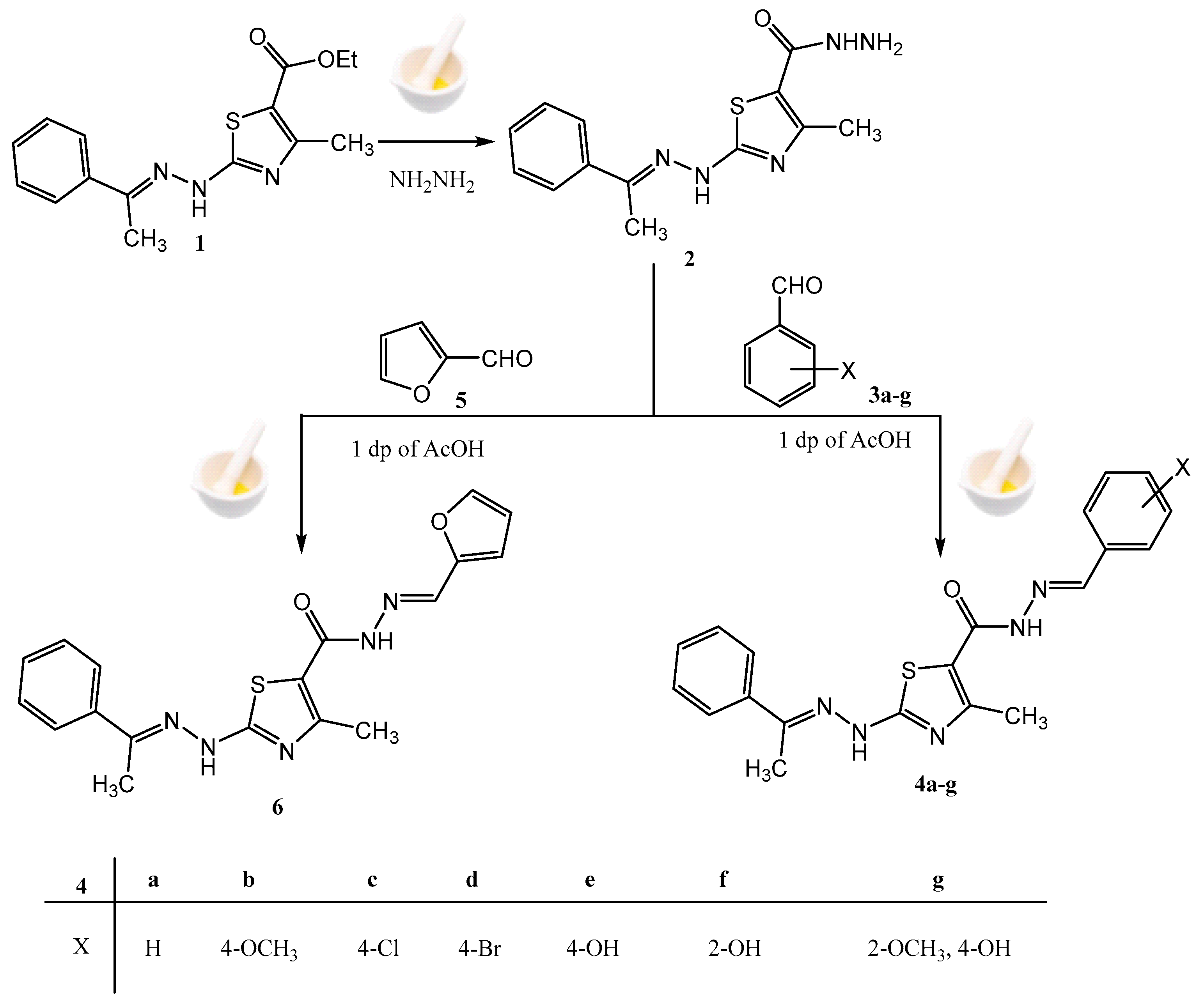
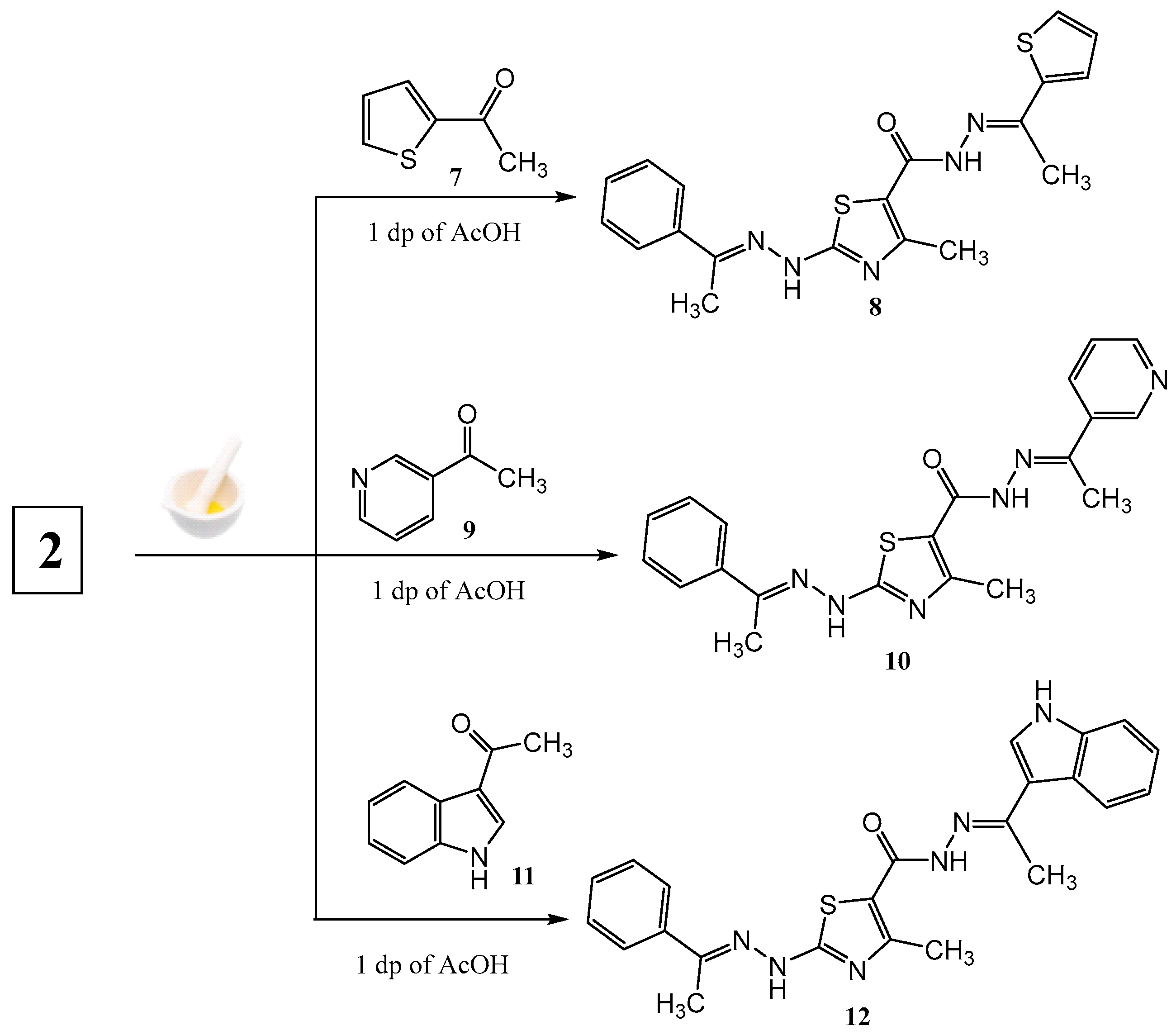
2.1. Chemistry
2.2. Molecular Docking Studies
2.3. Physicochemical Characteristics, Pharmacokinetics, and Drug-Likeness Profile; In Silico Prediction
3. Experimental Section
3.1. Chemistry
3.2. Docking Study
3.3. Target Optimization
3.4. Docking of the Target Molecules to SARS-CoV-2 RdRp Active Site
3.5. Physicochemical Properties, Pharmacokinetics, and Drug-Likeness Profile; In Silico Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Karhale, S.; Survase, D.; Bhat, R.; Ubale, P.; Helavi, V. A practical and green protocol for the synthesis of 2,3-dihydroquinazolin-4(1H)-ones using oxalic acid as organocatalyst. Res. Chem. Intermed. 2017, 43, 3915. [Google Scholar] [CrossRef]
- Gomha, S.M.; Badrey, M.G.; Arafa, W.A.A. DABCO-catalyzed green synthesis of thiazole and 1,3-thiazine derivatives linked to benzofuran. Heterocycles 2016, 92, 1450–1461. [Google Scholar]
- Khalil, K.D.; Riyadh, S.M.; Gomha, S.M.; Ali, I. Synthesis, characterization and application of copper oxide chitosan nanocomposite for green regioselective synthesis of [1,2,3]triazoles. Int. J. Biol. Macromol. 2019, 130, 928–937. [Google Scholar] [CrossRef]
- Brahmachari, G.; Banerjee, B. Facile and One-Pot Access to Diverse and Densely Functionalized 2-Amino-3-cyano-4H-pyrans and Pyran-Annulated Heterocyclic Scaffolds via an Eco-Friendly Multicomponent Reaction at Room Temperature Using Urea as a Novel Organo-Catalyst. ACS Sustain. Chem. Eng. 2014, 2, 411–422. [Google Scholar] [CrossRef]
- Alshabanah, L.A.; Al-Mutabagani, L.A.; Gomha, S.M.; Ahmed, H.A. Three-component synthesis of some new coumarin derivatives as anti-cancer agents. Front. Chem. 2022, 9, 762248. [Google Scholar] [CrossRef]
- Gomha, S.M.; Abdelaziz, M.R.; Abdel-Aziz, H.M.; Hassan, S.A. Green Synthesis and Molecular Docking of Thiazolyl-thiazole Derivatives as Potential Cytotoxic Agents. Mini Rev. Med. Chem. 2017, 17, 805–815. [Google Scholar] [CrossRef]
- Gomha, S.M.; Muhammad, Z.A.; Abdel-Aziz, H.M.; Matar, I.K.; El-Sayed, A.A. Green synthesis, molecular docking and anticancer activity of novel 1,4-dihydropyridine-3,5-Dicarbohydrazones under grind-stone chemistry. Green Chem. Lett. Rev. 2020, 13, 6–17. [Google Scholar] [CrossRef]
- Gomha, S.M.; Khalil, K.D.; El-Zanate, A.M.; Riyadh, S.M. A Facile green synthesis and anti-cancer activity of bis-arylhydrazononitriles, triazolo[5,1-c][1,2,4]triazine, and 1,3,4-thiadiazoline. Heterocycles 2013, 87, 1109–1120. [Google Scholar]
- Zhou, B.; Liu, Z.-C.; Qu, W.-W.; Yang, R.; Lin, X.-R.; Yan, S.-J.; Lin, J. An environmentally benign, mild, and catalyst-free reaction of quinones with heterocyclic ketene aminals in ethanol: Site-selective synthesis of rarely fused [1,2-a]indolone derivatives via an unexpected anti-Nenitzescu strategy. Green Chem. 2014, 16, 4359–4370. [Google Scholar] [CrossRef]
- Kiyani, H.; Darbandi, H.; Mosallanezhad, A.; Ghorbani, F. 2-Hydroxy-5-sulfobenzoic acid: An efficient organocatalyst for the three-component synthesis of 1-amidoalkyl-2-naphthols and 3,4-disubstituted isoxazol-5(4H)-ones. Res. Chem. Intermed. 2015, 41, 7561–7579. [Google Scholar] [CrossRef]
- Kiyani, H.; Ghorbani, F. Expeditious Green Synthesis of 3,4-disubstituted isoxazole-5(4H)-ones Catalyzed by Nano-MgO. Res. Chem. Intermed. 2016, 42, 6831–6844. [Google Scholar] [CrossRef]
- Sujatha, K.; Vedula, R.R. Novel one-pot expeditious synthesis of 2,4-disubstituted thiazoles through a three-component reaction under solvent free conditions. Synth. Commun. 2018, 48, 302–308. [Google Scholar] [CrossRef]
- Abdalla, M.A.; Gomha, S.M.; Abdelaziz, M.; Serag, N. Synthesis and antiviral evaluation of some novel thiazoles and 1,3-thiazines substituted with pyrazole moiety against rabies virus. Turk. J. Chem. 2016, 40, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Gomha, S.M.; Edrees, M.M.; Muhammad, Z.A.; El-Reedy, A.A.M. 5-(Thiophen-2-yl)-1,3,4-thiadiazole derivatives: Synthesis, molecular docking and in-vitro cytotoxicity evaluation as potential anticancer agents. Drug Des. Dev. Ther. 2018, 12, 1511–1523. [Google Scholar] [CrossRef] [Green Version]
- Nayak, S.; Gaonkar, S.L. A Review on recent synthetic strategies and pharmacological importance of 1,3-thiazole derivatives. Mini Rev. Med. Chem. 2019, 19, 215–238. [Google Scholar] [CrossRef]
- Kumar, S.; Aggarwal, R. Thiazole: A Privileged Motif in Marine Natural Products. Mini Rev. Org. Chem. 2019, 16, 26–34. [Google Scholar] [CrossRef]
- Gomha, S.M.; Salah, T.A.; Abdelhamid, A.O. Synthesis, characterization, and pharmacological evaluation of some novel thiadiazoles and thiazoles incorporating pyrazole moiety as anticancer agents. Monatsh. Chem. 2014, 146, 149–158. [Google Scholar] [CrossRef]
- Dos Santos Silva, T.D.; Bomfim, L.M.; da Cruz Rodrigues, A.C.B.; Dias, R.B.; Sales, C.B.S.; Rocha, C.A.G.; Soares, M.B.P.; Bezerra, D.P.; de Oliveira Cardoso, M.V.; Leite, A.C.; et al. Anti-liver cancer activity in vitro and in vivo induced by 2-pyridyl 2, 3-thiazole derivatives. Toxicol. Appl. Pharmacol. 2017, 329, 212–223. [Google Scholar] [CrossRef]
- Morigi, R.; Locatelli, A.; Leoni, A.; Rambaldi, M. Recent patents on thiazole derivatives endowed with antitumor activity. Recent Pat. Anticancer Drug Discov. 2015, 10, 280–297. [Google Scholar] [CrossRef]
- Abu-Melha, S.; Edrees, M.M.; Salem, H.H.; Kheder, N.A.; Gomha, S.M.; Abdelaziz, M.R. Synthesis and biological evaluation of some novel thiazole-based heterocycles as potential anticancer and antimicrobial agents. Molecules 2019, 24, 539. [Google Scholar] [CrossRef] [Green Version]
- Danilenko, G.I.; Rubalko, S.L.; Maksimov, Y.N.; Baklan, U.F.; Guzhova, S.V. Adamantane-1- and norbornane-2-carboxylic acid hydrazides as HIV inhibitors. Pharm. Chem. J. 2000, 34, 23–24. [Google Scholar] [CrossRef]
- Maniak, H.; Talma, M.; Giurg, M. Inhibitory Potential of New Phenolic Hydrazide-Hydrazones with a Decoy Substrate Fragment towards Laccase from a Phytopathogenic Fungus: SAR and Molecular Docking Studies. Int. J. Mol. Sci. 2021, 22, 12307. [Google Scholar] [CrossRef]
- Kratky, M.; Svrckova, K.; Vu, Q.A.; Stepankova, S.; Vinsova, J. Hydrazones of 4-(Trifluoromethyl)benzohydrazide as New Inhibitors of Acetyl- and Butyrylcholinesterase. Molecules 2021, 26, 989. [Google Scholar] [CrossRef]
- Javaid, S.; Saad, S.M.; Zafar, H.; Malik, R.; Khan, K.M.; Choudhary, M.I.; Rahman, A.-U. Thymidine phosphorylase and prostrate cancer cell proliferation inhibitory activities of synthetic 4-hydroxybenzohydrazides: In vitro, kinetic, and in silico studies. PLoS ONE 2020, 15, e0227549. [Google Scholar] [CrossRef] [Green Version]
- Constable, D.J.C.; Curzons, A.D.; Cunningham, V.L. Metrics to ‘Green’ Chemistry—Which Are the Best? Green Chem. 2002, 4, 521–527. [Google Scholar] [CrossRef]
- Saha, A.; Kumar, R.; Kumar, R.; Devakumar, C. Development and assessment of green synthesis of hydrazides. Indian J. Chem. 2010, 49, 526–531. [Google Scholar]
- Rudd, C.E. GSK-3 Inhibition as a Therapeutic Approach Against SARs CoV2: Dual Benefit of Inhibiting Viral Replication While Potentiating the Immune 412 Response. Front. Immunol. 2020, 11, 1638. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [Green Version]
- McKee, D.L.; Sternberg, A.; Stange, U.; Laufer, S.; Naujokat, C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020, 157, 104859. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.K.; Gadthula, S.; Chen, X.; Choo, H.; Olgen, S.; Barnard, D.L.; Sidwell, R.W. Antiviral Activity of Nucleoside Analogues against SARS-coronavirus (SARS-CoV). Antivir. Chem. Chemother. 2006, 17, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Makam, P.; Kankanala, R.; Prakash, A.; Kannan, T. 2-(2-Hydrazinyl)thiazole derivatives: Design, synthesis and in vitro antimycobacterial studies. Eur. J. Med. Chem. 2013, 69, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Rajanarendar, E.; Krishna, S.R.; Nagaraju, D.; Reddy, K.G.; Kishore, B.; Reddy, Y. Environmentally benign synthesis, molecular properties prediction and anti-inflammatory activity of novel isoxazolo[5,4-d]isoxazol-3-yl-aryl-methanones via vinylogous Henry nitroaldol adducts as synthons. Bioorg. Med. Chem. Lett. 2015, 25, 1630–1634. [Google Scholar] [CrossRef]
- Wetzel, S.; Schuffenhauer, A.; Roggo, S.; Ertl, P.; Waldmann, H. Cheminformatic analysis of natural products and their chemical space. CHIMIA Inter. J. Chem. 2007, 61, 355–360. [Google Scholar] [CrossRef]
- Arnautova, Y.A.; Abagyan, R.A.; Totrov, M. Development of a new physics-based internal coordinate mechanics force field and its application to protein loop modeling. Proteins Struct. Funct. Bioinform. 2011, 79, 477–498. [Google Scholar] [CrossRef] [Green Version]


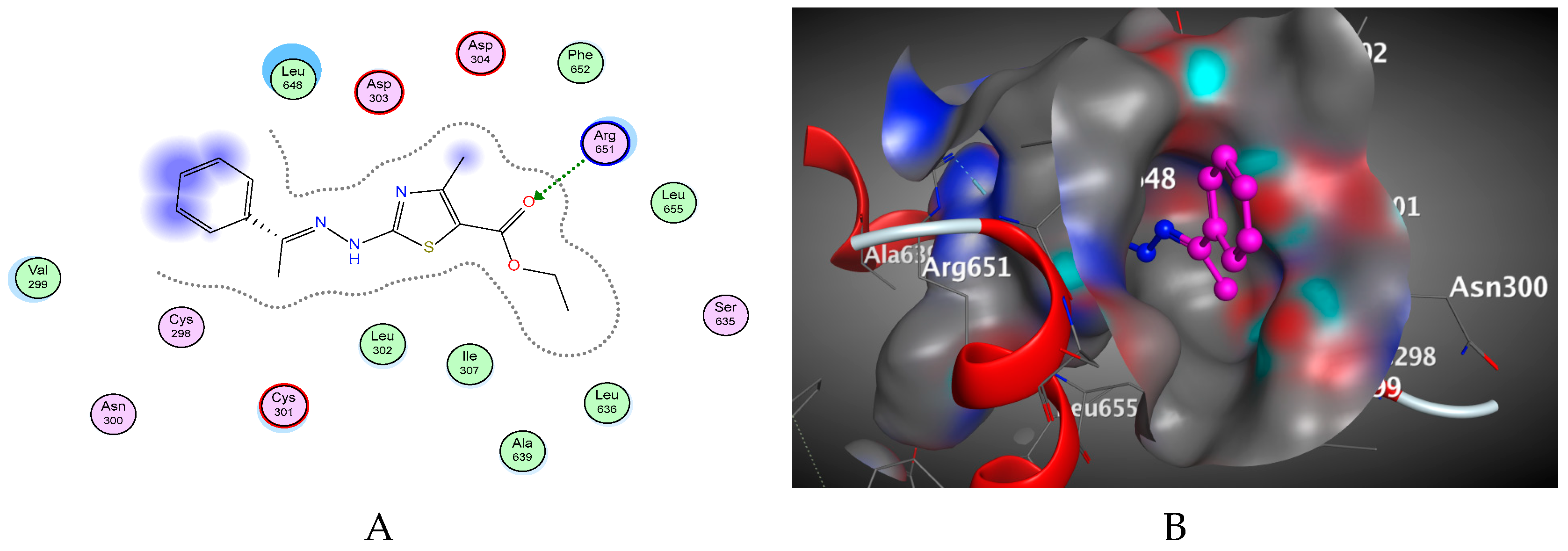
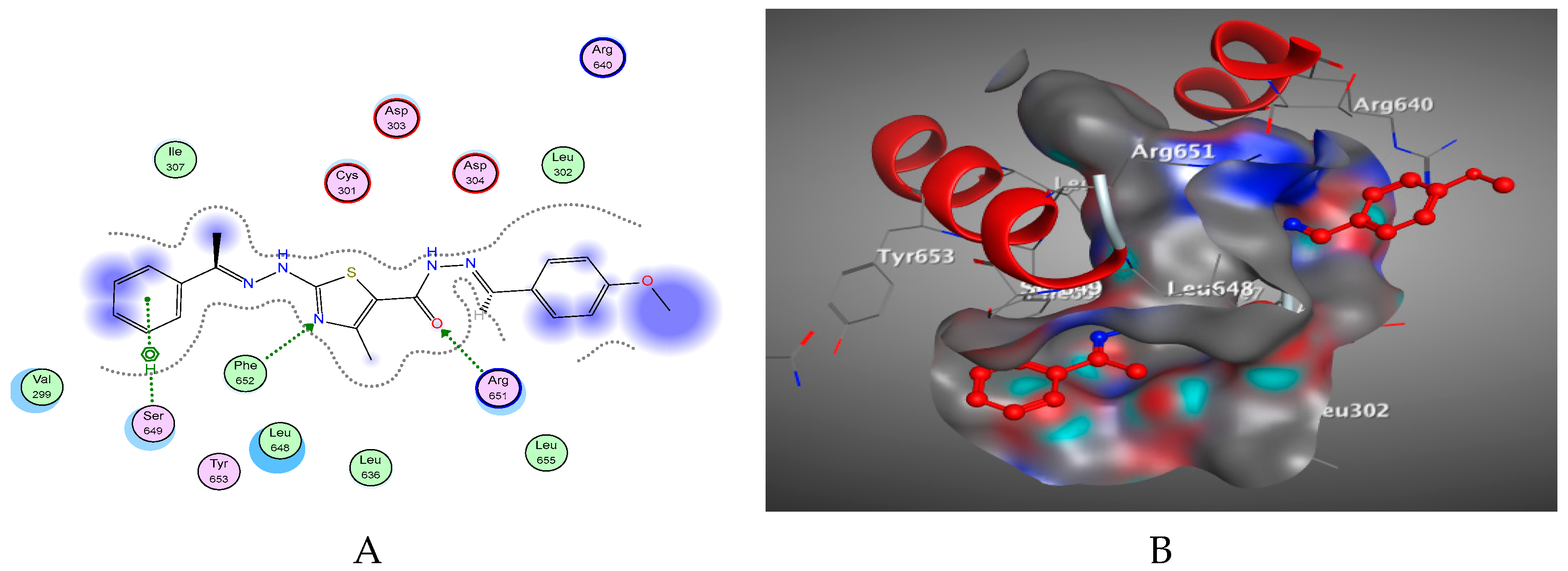
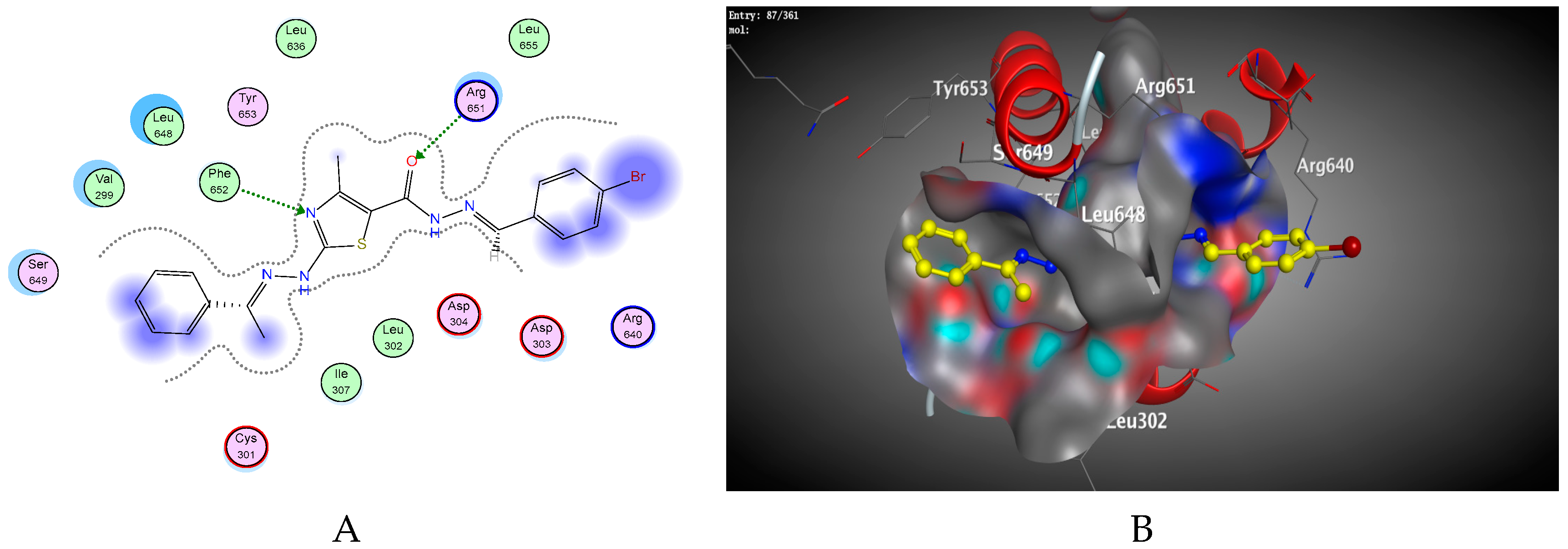
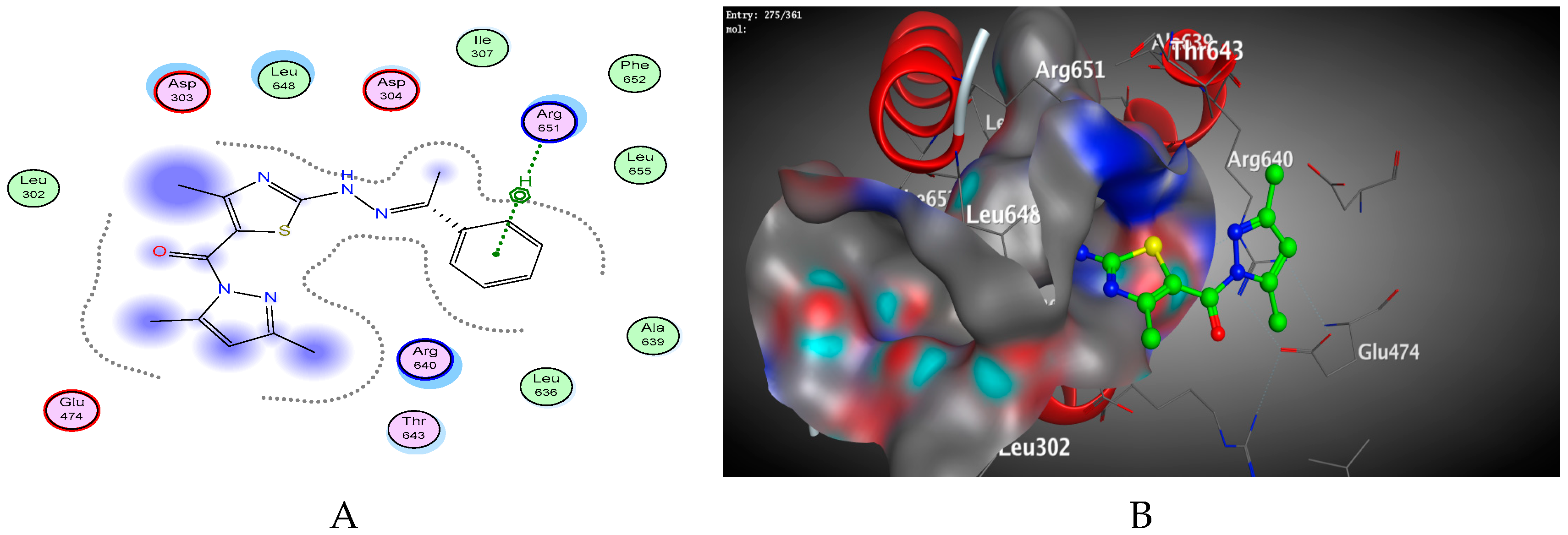
| Compound | Energy Score | Inhibition Constant Ki (µM) | ΔG (Kcal/mol) a | Ligand-Receptor Interactions | ||
|---|---|---|---|---|---|---|
| Residue | Type | Length (Å) | ||||
| 1 | −6.26 | 2.61 × 10−5 | −4.5 | ARG651 | H-acceptor | 2.73 |
| 2 | −5.91 | 4.72 × 10−5 | −1.2 −5.7 | ARG651 ARG651 | H-acceptor H-acceptor | 3.02 2.84 |
| 4a | −5.30 | 1.32 × 10−4 | −2.8 −0.9 −0.6 | ARG651 PHE652 SER649 | H-acceptor H-acceptor Pi-H | 2.93 3.14 3.62 |
| 4b | −5.94 | 4.48 × 10−5 | −2.8 −0.9 −0.6 | ARG651 PHE652 SER649 | H-acceptor H-acceptor Pi-H | 2.82 3.13 3.62 |
| 4c | −5.35 | 1.21 × 10−4 | −2.8 −0.9 | ARG651 PHE652 | H-acceptor H-acceptor | 2.82 3.13 |
| 4d | −5.38 | 1.15 × 10−4 | −2.8 −0.9 | ARG651 PHE652 | H-acceptor H-acceptor | 2.83 3.13 |
| 4e | −5.70 | 6.72 × 10−5 | −2.8 −0.9 | ARG651 PHE652 | H-acceptor H-acceptor | 2.82 3.13 |
| 4f | −5.85 | 5.22 × 10−5 | −1.7 −2.8 −0.8 −0.6 | ASP304 ARG651 PHE652 SER649 | H-donor H-acceptor H-acceptor Pi-H | 2.93 2.75 3.18 3.60 |
| 4g | −5.91 | 4.72 × 10−5 | −2.1 −2.5 −0.8 −0.6 | ARG651 ARG651 PHE652 SER649 | H-acceptor H-acceptor H-acceptor Pi-H | 2.97 2.69 3.20 3.60 |
| 6 | −6.63 | 1.4 × 10−5 | −2.1 −2.9 −0.9 | ARG651 ARG651 ASP303 | H-acceptor H-acceptor H-acceptor | 2.94 3.08 3.03 |
| 8 | −5.60 | 7.95 × 10−5 | −2.9 −0.8 −0.6 | ARG651 PHE652 SER649 | H-acceptor H-acceptor Pi-H | 2.77 3.19 3.62 |
| 10 | −5.57 | 8.36 × 10−5 | −2.3 | ARG651 | H-acceptor | 2.61 |
| 12 | −5.75 | 6.17 × 10−5 | −2.1 −0.6 | ARG651 SER649 | H-acceptor Pi-H | 2.62 3.60 |
| 14a | −4.52 | 4.91 × 10−4 | −0.9 | ARG651 | Pi-H | 3.73 |
| 14b | −5.08 | 1.91 × 10−4 | −2.9 −1.0 | CYS301 ARG651 | H-donor Pi-H | 3.32 3.67 |
| 16 | −4.71 | 3.56 × 10−4 | −0.8 | ARG651 | Pi-H | 3.73 |
| Remdesivir | −5.48 | 9.73 × 10−5 | −1.5 −2.3 | ARG651 ARG651 | H-acceptor H-acceptor | 3.19 3.12 |
| Physicochemical Properties | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | Lipophilicity Consensus log P | MW a g/mol | Heavy Atoms | Aromatic Heavy a Toms | Rot. Bond | HBA b | HBD c | MR d | TPSA e (A2) | % ABS f |
| 1 | 3.33 | 303.38 | 21 | 11 | 6 | 4 | 1 | 85.45 | 91.82 | 77.32 |
| 2 | 1.92 | 289.36 | 20 | 11 | 5 | 4 | 3 | 80.26 | 120.64 | 67.38 |
| 4a | 3.80 | 377.46 | 27 | 17 | 7 | 4 | 2 | 110.53 | 106.98 | 71.56 |
| 4b | 3.86 | 407.49 | 29 | 17 | 8 | 5 | 2 | 117.02 | 116.21 | 68.91 |
| 4c | 4.37 | 411.91 | 28 | 17 | 7 | 4 | 2 | 115.54 | 106.98 | 72.09 |
| 4d | 4.46 | 456.36 | 28 | 17 | 7 | 4 | 2 | 118.23 | 106.98 | 72.09 |
| 4e | 3.45 | 393.46 | 28 | 17 | 7 | 5 | 3 | 112.56 | 127.21 | 65.11 |
| 4f | 3.47 | 393.46 | 28 | 17 | 7 | 5 | 3 | 112.56 | 127.21 | 65.11 |
| 4g | 3.87 | 409.49 | 29 | 17 | 8 | 5 | 2 | 117.02 | 116.21 | 68.91 |
| 6 | 3.23 | 367.42 | 26 | 16 | 7 | 5 | 2 | 102.80 | 120.12 | 67.56 |
| 8 | ND g | 397.52 | 27 | 16 | 7 | 4 | 2 | 113.22 | 135.22 | 62.35 |
| 10 | ND | 392.48 | 28 | 17 | 7 | 5 | 2 | 113.13 | 119.87 | 67.64 |
| 12 | ND | 430.53 | 31 | 20 | 7 | 4 | 3 | 127.20 | 122.77 | 66.64 |
| 14a | 2.93 | 355.41 | 25 | 16 | 5 | 4 | 2 | 99.11 | 120.38 | 67.47 |
| 14b | 3.91 | 417.48 | 30 | 22 | 6 | 4 | 2 | 119.58 | 120.38 | 67.47 |
| 16 | 3.58 | 353.44 | 25 | 16 | 55 | 4 | 1 | 101.25 | 100.41 | 74.36 |
| Compound | S a (mg/L) | Drug-Likeness Model Score | Lipinski Violations | Bioavailability Score |
|---|---|---|---|---|
| 1 | 1.90 | 0.29 | 0 | 0.55 |
| 2 | 1.25 | −0.01 | 0 | 0.55 |
| 4a | 1.50 | 0.12 | 0 | 0.55 |
| 4b | 1.68 | 0.16 | 0 | 0.55 |
| 4c | 5.57 | 0.65 | 0 | 0.55 |
| 4d | 3.96 | 0.65 | 0 | 0.55 |
| 4e | 7.96 | 0.61 | 0 | 0.55 |
| 4f | 7.96 | 0.28 | 0 | 0.55 |
| 4g | 5.42 | −0.01 | 0 | 0.55 |
| 6 | 1.15 | 0.23 | 0 | 0.55 |
| 8 | ND | 0.80 | 0 | ND |
| 10 | ND | 0.04 | 0 | ND |
| 12 | 2.93 | −0.42 | 0 | 0.55 |
| 14a | 1.23 | 0.47 | 0 | 0.55 |
| 14b | 1.20 | 0.47 | 0 | 0.55 |
| 16 | 4.04 | −0.34 | 0 | 0.55 |
| Compound | Pharmacokinetics | |||||
|---|---|---|---|---|---|---|
| Caco-2 a | BBB b | MDCK c | HIA d | PPB e | CYP 2D6 f | |
| 1 | 21.25 | 0.12 | 10.49 | 96.60 | 85.13 | Non |
| 2 | 0.46 | 0.03 | 9.50 | 87.95 | 56.98 | Non |
| 4a | 14.36 | 0.04 | 14.23 | 94.87 | 91.92 | Non |
| 4b | 16.10 | 0.03 | 0.78 | 95.25 | 89.70 | Non |
| 4c | 14.38 | 0.09 | 0.92 | 95.15 | 89.12 | Non |
| 4d | 22.52 | 0.09 | 0.03 | 95.53 | 87.60 | Non |
| 4e | 1.99 | 0.08 | 0.79 | 92.26 | 87.62 | Non |
| 4f | 2.47 | 0.08 | 2.30 | 92.26 | 86.98 | Non |
| 4g | 17.97 | 0.03 | 0.716 | 95.25 | 88.24 | Non |
| 6 | 9.78 | 0.02 | 90.64 | 94.75 | 88.28 | Non |
| 8 | 11.54 | 0.03 | 5.08 | 95.86 | 86.89 | Non |
| 10 | 11.97 | 0.04 | 6.69 | 95.37 | 82.99 | Non |
| 12 | 14.75 | 0.44 | 0.27 | 91.31 | 88.24 | Non |
| 14a | 3.45 | 0.12 | 1.16 | 94.31 | 74.99 | Non |
| 14b | 9.85 | 0.07 | 0.22 | 96.19 | 94.04 | Non |
| 16 | 21.68 | 0.35 | 0.12 | 97.47 | 91.53 | Non |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Humaidi, J.Y.; Badrey, M.G.; Aly, A.A.; Nayl, A.A.; Zayed, M.E.M.; Jefri, O.A.; Gomha, S.M. Evaluation of the Binding Relationship of the RdRp Enzyme to Novel Thiazole/Acid Hydrazone Hybrids Obtainable through Green Synthetic Procedure. Polymers 2022, 14, 3160. https://doi.org/10.3390/polym14153160
Al-Humaidi JY, Badrey MG, Aly AA, Nayl AA, Zayed MEM, Jefri OA, Gomha SM. Evaluation of the Binding Relationship of the RdRp Enzyme to Novel Thiazole/Acid Hydrazone Hybrids Obtainable through Green Synthetic Procedure. Polymers. 2022; 14(15):3160. https://doi.org/10.3390/polym14153160
Chicago/Turabian StyleAl-Humaidi, Jehan Y., Mohamed G. Badrey, Ashraf A. Aly, AbdElAziz A. Nayl, Mohie E. M. Zayed, Ohoud A. Jefri, and Sobhi M. Gomha. 2022. "Evaluation of the Binding Relationship of the RdRp Enzyme to Novel Thiazole/Acid Hydrazone Hybrids Obtainable through Green Synthetic Procedure" Polymers 14, no. 15: 3160. https://doi.org/10.3390/polym14153160
APA StyleAl-Humaidi, J. Y., Badrey, M. G., Aly, A. A., Nayl, A. A., Zayed, M. E. M., Jefri, O. A., & Gomha, S. M. (2022). Evaluation of the Binding Relationship of the RdRp Enzyme to Novel Thiazole/Acid Hydrazone Hybrids Obtainable through Green Synthetic Procedure. Polymers, 14(15), 3160. https://doi.org/10.3390/polym14153160







