Microbial Degradation of Rubber: Actinobacteria
Abstract
1. Rubber—Polyisoprenes
1.1. Natural Rubber (NR)
1.2. Production and Usage
1.3. Synthetic Rubber (IR)
1.4. Rubber Wastes—Mitigation and Drawbacks
2. Biodegradation: Roles of Microbes
2.1. Rubber Degraders: Who, Where, How?
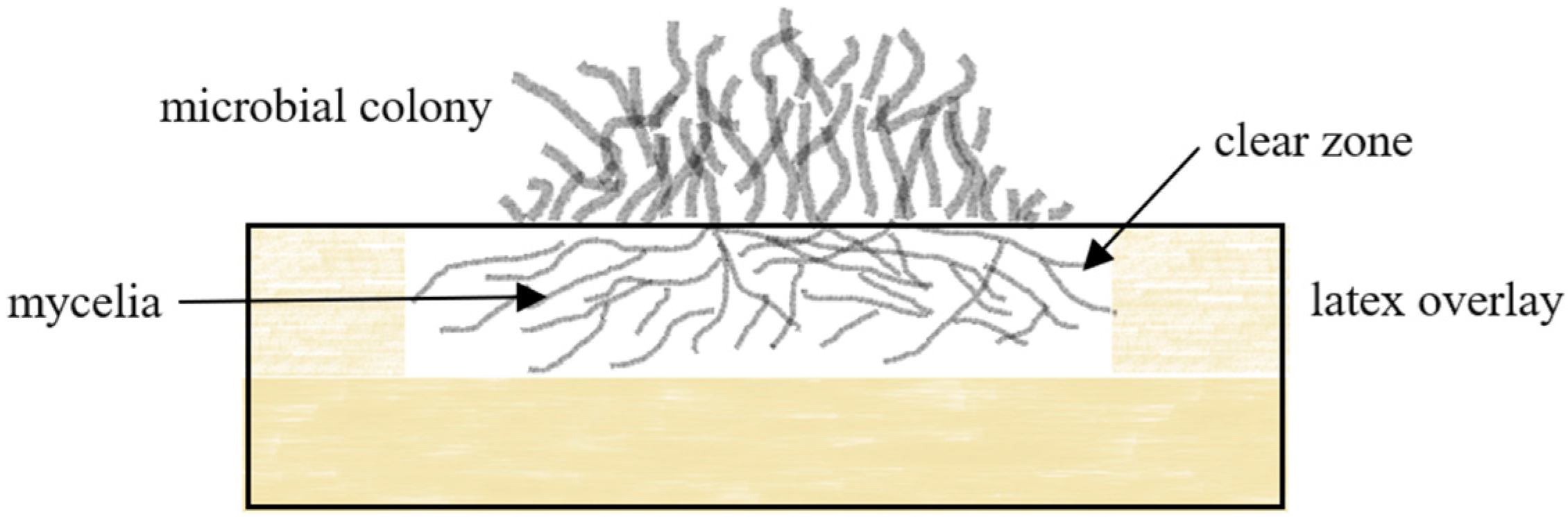
2.1.1. Clear Zone Formers
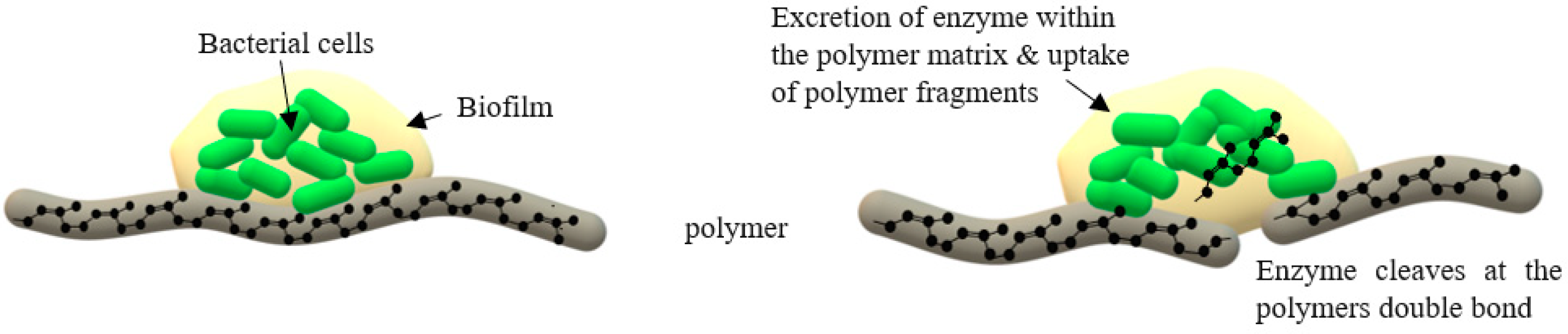
2.1.2. Adhesive Contact Strains
3. Rubber Degrading Enzymes: Rubber Oxygenase
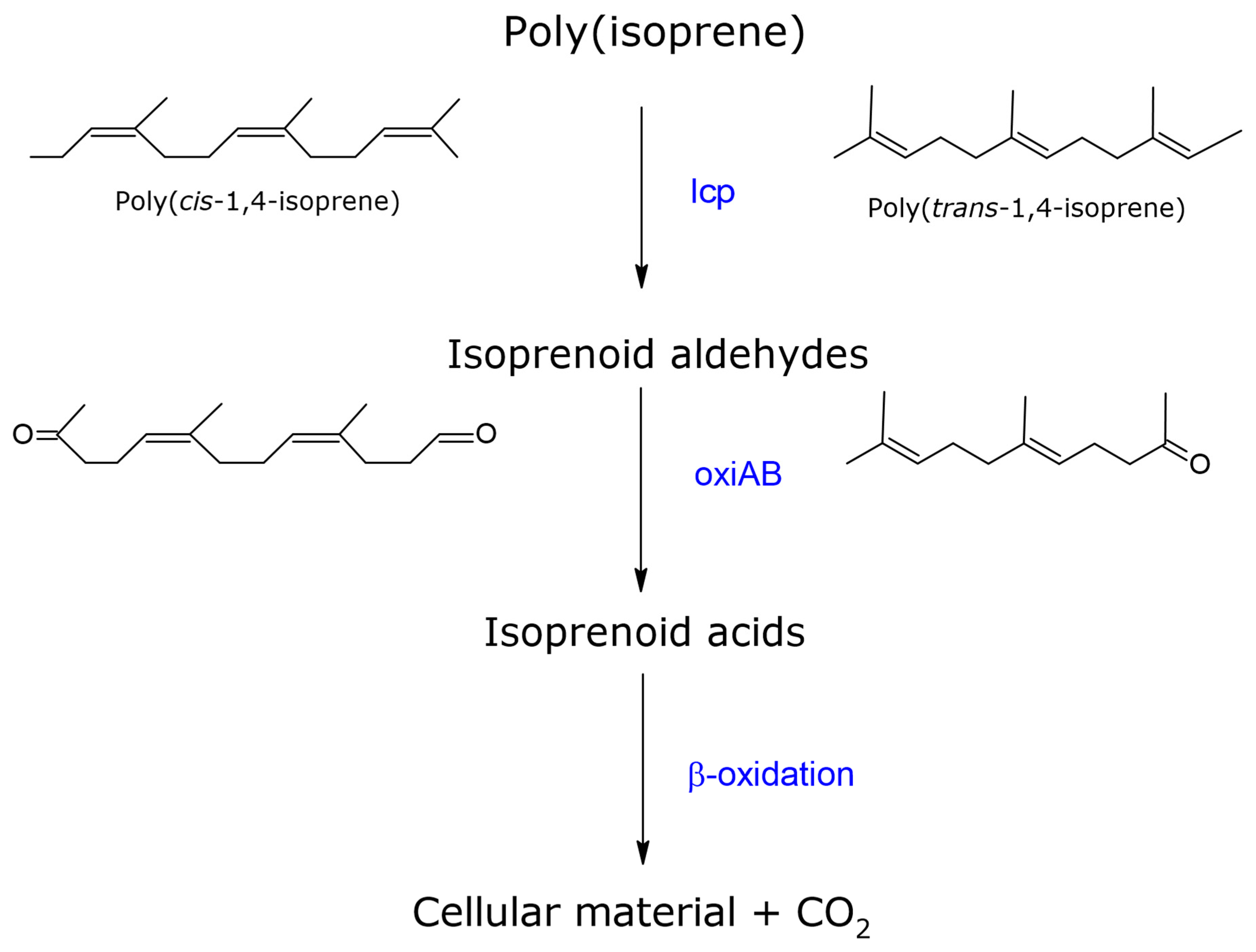
3.1. Latex Clearing Protein (lcp)
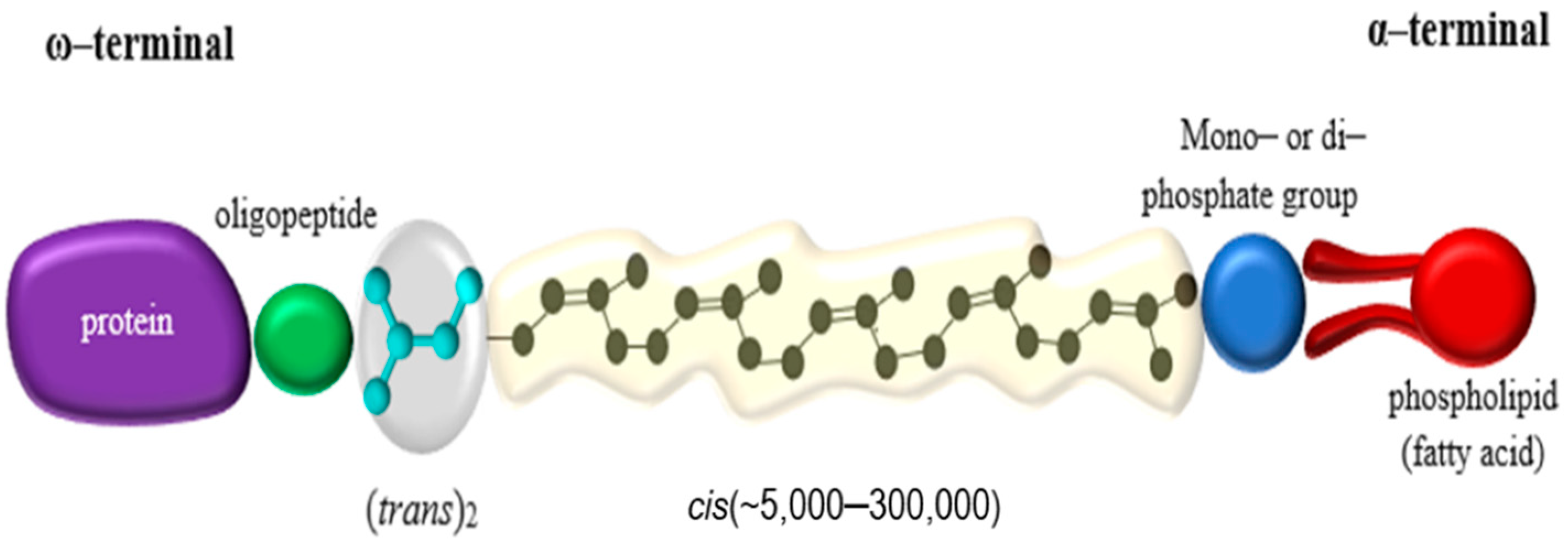
Rubber Degradation by Lcp Enzyme
3.2. RoxA and RoxB
3.3. Others: Enzyme Mediator Systems
3.4. Unknown: Rubber Degradation in Fungi
4. Lcp Gene and Its Pathway
4.1. Conserved Region
4.2. Lcp Operon & Rubber Degradation Pathway
4.3. Lcp Secretion Pathway
4.4. Lcp Regulator
5. Latex Clearing Protein: Synthesis
6. Actinobacteria and Their Potential for Biodegrading Rubber
6.1. Diversity
6.2. Distribution
6.3. Genomic Characteristics of Actinobacteria
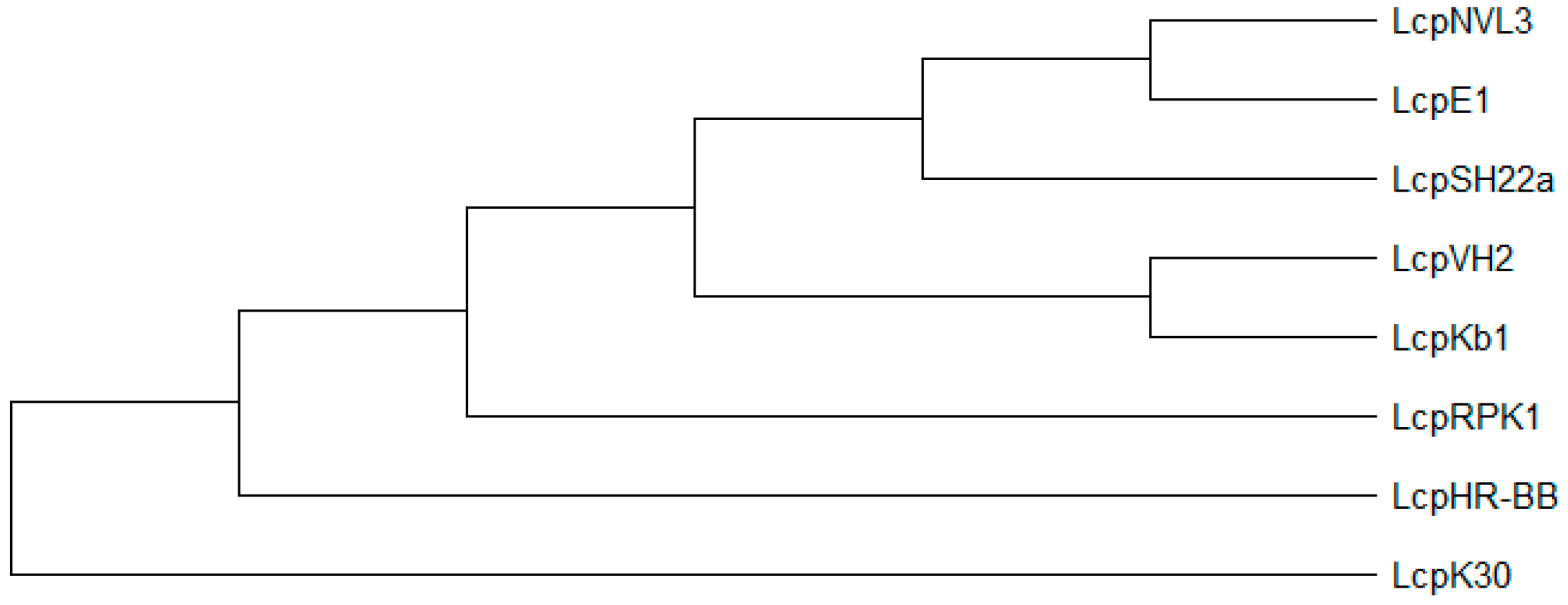
6.4. Latex Clearing Protein in the GenBank: Actinobacteria
7. Rubber Biodegradation
7.1. In Vivo Rubber Degradation Using Microbial Culture
7.1.1. Measurements for Rubber Degradation
7.1.2. Drawbacks
7.2. In Situ Rubber Degradation
7.2.1. Bioaugmentation and Consortia
7.2.2. Biotransformation
7.2.3. Bioreactors
7.2.4. Synthetic Biology
7.2.5. Drawbacks
7.3. Rubber Reclamation of Tyres by Microbes
8. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Birke, J.; Jendrossek, D. Rubber Oxygenase and Latex Clearing Protein Cleave Rubber to Different Products and Use Different Cleavage Mechanisms. Appl. Environ. Microbiol. 2014, 80, 5012–5020. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sakdapipanich, J.T. Chemical Structure and Occurrence of Natural Polyisoprenes. Biol. Chem. Biotechnol. Appl. 2005, 2. [Google Scholar] [CrossRef]
- Shah, A.A.; Hasan, F.; Shah, Z.; Kanwal, N.; Zeb, S. Biodegradation of natural and synthetic rubbers: A review. Int. Biodeterior. Biodegrad. 2013, 83, 145–157. [Google Scholar] [CrossRef]
- Ilcu, L.; Röther, W.; Birke, J.; Brausemann, A.; Einsle, O.; Jendrossek, D. Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Backhaus, R.A. Rubber Formation in Plants—A Mini-Review. Isr. J. Bot. 1985, 34, 283–293. [Google Scholar]
- Jendrossek, D.; Birke, J. Mini-Review Rubber Oxygenases. Appl. Microbiol. Biotechnol. 2018, 103, 125–142. [Google Scholar] [CrossRef]
- Rose, K.; Tenberge, K.B.; Steinbüchel, A. Identification and Characterization of Genes from Streptomyces sp. Strain K30 Responsible for Clear Zone Formation on Natural Rubber Latex and Poly(cis-1,4-isoprene) Rubber Degradation. Biomacromolecules 2005, 6, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Yikmis, M.; Steinbüchel, A. Historical and Recent Achievements in the Field of Microbial Degradation of Natural and Synthetic Rubber. Appl. Environ. Microbiol. 2012, 78, 4543–4551. [Google Scholar] [CrossRef] [PubMed]
- Cornish, K. Alternative Natural Rubber Crops: Why Should We Care? Technol. Innov. 2017, 18, 244–255. [Google Scholar] [CrossRef]
- Brüning, K. Natural Rubber. In Chemistry and Materials Science; Kobayashi, S.M.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Puskas, J.E.; Chiang, K.; Barkakaty, B. Natural Rubber (NR) Biosynthesis: Perspectives from Polymer Chemistry. In Chemistry, Manufacture and Applications of Natural Rubber; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 30–67. [Google Scholar] [CrossRef]
- Warneke, S.; Arenskötter, M.; Tenberge, K.B.; Steinbüchel, A. Bacterial degradation of poly(trans-1,4-isoprene) (gutta percha). Microbiology 2007, 153, 347–356. [Google Scholar] [CrossRef] [PubMed]
- OECD. Emission Scenario Document on Additives in Rubber Industry. Available online: https://www.oecd-ilibrary.org/docserver/9789264221192-en.pdf?expires=1622740209&id=id&accname=guest&checksum=6A03F078669879707636D40FA0CC9842 (accessed on 4 June 2020).
- White, J.L. First of a Series: Pioneering Polymer Industry Developments: Bayer and the First Synthetic Rubber First of a Series. Int. Polym. Process. 1999, 14, 114. [Google Scholar] [CrossRef]
- Statisca. Consumption of Natural and Synthetic Rubber Worldwide from 1990–2018 (in 1000 Metric Tons). Available online: https://www.statista.com/statistics/275399/world-consumption-of-natural-and-synthetic-caoutchouc/ (accessed on 12 March 2021).
- The Synthetic Rubber Production Process. AQUASEAL Rubber Limited. Available online: https://www.aquasealrubber.co.uk/uncategorized/the-synthetic-rubber-production-process/ (accessed on 4 June 2020).
- Tiseo, I. Rubber—Statistics; Facts. Available online: https://www.statista.com/topics/3268/rubber/ (accessed on 13 March 2021).
- Andrew, L. How Long it Takes 50 Common Items to Decompose. Available online: https://stacker.com/stories/2682/how-long-it-takes-50-common-items-decompose (accessed on 24 July 2020).
- Clark, T. Enhancing the Biodegradation of Waste Rubber Discarded Rubber Materials. International Latex Conference 2015. Available online: https://www.rubbernews.com/assets/PDF/RN10077285.PDF (accessed on 24 July 2020).
- Unciano, N.M. Microbial Processing of Natural Rubber Waste. Insight in the Microbial Technologies and Methods for Bioremediation. Available online: https://zenodo.org/record/202313/files/Chap_3_Insight_DBTex.pdf (accessed on 19 March 2020).
- Ward, J. Synthetic Biology: From Science to Bioeconomy Application: Opportunities for Challenge-led Innovation. Available online: https://www.changemakers.com/sites/default/files/b4_-_synthetic_biology_-_opportunities_for_challenge-led_innovation.pdf (accessed on 13 March 2020).
- Williams, P.T. Pyrolysis of Waste Tyres: A Review. Waste Manag. 2013, 33, 1714–1728. [Google Scholar] [CrossRef] [PubMed]
- EPA. Rubber and Leather: Material-Specific Data. Agency, United States Environmental Protection. Available online: https://www.epa.gov/facts-and-figures-about-materials-waste-and-recycling/rubber-and-leather-material-specific-data (accessed on 30 April 2020).
- WBSCD. Development for WBSCD. Stakeholders Launch Global Platform for Sustainable Natural Rubber. Available online: https://www.wbcsd.org/Sector-Projects/Tire-Industry-Project/News/Stakeholders-launch-Global-Platform-for-Sustainable-Natural-Rubber (accessed on 2 April 2019).
- Oriaku, E.C.; Agulanna, C.N.; Odenigbo, J.; Nnoruka, N. Waste to Wealth through the Incineration of Waste Tyres and Recovery of Carbon Black. Int. J. Multidiscip. Sci. Eng. 2013, 4, 30–36. [Google Scholar]
- Makitan, V. Waste Tyre Recycling: Current Status, Economic Analysis and Process Development. Ph.D. Thesis, Curtin University, Bentley, Australia, 2010. Available online: https://espace.curtin.edu.au/handle/20.500.11937/845 (accessed on 13 March 2021).
- EPA. Air Emissions from Scrap Tire Combustion. Agency, United States Environmental Protection. Available online: https://www3.epa.gov/ttncatc1/dir1/tire_eng.pdf (accessed on 13 March 2019).
- Wądrzyk, M.; Janus, R.; Rządzik, B.; Lewandowski, M.; Budzyń, S. Pyrolysis Oil from Scrap Tires as a Source of Fuel Components: Manufacturing, Fractionation, and Characterization. Energy Fuels 2020, 34, 5917–5928. [Google Scholar] [CrossRef]
- Yong, Z.J.; Bashir, M.J.; Ng, C.A.; Sethupathi, S.; Lim, J.W.; Show, P.L. Show Sustainable Waste-to-Energy Development in Malaysia: Appraisal of Environmental, Financial, and Public Issues Related with Energy Recovery from Municipal Solid Waste. Processes 2019, 7, 676. [Google Scholar] [CrossRef]
- EPA. Tire Fires. Agency, United States Environmental Protection. Available online: https://archive.epa.gov/epawaste/conserve/materials/tires/web/html/fires.html (accessed on 13 March 2019).
- A Study on Scrap Tyres Management for Peninsular Malaysia. Final Report (September 2011). Chemsain Konsultant Sdn. Bhd. Available online: https://jpspn.kpkt.gov.my/resources/index/user_1/Sumber_Rujukan/kajian/Tyre%20Study_Final%20Report_Eng%20Version.pdf (accessed on 13 March 2020).
- Holdings, D. Waste Tyre Solution. Available online: https://www.wasteoiltodieseloil.com/pyrolysis_oil_solutions/ (accessed on 30 April 2020).
- Halle, L.L.; Palmqvist, A.; Kampmann, K.; Khan, F.R. Ecotoxicology of micronized tire rubber: Past, present and future considerations. Sci. Total. Environ. 2020, 706, 135694. [Google Scholar] [CrossRef]
- Chatterjee, S.; Sharma, S. Microplastics in Our Oceans and Marine Health. Field Actions Sci. Reports. J. Field Actions 2019, 19, 54–61. [Google Scholar]
- Tamma, P. Tires Tread on the Environment. Available online: https://www.politico.eu/article/tires-plastic-circular-economy-waste-ocean-pollution/ (accessed on 12 March 2020).
- Sieber, R.; Kawecki, D.; Nowack, B. Dynamic probabilistic material flow analysis of rubber release from tires into the environment. Environ. Pollut. 2020, 258, 113573. [Google Scholar] [CrossRef]
- TheStar. Car Tyres a Likely Source of Microplastics in Coastal Waters. Available online: https://www.thestar.com.my/lifestyle/living/2020/02/20/car-tyres-a-likely-source-of-microplastics-in-coastal-waters (accessed on 12 March 2020).
- Wiess, K.R. The Pileup of Plastic Debris Is More than Ugly Ocean Litter. Available online: https://knowablemagazine.org/article/sustainability/2017/pileup-plastic-debris-more-ugly-ocean-litter (accessed on 12 March 2020).
- UEG. UEG Week: Microplastics Discovered in Human Stools across the Globe in First Study of Its Kind. Gastroenterology, United European. Available online: https://ueg.eu/a/39 (accessed on 30 April 2020).
- Serumgard, J. Internalization of Scrap Tire Management Costs: A Review of the North American Experience. In Rubber and the Environment, Proceedings of the Joint Workshop of the Secretariat of the United Nations Conference on Trade and Development and the International Rubber Study Group on Rubber and the Environment, Bali, Indonesia, 30 October 1998; United Nations Conference on Trade and Development: Geneva, Switzerland.
- Montazer, Z.; Najafi, M.B.H.; Levin, D.B. Challenges with Verifying Microbial Degradation of Polyethylene. Polymers 2020, 12, 123. [Google Scholar] [CrossRef]
- Joutey, N.T.; Bahafid, W. Biodegradation: Involved Microorganisms and Genetically Engineered Microorganisms. Biodegrad. Life Sci. 2013, 289–320. [Google Scholar] [CrossRef]
- Taysum, D.H. In Microbiological Deterioration in the Tropics. In Monograh No. 23; Society of Chemical Industry: London, UK, 1966. [Google Scholar]
- Kasai, D. Poly(cis-1,4-isoprene)-cleavage enzymes from natural rubber-utilizing bacteria. Biosci. Biotechnol. Biochem. 2020, 84, 1089–1097. [Google Scholar] [CrossRef]
- Tsuchii, A.; Suzuki, T.; Takeda, K. Microbial Degradation of Natural Rubber Vulcanizates. Appl. Environ. Microbiol. 1985, 50, 965–970. [Google Scholar] [CrossRef]
- Luo, Q.; Hiessl, S.; Poehlein, A.; Daniel, R.; Steinbüchel, A. Insights into the Microbial Degradation of Rubber and Gutta-Percha by Analysis of the Complete Genome of Nocardia nova SH22a. Appl. Environ. Microbiol. 2014, 80, 3895–3907. [Google Scholar] [CrossRef]
- Gibu, N.; Arata, T.; Kuboki, S.; Linh, D.V.; Fukuda, M.; Steinbüchel, A.; Kasai, D. Characterization of the genes responsible for rubber degradation in Actinoplanes sp. strain OR16. Appl. Microbiol. Biotechnol. 2020, 104, 7367–7376. [Google Scholar] [CrossRef]
- Röther, W.; Birke, J.; Grond, S.; Beltran, J.M.; Jendrossek, D. Production of functionalized oligo-isoprenoids by enzymatic cleavage of rubber. Microb. Biotechnol. 2017, 10, 1426–1433. [Google Scholar] [CrossRef]
- Watcharakul, S.; Röther, W.; Birke, J.; Umsakul, K.; Hodgson, B.; Jendrossek, D. Biochemical and spectroscopic characterization of purified Latex Clearing Protein (Lcp) from newly isolated rubber degrading Rhodococcus rhodochrous strain RPK1 reveals novel properties of Lcp. BMC Microbiol. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Jendrossek, D.; Tomasi, G.; Kroppenstedt, R.M. Bacterial Degradation of Natural Rubber: A Privilege of Actinomycetes? FEMS Microbiol. Lett. 1997, 150, 179–188. [Google Scholar] [CrossRef]
- Tsuchii, A.; Takeda, K. Rubber-Degrading Enzyme from a Bacterial Culture. Appl. Environ. Microbiol. 1990, 56, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Linos, A.; Berekaa, M.M.; Reichelt, R.; Keller, U.; Schmitt, J.; Flemming, H.-C.; Kroppenstedt, R.M.; Steinbüchel, A. Biodegradation of cis-1,4-Polyisoprene Rubbers by Distinct Actinomycetes: Microbial Strategies and Detailed Surface Analysis. Appl. Environ. Microbiol. 2000, 66, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Arenskötter, M.; Bröker, D.; Steinbüchel, A. Biology of the Metabolically Diverse Genus Gordonia. Appl. Environ. Microbiol. 2004, 70, 3195–3204. [Google Scholar] [CrossRef] [PubMed]
- Hiessl, S.; Böse, D.; Oetermann, S.; Eggers, J.; Pietruszka, J.; Steinbüchel, A. Latex Clearing Protein—An Oxygenase Cleaving Poly(cis-1,4-Isoprene) Rubber at the cis Double Bonds. Appl. Environ. Microbiol. 2014, 80, 5231–5240. [Google Scholar] [CrossRef]
- Watcharakul, S. Isolation of a Novel Rubber Degrading Bacterium from a Consortium and Characterization of Its lcp Gene Products Sirimaporn Watcharakul A Thesis Submitted in Fulfillment of the Requirements for the Doctor of Philosophy in Microbiology. Ph.D. Thesis, Prince of Songkla University, Songkhla, Thailand, 2017. [Google Scholar]
- Altenhoff, A.-L.; de Witt, J.; Andler, R.; Steinbüchel, A. Impact of additives of commercial rubber compounds on the microbial and enzymatic degradation of poly(cis-1,4-isoprene). Biodegradation 2019, 30, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Yikmis, M.; Arenskӧtter, M.; Rose, K.; Lange, N.; Wernsmann, H.; Wiefel, L.; Steinbüchel, A. Secretion and Transcriptional Regulation of the Latex-Clearing Protein, Lcp, by the Rubber-Degrading Bacterium Streptomyces sp. Strain K30. Appl. Environ. Microbiol. 2008, 74, 5373–5382. [Google Scholar] [CrossRef][Green Version]
- Yikmis, M.; Steinbüchel, A. Importance of the Latex-clearing protein (Lcp) for Poly(cis-1,4-isoprene) Rubber Cleavage in Streptomyces sp. K30. Microbiol. Open 2012, 1, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Brӧker, D.; Arenskӧtter, M.; Legatzki, A.; Nies, D.H.; Steinbüchel, A. Characterization of the 101-Kilobase-Pair Megaplasmid pKB1, Isolated from the Rubber-Degrading Bacterium Gordonia westfalica Kb1. J. Bacteriol. 2004, 186, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Hiessl, S.; Schuldes, J.; Thürmer, A.; Halbsguth, T.; Bröker, D.; Angelov, A.; Liebl, W.; Daniel, R.; Steinbüchel, A. Involvement of Two Latex-Clearing Proteins during Rubber Degradation and Insights into the Subsequent Degradation Pathway Revealed by the Genome Sequence of Gordonia polyisoprenivorans Strain VH2. Appl. Environ. Microbiol. 2012, 78, 2874–2887. [Google Scholar] [CrossRef]
- Linh, D.V.; Huong, N.L.; Tabata, M.; Imai, S.; Iijima, S.; Kasai, D.; Anh, T.K.; Fukuda, M. Characterization and functional expression of a rubber degradation gene of a Nocardia degrader from a rubber-processing factory. J. Biosci. Bioeng. 2017, 123, 412–418. [Google Scholar] [CrossRef]
- Brӧker, D.; Dietz, D.; Arenskӧtter, M.; Steinbüchel, A. The Genomes of the Non-Clearing-Zone-Forming and Natural-Rubber- Degrading Species Gordonia polyisoprenivorans and Gordonia westfalica Harbor Genes Expressing Lcp Activity in Streptomyces Strains. Appl. Environ. Microbiol. 2008, 74, 2288–2297. [Google Scholar] [CrossRef]
- Birke, J.; Jendrossek, D. Solimonas fluminis has an active latex-clearing protein. Appl. Microbiol. Biotechnol. 2019, 103, 8229–8239. [Google Scholar] [CrossRef] [PubMed]
- Nawong, C.; Umsakul, K.; Sermwittayawong, N. Rubber gloves biodegradation by a consortium, mixed culture and pure culture isolated from soil samples. Braz. J. Microbiol. 2018, 49, 481–488. [Google Scholar] [CrossRef]
- Jendrossek, D.; Reinhardt, S. Sequence Analysis of a Gene Product Synthesized by Xanthomonas sp. during Growth on Natural Rubber Latex. FEMS Microbiol. Lett. 2003, 224, 61–65. [Google Scholar] [CrossRef]
- Sharma, V.; Siedenburg, G.; Birke, J.; Mobeen, F.; Jendrossek, D.; Prakash, T. Metabolic and Taxonomic Insights into the Gram-negative Natural Rubber Degrading Bacterium Steroidobacter cummioxidans sp. nov., strain 35Y. PLoS ONE 2018, 13, 1–20. [Google Scholar]
- Birke, J.; Röther, W.; Jendrossek, D. Rhizobacter gummiphilus NS21 has two rubber oxygenases (RoxA and RoxB) acting synergistically in rubber utilisation. Appl. Microbiol. Biotechnol. 2018, 102, 10245–10257. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Zeeck, A.; Pluckhahn, K.; Jendrossek, D. Physiological and Chemical Investigations into Microbial Degradation of Synthetic Poly(cis-1,4-isoprene). Appl. Environ. Microbiol. 2000, 66, 3722–3726. [Google Scholar] [CrossRef]
- Chengalroyen, M.D.; Dabbs, E.R. The Biodegradation of Latex Rubber: A Mini review. J. Polym. Environ. 2013, 21, 874–880. [Google Scholar] [CrossRef]
- Imai, S.; Ichikawa, K.; Kasai, D.; Masai, E.; Fukuda, M. Isolation and characterization of rubber-degrading bacteria. J. Biotechnol. 2010, 150, 237. [Google Scholar] [CrossRef]
- Kanwal, N.; Shah, A.A.; Qayyum, S.; Hasan, F. Optimization of pH and temperature for degradation of tyre rubber by Bacillus sp. strain S10 isolated from sewage sludge. Int. Biodeterior. Biodegrad. 2015, 103, 154–160. [Google Scholar] [CrossRef]
- Krishnaswamy, V.; Ahongsangbam, N.A. Study on Mineralisation of Poly(cis-1,4-isoprene) and Synthetic Rubber Gloves (SRG) by the Bacterial Consortium. Ann. Appl. Microbiol. Biotechnol. J. 2017, 1, 1–10. [Google Scholar] [CrossRef]
- Muralidharan, M.; Krishnaswamy, V. Artificial rubber mineralization by co-cultured bacterial strains. Int. J. Biol. Res. 2016, 4, 105. [Google Scholar] [CrossRef]
- Birke, J.; Röther, W.; Schmitt, G.; Jendrossek, D. Functional Identification of Rubber Oxygenase (RoxA) in Soil and Marine Myxobacteria. Appl. Environ. Microbiol. 2013, 79, 6391–6399. [Google Scholar] [CrossRef]
- Enoki, M.; Doi, Y.; Iwata, T. Oxidative Degradation ofcis-andtrans-1,4-Polyisoprenes and Vulcanized Natural Rubber with Enzyme-Mediator Systems. Biomacromolecules 2003, 4, 314–320. [Google Scholar] [CrossRef]
- Rose, K.; Steinbüchel, A. Biodegradation of Natural Rubber and Related Compounds: Recent Insights into a Hardly Understood Catabolic Capability of Microorganisms. Appl. Environ. Microbiol. 2005, 71, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Oxygenases without requirement for cofactors or metal ions. Appl. Microbiol. Biotechnol. 2002, 60, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, N.H.; Ismail, M.A.; Abdel-Mageed, W.M.; Shoreit, A.A. Biodegradation of Natural Rubber Latex of Calotropis procera by Two Endophytic Fungal Species. J. Bioremediation Biodegrad. 2017, 8. [Google Scholar] [CrossRef]
- Oghenekaro, A.O.; Kovalchuk, A.; Raffaello, T.; Camarero, S.; Gressler, M.; Henrissat, B.; Lee, J.; Liu, M.; Martínez, A.T.; Miettinen, O.; et al. Genome sequencing of Rigidoporus microporus provides insights on genes important for wood decay, latex tolerance and interspecific fungal interactions. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Röther, W.; Austen, S.; Birke, J.; Jendrossek, D. Molecular Insights in the Cleavage of Rubber by the Latex Clearing Protein (Lcp) of Streptomyces sp. Strain K30: Molecular Insights. Appl. Environ. Microbiol. 2016, 82, 6593–6602. [Google Scholar] [CrossRef]
- Oetermann, S.; Vivod, R.; Hiessl, S.; Hogeback, J.; Holtkamp, M.; Karst, U.; Steinbüchel, A. Histidine at Position 195 is Essential for Association of Heme-b in Lcp1VH2. Earth Syst. Environ. 2018, 2, 5–14. [Google Scholar] [CrossRef]
- Bode, H.B.; Kerkhoff, K.; Jendrossek, D. Bacterial Degradation of Natural and Synthetic Rubber. Biomacromolecules 2001, 2, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Vivod, R.; Oetermann, S.; Hiessl, S.; Gutsche, S.; Remmers, N.; Meinert, C.; Riedel, K.; Steinbüchel, A.; Voigt, B. Oligo(cis-1,4-isoprene) aldehyde-oxidizing dehydrogenases of the rubber-degrading bacterium Gordonia polyisoprenivorans VH2. Appl. Microbiol. Biotechnol. 2017, 101, 7945–7960. [Google Scholar] [CrossRef] [PubMed]
- Birke, J.; Röther, W.; Jendrossek, D. Latex clearing protein (Lcp) of Streptomyces sp. strain K30 is a b-type Cytochrome and Differs from Rubber oxygenase A (RoxA) in its Biophysical Properties. Appl. Environ. Microbiol. 2015, 81, 3793–3799. [Google Scholar] [CrossRef]
- Lee, P.A.; Tullman-Ercek, D.; Georgiou, G. The Bacterial Twin-Arginine Translocation Pathway. Annu. Rev. Microbiol. 2006, 60, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Coenen, A.; Oetermann, S.; Steinbüchel, A. Identification of LcpRBA3(2), a novel regulator of lcp expression in Streptomyces coelicolor A3(2). Appl. Microbiol. Biotechnol. 2019, 103, 5715–5726. [Google Scholar] [CrossRef]
- De Witt, J.; Oetermann, S.; Parise, M.; Parise, D.; Baumbach, J.; Steinbüchel, A. Global regulator of rubber degradation in Gordonia polyisoprenivorans VH2: Identification and involvement in the regulation network. Appl. Environ. Microbiol. 2020, 86, 1–45. [Google Scholar] [CrossRef]
- Vivod, R.; Andler, R.; Oetermann, S.; Altenhoff, A.-L.; Seipel, N.; Holtkamp, M.; Hogeback, J.; Karst, U.; Steinbüchel, A. Characterization of the latex clearing protein of the poly(cis-1,4-isoprene) and poly(trans-1,4-isoprene) degrading bacterium Nocardia nova SH22a. J. Gen. Appl. Microbiol. 2019, 65, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E.M.A.; Arenskötter, M.; Luftmann, H.; Steinbüchel, A. Identification of Poly(cis-1,4-Isoprene) Degradation Intermediates during Growth of Moderately Thermophilic Actinomycetes on Rubber and Cloning of a Functional Lcp Homologue from Nocardia farcinica Strain E1 Downloaded from. Appl. Environ. Microbiol. 2006, 72, 3375–3382. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Andler, R.; Steinbüchel, A. A simple, rapid and cost-effective process for production of latex clearing protein to produce oligopolyisoprene molecules. J. Biotechnol. 2017, 241, 184–192. [Google Scholar] [CrossRef]
- Korz, D.; Rinas, U.; Hellmuth, K.; Sanders, E.; Deckwer, W.-D. Simple fed-batch technique for high cell density cultivation of Escherichia coli. J. Biotechnol. 1995, 39, 59–65. [Google Scholar] [CrossRef]
- Birke, J.; Hambsch, N.; Schmitt, G.; Altenbuchner, J.; Jendrossek, D. Phe317 is Essential for Rubber Oxygenase RoxA Activity. Appl. Environ. Microbiol. 2012, 78, 7876–7883. [Google Scholar] [CrossRef]
- Hambsch, N.; Schmitt, G.; Jendrossek, D. Development of a homologous expression system for rubber oxygenase RoxA from Xanthomonas sp. J. Appl. Microbiol. 2010, 109, 1067–1075. [Google Scholar] [CrossRef]
- Goodfellow, M.; Kämpfer, P.; Busse, H.J.; Trujillo, M.E.; Suzuki, K.I.; Ludwig, W.B.; Whitman, W. Bergey’s Manual of Systematic Bacteriology: Actinobacteria, 2nd ed.; Springer: New York, NY, USA, 2012. [Google Scholar]
- Atlas, R.M. Principles of Microbiology; WCB McGraw-Hill: Boston, MA, USA, 1997. [Google Scholar]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and Ecology of Actinobacteria and their Bioenergy Applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef]
- Tischler, D.; van Berkel, W.J.H.; Fraaije, M.W. Editorial: Actinobacteria, A Source of Biocatalytic Tools. Front. Microbiol. 2019, 10, 1–4. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Goodfellow, M.; Williams, S.T. Ecology of Actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- Stach, E.M.; Bull, A.T. Estimating and comparing the Diversity of Marine Actinobacteria. Antonie Leeuwenhoek 2005, 87, 3–9. [Google Scholar] [CrossRef]
- Ananda, R.; Dharumadurai, D.; Manogaran, G.P. An Introduction to Actinobacteria. In Basics and Biotechnological Applications; InTechopen: London, UK, 2016; pp. 3–38. Available online: http://www.intechopen.com/books/actinobacteria-basics-and-biotechnological-applications/an-introduction-to-actinobacteria (accessed on 17 July 2020). [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Brunecky, R.; Chung, D.; Sarai, N.S.; Hengge, N.; Russell, J.F.; Young, J.; Mittal, A.; Pason, P.; Wall, T.V.; Michener, W.; et al. High activity CAZyme cassette for improving biomass degradation in thermophiles. Biotechnol. Biofuels 2018, 11, 1–12. [Google Scholar] [CrossRef]
- Kirby, R. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol. Lett. 2011, 319, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bohlin, J.; Eldholm, V.; Pettersson, J.H.O.; Brynildsrud, O.; Snipen, L. The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC Genom. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Goshi, K.; Uchida, T.; Lezhava, A.; Yamasaki, M.; Hiratsu, K.; Shinkawa, H.; Kinashi, H. Cloning and Analysis of the Telomere and Terminal Inverted Repeat of the Linear Chromosome of Streptomyces griseus. J. Bacteriol. 2002, 184, 3411–3415. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Ōmura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef]
- Bentley, S.D.; Brosch, R.; Gordon, S.; Hopwood, D.; Cole, S. Genomics of Actinobacteria, the High G+C Gram-positive Bacteria. In Microbial Genomics; Fraser, C.M., Read, T., Nelson, K.E., Eds.; Humana Press: Totowa, NJ, USA, 2004; pp. 333–360. [Google Scholar]
- Lin, Y.-S.; Kieser, H.M.; Hopwood, D.A.; Chen, C.W. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 1993, 10, 923–933. [Google Scholar] [CrossRef]
- Reeves, A.R.; Post, D.A.; Boom, T.J.V. Physical-genetic map of the erythromycin-producing organism Saccharopolyspora erythraea. Microbiology 1998, 144, 2151–2159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Redenbach, M.; Scheel, J.; Schmidt, U. Chromosome topology and genome size of selected actinomycetes species. Antonie van Leeuwenhoek 2000, 78, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-W.; Zhi, X.-Y.; Yao, J.-C.; Zhou, Y.; Tang, S.-K.; Klenk, H.-P.; Zhao, J.; Li, W.-J. Comparative Genomic Analysis of the Genus Nocardiopsis Provides New Insights into Its Genetic Mechanisms of Environmental Adaptability. PLoS ONE 2013, 8, e61528. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Soil to Genomics: The Streptomyces Chromosome. Annu. Rev. Genet. 2006, 40, 1–23. [Google Scholar] [CrossRef]
- Chen, W.; He, F.; Zhang, X.; Chen, Z.; Wen, Y.; Li, J. Chromosomal instability in Streptomyces avermitilis: Major deletion in the central region and stable circularized chromosome. BMC Microbiol. 2010, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Al Doghaither, H.; Gull, M. Plasmids as Genetic Tools and Their Applications in Ecology and Evolution. Plasmid 2019, 32, 137–144. [Google Scholar] [CrossRef]
- Cornell, C.R.; Marasini, D.; Fakhr, M.K. Molecular Characterization of Plasmids Harbored by Actinomycetes Isolated from the Great Salt Plains of Oklahoma Using PFGE and Next Generation Whole Genome Sequencing. Front. Microbiol. 2018, 9, 2282. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Euzéby, J.P. List of Bacterial Names with Standing in Nomenclature: A Folder Available on the Internet. Int. J. Syst. Bacteriol. 1997, 47, 590–592. [Google Scholar] [CrossRef]
- Kim, D.; Choi, K.Y.; Yoo, M.; Zylstra, G.J.; Kim, E. Biotechnological Potential of Rhodococcus Biodegradative Pathways. J. Microbiol. Biotechnol. 2018, 28, 1037–1051. [Google Scholar] [CrossRef]
- Shivlata, L.; Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1–29. [Google Scholar] [CrossRef]
- Delegan, Y.A.; Vetrova, A.A.; Akimov, V.N.; Titok, M.A.; Filonov, A.E.; Boronin, A.M. Thermotolerant Oil-degrading Bacteria Isolated from Soil and Water of Geographically Distant Regions. Prikl. Biokhim. Mikrobiol. 2016, 54, 383–391. [Google Scholar] [CrossRef]
- Gallert, C. Degradation of Latex and of Natural Rubber by Streptomyces Strain La 7. Syst. Appl. Microbiol. 2000, 23, 433–441. [Google Scholar] [CrossRef]
- Berekaa, M.M.; Linos, A.; Reichelt, R.; Keller, U.; Steinbüchel, A. Effect of pretreatment of rubber material on its biodegradability by various rubber degrading bacteria. FEMS Microbiol. Lett. 2000, 184, 199–206. [Google Scholar] [CrossRef]
- Tsuchii, A.; Tokiwa, Y. Microbial degradation of tyre rubber particles. Biotechnol. Lett. 2001, 23, 963–969. [Google Scholar] [CrossRef]
- Heisey, R.M.; Papadatos, S. Isolation of Microorganisms able to Metabolize Purified Natural Rubber. Appl. Environ. Microbiol. 1995, 61, 3092–3097. [Google Scholar] [CrossRef] [PubMed]
- Watcharakul, S.; Umsakul, K.; Hodgson, B.; Chumeka, W.; Tanrattanakul, V. Biodegradation of a Blended Starch/natural Rubber Foam Biopolymer and Rubber Gloves by Streptomyces coelicolor CH13. Electron. J. Biotechnol. 2012, 15, 1–14. [Google Scholar]
- Onyeagoro, G.N.; Ohaeri, E.G.; Timothy, U.J. Studies on Microbial Degradation of Natural Rubber Using Dilute Solution Viscosity Measurement and Weight Loss Techniques. Int. J. Basic Appl. Sci. 2012, 1, 434–446. [Google Scholar] [CrossRef]
- Adzami, N.S.; Tajarudin, H.A. Biodegradation of Natural Rubber Latex Film added with Metroxylan sagu Pith form by Bacillus cereus ATCC 14579. Malays. J. Microbiol. 2018, 14, 101–107. [Google Scholar] [CrossRef]
- Rashid, A.A.; Misman, M.A.; Ahmad, H.I. Mechanical and Biodegradation Properties of Sago Starch Natural Rubber Latex Composites. In Proceedings of the 7th Asian-Australasian Conference on Composite Materials, Taibei, Taiwan, 15–18 November 2010. [Google Scholar]
- Vudjung, C.; Chaisuwan, U.; Pangan, U.; Chaiougdee, N.; Boonyod, S.; Santawitee, O.; Saengsuwan, S. Effect of Natural Rubber Contents on Biodegradation and Water Absorption of Interpenetrating Polymer Network (IPN) Hydrogel from Natural Rubber and Cassava Starch. Energy Procedia 2014, 56, 255–263. [Google Scholar] [CrossRef]
- Aboelkheir, M.G.; Bedor, P.B.; Leite, S.G.; Pal, K.; Toledo Filho, R.D.; Gomes de Souza, F. Biodegradation of Vulcanized SBR: A Comparison between Bacillus subtilis, Pseudomonas aeruginosa and Streptomyces sp. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhao, S.; Li, C.; Wang, B.; Fu, Y.; Wang, Y. Biodesulfurization of Vulcanized Rubber by Enzymes induced from Gordonia amicalisa. Polym. Degrad. Stab. 2016, 128, 8–14. [Google Scholar] [CrossRef]
- Ghavipanjeh, F.; Ziaei Rad, Z.; Pazouki, M. Devulcanization of Ground Tires by Different Strains of Bacteria: Optimization of Culture Condition by Taguchi Method. J. Polym. Environ. 2018, 26, 3168–3175. [Google Scholar] [CrossRef]
- Christofi, N.; Geoffrey, J.; Edward, D. WO/2004/076492 Rubber Treatment Method (2010). Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2004076492 (accessed on 4 June 2021).
- Aielo, P.B.; Borges, F.A.; Romeira, K.M.; Miranda, M.C.R.; Arruda, L.B.D.; Filho, P.N.L.; Drago, B.D.C.; Herculano, R.D. Evaluation of sodium diclofenac release using natural rubber latex as carrier. Mater. Res. 2014, 17, 146–152. [Google Scholar] [CrossRef]
- Horikx, M. Chain Scissions in a Polymer Network. J. Polym. Sci. 1956, 19, 445–454. [Google Scholar] [CrossRef]
- Zeb, S. Vulcanized Rubber Degrading Paecilomyces variotii Strain SFA-25 Isolated from Hot Spring in Pakistan. Ph.D. Thesis, Quaid-i-Azam University, Islamabad, Pakistan, 2009. [Google Scholar]
- Tribedi, P.; Goswami, M.; Chakraborty, P.; Mukherjee, K.; Mitra, G.; Dey, S. Bioaugmentation and Biostimulation: A Potential Strategy for Environmental Remediation. J. Microbiol. Exp. 2018, 6, 223–231. [Google Scholar]
- Mrozik, A.; Piotrowska-Seget, Z. Bioaugmentation as a Strategy for Cleaning Up of Soils Contaminated with Aromatic Compounds. Microbiol. Res. 2010, 165, 363–375. [Google Scholar] [CrossRef]
- Nzila, A.; Razzak, S.A.; Zhu, J. Bioaugmentation: An Emerging Strategy of Industrial Wastewater Treatment for Reuse and Discharge. Int. J. Environ. Res. Public Health 2016, 13, 846. [Google Scholar] [CrossRef]
- Adams, G.O.; Fufeyin, P.T.; Okoro, S.E.; Ehinomen, I. Bioremediation, Biostimulation and Bioaugmention: A Review. Int. J. Environ. Bioremediation Biodegrad. 2015, 3, 28–39. [Google Scholar] [CrossRef]
- Akkharabunditsakul, V.; Tanrattanakul, V.; Umsakul, K. Biodegradation of Synthetic Rubbers by a Mixed Culture Isolated from Various Rubber Factory Soils in Songkhla, Thailand. Int. J. Adv. Agric. Environ. Eng. 2017, 4, 1–5. [Google Scholar]
- Rahman, M.F.A.; Rusli, A.; Adzami, N.S.; Azura, A.R. Studies on the Influence of Mixed Culture from Buried Soil Sample for Biodegradation of Sago Starch Filled Natural Rubber Latex Gloves. IOP Conf. Ser. Mater. Sci. Eng. 2019, 548, 012018. [Google Scholar] [CrossRef]
- Andler, R.; Vivod, R.; Steinbüchel, A. Synthesis of Polyhydroxyalkanoates through the Biodegradation of Poly(cis-1,4-isoprene) Rubber. J. Biosci. Bioeng. 2019, 127, 360–365. [Google Scholar] [CrossRef]
- Yao, C.; Zhao, S.; Wang, Y.; Wang, B.; Wei, M.; Hu, M. Microbial Desulfurization of Waste Latex Rubber with Alicyclobacillus sp. Polym. Degrad. Stab. 2013, 98, 1724–1730. [Google Scholar] [CrossRef]
- Stevenson, K.; Stallwood, B.; Hart, A.G. Tire Rubber Recycling and Bioremediation: A Review. Bioremediat. J. 2008, 12, 1–11. [Google Scholar] [CrossRef]
- Bhanjadeo, M.M.; Rath, K.; Gupta, D.; Pradhan, N.; Biswal, S.K.; Mishra, B.K.; Subudhi, U. Differential Desulfurization of Dibenzothiophene by Newly Identified MTCC Strains: Influence of Operon Array. PLoS ONE 2018, 13, e0192536. [Google Scholar] [CrossRef] [PubMed]
- Karimi, E.; Jeffreys, C.; Yazdian, F.; Sepahi, A.A.; Hatamian, A.; Rasekh, B.; Omidi, M.; Ebrahim-Habibi, M.-B.; Asharafi, S.J. DBT Desulfurization by Decorating Rhodococcus erythropolis IGTS8 using Magnetic Fe3O4 Nanoparticles in a Bioreactor. Life Sci. 2017, 17, 528–535. [Google Scholar]
- Crow, J.M. Inside the Synthetic Biology Revolution. The Science of Everything COSMOS. 2018. Available online: https://cosmosmagazine.com/biology/life-2-0-inside-the-synthetic-biology-revolution/ (accessed on 17 July 2020).
- El Karoui, M.; Hoyos-Flight, M.; Fletcher, L. Future Trends in Synthetic Biology-A Report. Front. Bioeng. Biotechnol. 2019, 1, 175. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chang, A.Y.; Novosad, V.; Chupin, V.V.; Schaller, R.D.; Rozhkova, E.A. Cell-Free Synthetic Biology Chassis for Nanocatalytic Photon-to-Hydrogen Conversion. ACS Nano 2017, 11, 6739–6745. [Google Scholar] [CrossRef] [PubMed]
- Karzbrun, E.; Tayar, A.M.; Noireaux, V.; Bar-Ziv, R.H. Programmable On-chip DNA Compartments as Artificial Cells. Science 2014, 80, 829–832. [Google Scholar] [CrossRef] [PubMed]
- McMillan, F.; Ilsley, J.; Adam, B.; Heinemann, J. The Littlest Factories. The University of Queensland. 2019. Available online: https://aibn.uq.edu.au/littlest-factories (accessed on 17 July 2020).
- McMillan, F.; Ilsley, J.; Adam, B.; Heinemann, J. Making Tyres from Microbes and Plants. The University of Queensland. 2019. Available online: https://aibn.uq.edu.au/making-tyres-microbes-and-plants (accessed on 28 May 2021).
- Achal, V.; Fayolle, F.; Shin, S.G.; Ercole, C.; Matteucci, F.; Del Gallo, M. A Study of Chlorinated Solvent Contamination of the Aquifers of an Industrial Area in Central Italy: A Possibility of Bioremediation. Front. Microbiol. 2015, 6, 924. [Google Scholar] [CrossRef]
- ACS. Reducing Tire Waste by Using Completely Degradable, Synthetic Rubber. American Chemical Society. Available online: https://www.acs.org/content/acs/en/pressroom/newsreleases/2016/august/reducing-tire-waste-by-using-completely-degradable-synthetic-rubber.html (accessed on 4 June 2021).
- Owana, N. Michelin Takes Wraps Off Connected Tire Concept, 3-D Printing, Bio-Sourced Materials in the Mix. Available online: https://techxplore.com/news/2017-06-michelin-concept-d-bio-sourced-materials.html (accessed on 4 June 2021).
- Harper, J. Goodyear’s Biodegradable Concept Tire Regenerates Its Tread. Available online: https://techxplore.com/news/2020-03-goodyear-biodegradable-concept-regenerates.html (accessed on 4 June 2021).
| Lcp | RoxB | RoxA | |
|---|---|---|---|
| First Identified From | Streptomyces sp. strain K30 | Steroidobacter cummioxidans 35Y (Xanthomonas sp. 35Y) | |
| Accession no. | AY387589 | KY498024 | KC980911 |
| Metabolite Products |
Tetra C20, oligoisoprene/ aldehyde and ketone terminal | ODTDs | |
| Bacteria | Mostly Gram-positive | Gram-negative only | |
| Gene Length (bp) | ~1224–1227 | ~2037–2046 | |
| Size (kDa) | 43 | 73–74 | |
| Pathway | TAT | SEC | |
| Co-factor | b-type heme | c-type heme | |
| Mechanism to Cleavage Isoprene | Endo-type | Endo and Exo-type | |
| Major Metal Atoms in Protein Molecule | 1 Fe | 2 Fe | |
| Oxidation State of Iron | Fe3+ | Fe3+----O2 | |
| Family | Genus | Genus | Species | Lcp Genes | Species Diversity |
|---|---|---|---|---|---|
| Actinosynnemataceae | 1 | Actinosynnema | 2 | 3 | 2 |
| Corynebacterineae | 1 | Williamsia | 3 | 6 | 2 |
| Frankiaceae | 1 | Frankia | 3 | 7 | 2 |
| Gordoniaceae | 1 | Gordonia | 12 | 23 | 2 |
| Microbacteriaceae | 1 | Microbacterium | 3 | 3 | 1 |
| Micrococcaceae | 1 | Psychromicrobium | 1 | 1 | 1 |
| Micromonosporaceae | 2 | Actinoplanes | 2 | 4 | 2 |
| Micromonospora | 4 | 7 | 2 | ||
| Mycobacteriaceae | 3 | Mycobacterium | 3 | 6 | 2 |
| Mycobacteroides | 5 | 43 | 9 | ||
| Mycolicibacterium | 5 | 5 | 1 | ||
| Nocardiaceae | 2 | Nocardia | 12 | 29 | 2 |
| Rhodococcus | 9 | 15 | 2 | ||
| Nocardioidaceae | 2 | Mumia | 1 | 3 | 3 |
| Nocardioides | 4 | 4 | 1 | ||
| Nocardiopsaceae | 2 | Streptomonospora | 1 | 2 | 2 |
| Nocardiopsis | 1 | 1 | 1 | ||
| Pseudonocardiaceae | 7 | Actinoalloteichus | 1 | 1 | 1 |
| Amycolatopsis | 11 | 16 | 1 | ||
| Kibdelosporangium | 1 | 2 | 2 | ||
| Kutzneria | 1 | 1 | 1 | ||
| Prauserella | 1 | 2 | 2 | ||
| Pseudonocardia | 1 | 1 | 1 | ||
| Saccharomonospora | 1 | 1 | 1 | ||
| Streptomycetaceae | 1 | Streptomyces | 39 | 51 | 1 |
| Streptosporangiaceae | 2 | Nonomuraea | 1 | 1 | 1 |
| Streptosporangium | 1 | 1 | 1 | ||
| Thermomonosporaceae | 1 | Actinomadura | 2 | 2 | 1 |
| Tsukamurellaceae | 1 | Tsukamurella | 3 | 7 | 2 |
| TOTAL | 29 | 134 | 248 |
| Culture | Duration (Weeks) | Weight Loss (%) | Material | Pre-Treatment | Reference |
|---|---|---|---|---|---|
| Nocardia sp. strain 835A | 8 | 100 | NR | – | [45] |
| 90 | Latex glove | Acetone & CHCl3 | |||
| S. cummioxidans 35Y | 1 | 60 | NR | NS | [51] |
| Amycolatopsis S1A | 6 | 11 | Rubber coated slides (absence of organic nutrients) | Soxhlet purification | [126] |
| Amycolatopsis S1D | 12 | ||||
| Nocardia S3F | 13 | ||||
| Streptomyces S1G | 44 | ||||
| Streptomyces S3D | 21 | ||||
| Streptomyces S4C | 26 | ||||
| Streptomyces S4D | 43 | ||||
| Streptomyces S4E | 37 | ||||
| Streptomyces S4F | 43 | ||||
| Streptomyces S4G | 38 | ||||
| S. sp. strain LA7 | 8.6 | 80 | Emulgated latex | Dialysed latex | [123] |
| 3.3 | 60 | ||||
| 10 | 29.4 | Latex glove | Acetone & CHCl3 | ||
| 10 | 31.3 | Latex glove + Triton X (0.1%, w/w) | Acetone & CHCl3 | ||
| S. coelicolor CH13 | 6 | 14 | Latex glove | – | [127] |
| 92 | Latex glove + starch (65%) | NS | |||
| Nocardia sp. strain 385A | 10 | 5.5 | Fresh latex | NS | [128] |
| Bacillus cereus | 2 | 3.5 | Tyre | NS | [129] |
| 7.8 | NR | Acetone | |||
| 12.3 | Latex film of MR + metroxylan sago pith waste | ||||
| R. pyridinivorans strain F5 | 4 | 9.36 | Latex glove | – | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basik, A.A.; Sanglier, J.-J.; Yeo, C.T.; Sudesh, K. Microbial Degradation of Rubber: Actinobacteria. Polymers 2021, 13, 1989. https://doi.org/10.3390/polym13121989
Basik AA, Sanglier J-J, Yeo CT, Sudesh K. Microbial Degradation of Rubber: Actinobacteria. Polymers. 2021; 13(12):1989. https://doi.org/10.3390/polym13121989
Chicago/Turabian StyleBasik, Ann Anni, Jean-Jacques Sanglier, Chia Tiong Yeo, and Kumar Sudesh. 2021. "Microbial Degradation of Rubber: Actinobacteria" Polymers 13, no. 12: 1989. https://doi.org/10.3390/polym13121989
APA StyleBasik, A. A., Sanglier, J.-J., Yeo, C. T., & Sudesh, K. (2021). Microbial Degradation of Rubber: Actinobacteria. Polymers, 13(12), 1989. https://doi.org/10.3390/polym13121989






