Studies of Cellulose and Starch Utilization and the Regulatory Mechanisms of Related Enzymes in Fungi
Abstract
1. Introduction
2. The Fungi and Their Potential in the Utilization of Plant Polysaccharides
3. Cellulolytic Enzymes and Their Regulatory Mechanisms in Fungi
3.1. Classification of Cellulolytic Enzymes
3.2. Mechanism of Cellulase Induction
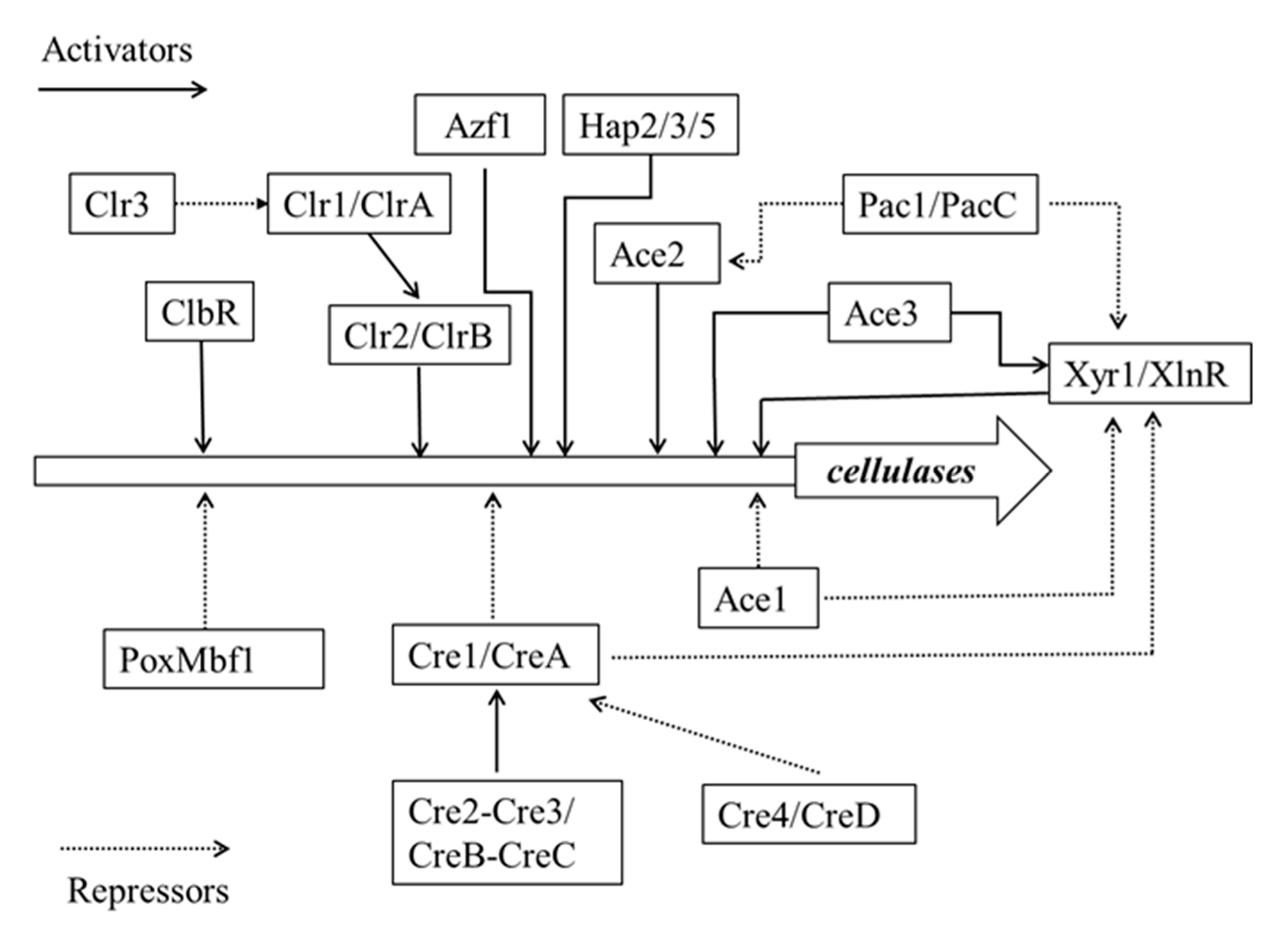
3.3. Molecular Regulation Mechanisms of Cellulase Gene Expression
4. Amylolytic Enzymes and Their Regulatory Mechanisms in Fungi
4.1. Amylolytic Enzymes
4.2. Molecular Regulation Mechanism of Amylase Gene Expression
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gorshkova, T.A.; Kozlova, L.V.; Mikshina, P.V. Spatial structure of plant cell wall polysaccharides and its functional significance. Biochemistry 2013, 78, 836–853. [Google Scholar] [CrossRef] [PubMed]
- Voiniciuc, C.; Pauly, M.; Usadel, B. Monitoring Polysaccharide Dynamics in the Plant Cell Wall. Plant Physiol. 2018, 176, 2590–2600. [Google Scholar] [CrossRef] [PubMed]
- Lovegrove, A.; Edwards, C.H.; De Noni, I.; Patel, H.; El, S.N.; Grassby, T.; Zielke, C.; Ulmius, M.; Nilsson, L.; Butterworth, P.J.; et al. Role of polysaccharides in food, digestion, and health. Crit. Rev. Food Sci. Nutr. 2017, 57, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Meents, M.J.; Watanabe, Y.; Samuels, A.L. The cell biology of secondary cell wall biosynthesis. Ann. Bot. 2018, 121, 1107–1125. [Google Scholar] [CrossRef]
- Chen, F.; Srinivasa Reddy, M.S.; Temple, S.; Jackson, L.; Shadle, G.; Dixon, R.A. Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J. 2006, 48, 113–124. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef]
- Showalter, A.M.; Basu, D. Extensin and Arabinogalactan-Protein Biosynthesis: Glycosyltransferases, Research Challenges, and Biosensors. Front. Plant Sci. 2016, 7, 814. [Google Scholar] [CrossRef]
- Park, S.; Baker, J.O.; Himmel, M.E.; Parilla, P.A.; Johnson, D.K. Cellulose crystallinity index: Measurement techniques and their impact on interpreting cellulase performance. Biotechnol. Biofuels 2010, 3, 10. [Google Scholar] [CrossRef]
- Moreira, L.R.; Filho, E.X. Insights into the mechanism of enzymatic hydrolysis of xylan. Appl. Microbiol. Biotechnol. 2016, 100, 5205–5214. [Google Scholar] [CrossRef]
- Park, J.; Kim, B.; Son, J.; Lee, J.W. Solvo-thermal in situ transesterification of wet spent coffee grounds for the production of biodiesel. Bioresour. Technol. 2018, 249, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.P.; Yan, R.; Chen, H.P.; Lee, D.H.; Zheng, C.G. Characteristics of hemicellulose, cellulose and lignin pyrolysis. Fuel 2007, 86, 1781–1788. [Google Scholar] [CrossRef]
- Kumar, P.; Barrett, D.M.; Delwiche, M.J.; Stroeve, P. Methods for Pretreatment of Lignocellulosic Biomass for Efficient Hydrolysis and Biofuel Production. Ind. Eng. Chem. Res. 2009, 48, 3713–3729. [Google Scholar] [CrossRef]
- Taher, H.; Al-Zuhair, S.; Al-Marzouqi, A.H.; Haik, Y.; Farid, M. Effective extraction of microalgae lipids from wet biomass for biodiesel production. Biomass Bioenergy 2014, 66, 159–167. [Google Scholar] [CrossRef]
- Fatma, S.; Hameed, A.; Noman, M.; Ahmed, T.; Shahid, M.; Tariq, M.; Sohail, I.; Tabassum, R. Lignocellulosic Biomass: A Sustainable Bioenergy Source for the Future. Protein Pept. Lett. 2018, 25, 148–163. [Google Scholar] [CrossRef]
- Lu, Y.; Li, G.S.; Lu, Y.C.; Fan, X.; Wei, X.Y. Analytical Strategies Involved in the Detailed Componential Characterization of Biooil Produced from Lignocellulosic Biomass. Int. J. Anal. Chem. 2017, 2017, 19. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Lee, W.H.; Jin, Y.S. Evaluation of Ethanol Production Activity by Engineered Saccharomyces cerevisiae Fermenting Cellobiose through the Phosphorolytic Pathway in Simultaneous Saccharification and Fermentation of Cellulose. J. Microbiol. Biotechnol. 2017, 27, 1649–1656. [Google Scholar] [CrossRef]
- Xu, H.P.; Li, Y.; Hua, D.L.; Mu, H.; Zhao, Y.X.; Chen, G.Y. Methane production from the anaerobic digestion of substrates from corn stover: Differences between the stem bark, stem pith, and leaves. Sci. Total Environ. 2019, 694, 133641. [Google Scholar] [CrossRef]
- Gaworski, M.; Jablonski, S.; Pawlaczyk-Graja, I.; Ziewiecki, R.; Rutkowski, P.; Wieczynska, A.; Gancarz, R.; Lukaszewicz, M. Enhancing biogas plant production using pig manure and corn silage by adding wheat straw processed with liquid hot water and steam explosion. Biotechnol. Biofuels 2017, 10, 259. [Google Scholar] [CrossRef]
- Chandra, R.; Takeuchi, H.; Hasegawa, T. Methane production from lignocellulosic agricultural crop wastes: A review in context to second generation of biofuel production. Renew. Sust. Energy Rev. 2012, 16, 1462–1476. [Google Scholar] [CrossRef]
- Yang, B.; Wyman, C.E. Pretreatment: The key to unlocking low-cost cellulosic ethanol. Biofuel. Bioprod. Bior. 2008, 2, 26–40. [Google Scholar] [CrossRef]
- Amin, F.R.; Khalid, H.; Zhang, H.; Rahman, S.U.; Zhang, R.; Liu, G.; Chen, C. Pretreatment methods of lignocellulosic biomass for anaerobic digestion. AMB Express 2017, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Taherzadeh, M.J.; Karimi, K. Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: A review. Int. J. Mol. Sci. 2008, 9, 1621–1651. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.J.; Hobbs, P.J.; Holliman, P.J.; Jones, D.L. Optimisation of the anaerobic digestion of agricultural resources. Bioresour. Technol. 2008, 99, 7928–7940. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Jarboe, L.; Brown, R.; Wen, Z. A thermochemical-biochemical hybrid processing of lignocellulosic biomass for producing fuels and chemicals. Biotechnol. Adv. 2015, 33, 1799–1813. [Google Scholar] [CrossRef]
- Xia, Y.; Wang, Y.; Fang, H.H.; Jin, T.; Zhong, H.; Zhang, T. Thermophilic microbial cellulose decomposition and methanogenesis pathways recharacterized by metatranscriptomic and metagenomic analysis. Sci. Rep. 2014, 4, 6708. [Google Scholar] [CrossRef]
- Sarparanta, M.; Pourat, J.; Carnazza, K.E.; Tang, J.; Paknejad, N.; Reiner, T.; Kostiainen, M.A.; Lewis, J.S. Multimodality labeling strategies for the investigation of nanocrystalline cellulose biodistribution in a mouse model of breast cancer. Nucl. Med. Biol. 2019, 80, 1–12. [Google Scholar] [CrossRef]
- Sharma, A.; Thakur, M.; Bhattacharya, M.; Mandal, T.; Goswami, S. Commercial application of cellulose nano-composites-A review. Biotechnol. Rep. 2019, 21, e00316. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Sun, Y.; Zheng, Y.D.; He, W.; Yang, Y.Y.; Xie, Y.J.; Feng, Z.X.; Qiao, K. A biocompatible bacterial cellulose/tannic acid composite with antibacterial and anti-biofilm activities for biomedical applications. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 106, 110249. [Google Scholar] [CrossRef]
- Xun, Z.; Ni, S.; Gao, Z.; Zhang, Y.; Gu, J.; Huo, P. Construction of Polymer Electrolyte Based on Soybean Protein Isolate and Hydroxyethyl Cellulose for a Flexible Solid-State Supercapacitor. Polymers 2019, 11, 1895. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.M.; He, X.X.; Cai, C.; Lan, T.Q.; Pang, Y.X.; Zhou, H.F.; Qiu, X.Q. Enhancement and Mechanism of a Lignin Amphoteric Surfactant on the Production of Cellulosic Ethanol from a High-Solid Corncob Residue. J. Agric. Food Chem. 2019, 67, 6248–6256. [Google Scholar] [CrossRef] [PubMed]
- Nghiem, N.P.; Senske, G.E.; Kim, T.H. Pretreatment of Corn Stover by Low Moisture Anhydrous Ammonia (LMAA) in a Pilot-Scale Reactor and Bioconversion to Fuel Ethanol and Industrial Chemicals. Appl. Biochem. Biotechnol. 2016, 179, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.O.; Bischoff, K.M.; Leathers, T.D.; Anderson, A.M.; Liu, S.; Skory, C.D. Resolving bacterial contamination of fuel ethanol fermentations with beneficial bacteria-An alternative to antibiotic treatment. Bioresour. Technol. 2018, 247, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, I.; Shinohara, Y.; Oguma, T.; Koyama, Y. Survival strategy of the salt-tolerant lactic acid bacterium, Tetragenococcus halophilus, to counteract koji mold, Aspergillus oryzae, in soy sauce brewing. Biosci. Biotechnol. Biochem. 2018, 82, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: Role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef]
- Kim, H.Y.; Park, S.S.; Lim, S.T. Preparation, characterization and utilization of starch nanoparticles. Colloids Surf. B Biointerfaces 2015, 126, 607–620. [Google Scholar] [CrossRef]
- Kitamoto, K. Cell biology of the Koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2015, 79, 863–869. [Google Scholar] [CrossRef]
- Papagianni, M. Advances in citric acid fermentation by Aspergillus niger: Biochemical aspects, membrane transport and modeling. Biotechnol. Adv. 2007, 25, 244–263. [Google Scholar] [CrossRef]
- Kitamoto, K. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 2002, 51, 129–153. [Google Scholar] [CrossRef]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Yamada, O.; Gomi, K. Genomics of Aspergillus oryzae: Learning from the History of Koji Mold and Exploration of Its Future. DNA Res. 2008, 15, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Abarca, M.L.; Accensi, F.; Cano, J.; Cabanes, F.J. Taxonomy and significance of black aspergilli. Antonie Leeuwenhoek Int. J. G 2004, 86, 33–49. [Google Scholar] [CrossRef]
- Jin, F.J.; Watanabe, T.; Juvvadi, P.R.; Maruyama, J.I.; Arioka, M.; Kitamoto, K. Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2007, 76, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Joosten, V.; Gouka, R.J.; van den Hondel, C.A.M.J.J.; Verrips, C.T.; Lokman, B.C. Expression and production of llama variable heavy-chain antibody fragments (V(HH)s) by Aspergillus awamori. Appl. Microbiol. Biotechnol. 2005, 66, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Katayama, T.; Maruyama, J.; Kitamoto, K. Comparative genomic analysis identified a mutation related to enhanced heterologous protein production in the filamentous fungus Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2016, 100, 9163–9174. [Google Scholar] [CrossRef]
- Kim, S.K.; Jo, J.H.; Jin, Y.S.; Seo, J.H. Enhanced ethanol fermentation by engineered Saccharomyces cerevisiae strains with high spermidine contents. Bioprocess Biosyst. Eng. 2017, 40, 683–691. [Google Scholar] [CrossRef]
- Myburgh, M.W.; Cripwell, R.A.; Favaro, L.; van Zyl, W.H. Application of industrial amylolytic yeast strains for the production of bioethanol from broken rice. Bioresour. Technol. 2019, 294, 122222. [Google Scholar] [CrossRef]
- Ruchala, J.; Kurylenko, O.O.; Dmytruk, K.V.; Sibirny, A.A. Construction of advanced producers of first- and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha). J. Ind. Microbiol. Biotechnol. 2019, 47, 109–132. [Google Scholar] [CrossRef]
- Beltran, G.; Torija, M.J.; Novo, M.; Ferrer, N.; Poblet, M.; Guillamon, J.M.; Rozes, N.; Mas, A. Analysis of yeast populations during alcoholic fermentation: A six year follow-up study. Syst. Appl. Microbiol. 2002, 25, 287–293. [Google Scholar] [CrossRef]
- Torija, M.J.; Rozes, N.; Poblet, M.; Guillamon, J.M.; Mas, A. Yeast population dynamics in spontaneous fermentations: Comparison between two different wine-producing areas over a period of three years. Antonie Leeuwenhoek Int. J. G 2001, 79, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Xufre, A.; Albergaria, H.; Inacio, J.; Spencer-Martins, I.; Girio, F. Application of fluorescence in situ hybridisation (FISH) to the analysis of yeast population dynamics in winery and laboratory grape must fermentations. Int. J. Food Microbiol. 2006, 108, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.M. Effect of Sugar Transport Inactivation in Saccharomyces cerevisiae on Sluggish and Stuck Enological Fermentations. Appl. Environ. Microbiol. 1989, 55, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Talamantes, D.; Biabini, N.; Dang, H.; Abdoun, K.; Berlemont, R. Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol Biofuels 2016, 9, 133. [Google Scholar] [CrossRef]
- Mandels, M.; Reese, E.T. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 1957, 73, 269–278. [Google Scholar] [CrossRef]
- Ilmen, M.; Saloheimo, A.; Onnela, M.L.; Penttila, M.E. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 1997, 63, 1298–1306. [Google Scholar] [CrossRef]
- Nogawa, M.; Goto, M.; Okada, H.; Morikawa, Y. L-Sorbose induces cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr. Genet. 2001, 38, 329–334. [Google Scholar] [CrossRef]
- Aro, N.; Saloheimo, A.; Ilmen, M.; Penttila, M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 2001, 276, 24309–24314. [Google Scholar] [CrossRef]
- Hakkinen, M.; Valkonen, M.J.; Westerholm-Parvinen, A.; Aro, N.; Arvas, M.; Vitikainen, M.; Penttila, M.; Saloheimo, M.; Pakula, T.M. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol. Biofuels 2014, 7, 14. [Google Scholar] [CrossRef]
- Shida, Y.; Furukawa, T.; Ogasawara, W. Deciphering the molecular mechanisms behind cellulase production in Trichoderma reesei, the hyper-cellulolytic filamentous fungus. Biosci. Biotechnol. Biochem. 2016, 80, 1712–1729. [Google Scholar] [CrossRef][Green Version]
- Mantyla, A.L.; Rossi, K.H.; Vanhanen, S.A.; Penttila, M.E.; Suominen, P.L.; Nevalainen, K.M. Electrophoretic karyotyping of wild-type and mutant Trichoderma longibrachiatum (reesei) strains. Curr. Genet. 1992, 21, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Porciuncula Jde, O.; Furukawa, T.; Mori, K.; Shida, Y.; Hirakawa, H.; Tashiro, K.; Kuhara, S.; Nakagawa, S.; Morikawa, Y.; Ogasawara, W. Single nucleotide polymorphism analysis of a Trichoderma reesei hyper-cellulolytic mutant developed in Japan. Biosci. Biotechnol. Biochem. 2013, 77, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Shida, Y.; Yamaguchi, K.; Nitta, M.; Nakamura, A.; Takahashi, M.; Kidokoro, S.; Mori, K.; Tashiro, K.; Kuhara, S.; Matsuzawa, T.; et al. The impact of a single-nucleotide mutation of bgl2 on cellulase induction in a Trichoderma reesei mutant. Biotechnol. Biofuels 2015, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Suzuki, M.R.; Hammel, K.E. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 2005, 71, 2412–2417. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Larrondo, L.F.; Putnam, N.; Gelpke, M.D.; Huang, K.; Chapman, J.; Helfenbein, K.G.; Ramaiya, P.; Detter, J.C.; Larimer, F.; et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 2004, 22, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.T.; Cajthaml, T.; Snajdr, J.; Baldrian, P. Differential degradation of oak (Quercus petraea) leaf litter by litter-decomposing basidiomycetes. Res. Microbiol. 2007, 158, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P.; Valaskova, V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 2008, 32, 501–521. [Google Scholar] [CrossRef]
- Stursova, M.; Zifcakova, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose utilization in forest litter and soil: Identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An important phylum of cellulose-degrading bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Jiang, Y.P.; Duarte, A.V.; van den Brink, J.; Wiebenga, A.; Zou, G.; Wang, C.S.; de Vries, R.P.; Zhou, Z.H.; Benoit, I. Enhancing saccharification of wheat straw by mixing enzymes from genetically-modified Trichoderma reesei and Aspergillus niger. Biotechnol. Lett. 2016, 38, 65–70. [Google Scholar] [CrossRef]
- Kolasa, M.; Ahring, B.K.; Lubeck, P.S.; Lubeck, M. Co-cultivation of Trichoderma reesei RutC30 with three black Aspergillus strains facilitates efficient hydrolysis of pretreated wheat straw and shows promises for on-site enzyme production. Bioresour. Technol. 2014, 169, 143–148. [Google Scholar] [CrossRef] [PubMed]
- van den Brink, J.; de Vries, R.P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 2011, 91, 1477–1492. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Tewari, R.; Rana, S.S.; Soni, R.; Soni, S.K. Cellulases: Classification, Methods of Determination and Industrial Applications. Appl. Biochem. Biotechnol. 2016, 179, 1346–1380. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Navarro Poulsen, J.C.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef]
- Morgenstern, I.; Powlowski, J.; Tsang, A. Fungal cellulose degradation by oxidative enzymes: From dysfunctional GH61 family to powerful lytic polysaccharide monooxygenase family. Brief. Funct. Genom. 2014, 13, 471–481. [Google Scholar] [CrossRef]
- Cuervo-Soto, L.I.; Valdes-Garcia, G.; Batista-Garcia, R.; del Rayo Sanchez-Carbente, M.; Balcazar-Lopez, E.; Lira-Ruan, V.; Pastor, N.; Folch-Mallol, J.L. Identification of a novel carbohydrate esterase from Bjerkandera adusta: Structural and function predictions through bioinformatics analysis and molecular modeling. Proteins 2015, 83, 533–546. [Google Scholar] [CrossRef]
- Santos, C.A.; Ferreira-Filho, J.A.; O’Donovan, A.; Gupta, V.K.; Tuohy, M.G.; Souza, A.P. Production of a recombinant swollenin from Trichoderma harzianum in Escherichia coli and its potential synergistic role in biomass degradation. Microb. Cell Fact. 2017, 16, 83. [Google Scholar] [CrossRef]
- Carle-Urioste, J.C.; Escobar-Vera, J.; El-Gogary, S.; Henrique-Silva, F.; Torigoi, E.; Crivellaro, O.; Herrera-Estrella, A.; El-Dorry, H. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 1997, 272, 10169–10174. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Messner, R.; Gruber, F.; Mandels, M.; Kubicek-Pranz, E.M. Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. Involvement of a constitutive, sophorose-inducible, glucose-inhibited beta-diglucoside permease. J. Biol. Chem. 1993, 268, 19364–19368. [Google Scholar]
- Zhou, Q.X.; Xu, J.T.; Kou, Y.B.; Lv, X.X.; Zhang, X.; Zhao, G.L.; Zhang, W.X.; Chen, G.J.; Liu, W.F. Differential Involvement of beta-Glucosidases from Hypocrea jecorina in Rapid Induction of Cellulase Genes by Cellulose and Cellobiose. Eukaryot. Cell 2012, 11, 1371–1381. [Google Scholar] [CrossRef]
- Vazquez-Montoya, E.L.; Castro-Ochoa, L.D.; Maldonado-Mendoza, I.E.; Luna-Suarez, S.; Castro-Martinez, C. Moringa straw as cellulase production inducer and cellulolytic fungi source. Rev. Argent. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lafon, A.; Seo, J.A.; Han, K.H.; Yu, J.H.; d’Enfert, C. The heterotrimeric G-protein GanB(alpha)-SfaD(beta)-GpgA(gamma) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 2005, 171, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, T.F.; Nitsche, B.M.; de Lima, P.B.; de Assis, L.J.; Mellado, L.; Harris, S.D.; Meyer, V.; Dos Santos, R.A.; Riano-Pachon, D.M.; Ries, L.N.; et al. The low affinity glucose transporter HxtB is also involved in glucose signalling and metabolism in Aspergillus nidulans. Sci. Rep. 2017, 7, 45073. [Google Scholar] [CrossRef]
- Rutter, J.; Probst, B.L.; McKnight, S.L. Coordinate regulation of sugar flux and translation by PAS kinase. Cell 2002, 111, 17–28. [Google Scholar] [CrossRef]
- el-Gogary, S.; Leite, A.; Crivellaro, O.; Eveleigh, D.E.; el-Dorry, H. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proc. Natl. Acad. Sci. USA 1989, 86, 6138–6141. [Google Scholar] [CrossRef] [PubMed]
- Aro, N.; Pakula, T.; Penttila, M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 719–739. [Google Scholar] [CrossRef]
- Lin, L.C.; Chen, Y.; Li, J.G.; Wang, S.S.; Sun, W.L.; Tian, C.G. Disruption of non-anchored cell wall protein NCW-1 promotes cellulase production by increasing cellobiose uptake in Neurospora crassa. Biotechnol. Lett. 2017, 39, 545–551. [Google Scholar] [CrossRef]
- Parisutham, V.; Chandran, S.P.; Mukhopadhyay, A.; Lee, S.K.; Keasling, J.D. Intracellular cellobiose metabolism and its applications in lignocellulose-based biorefineries. Bioresour. Technol. 2017, 239, 496–506. [Google Scholar] [CrossRef]
- Hsieh, C.W.C.; Cannella, D.; Jorgensen, H.; Felby, C.; Thygesen, L.G. Cellulase Inhibition by High Concentrations of Monosaccharides. J. Agric. Food Chem. 2014, 62, 3800–3805. [Google Scholar] [CrossRef]
- Fowler, T.; Brown, R.D., Jr. The bgl1 gene encoding extracellular beta-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 1992, 6, 3225–3235. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- van Peij, N.N.; Visser, J.; de Graaff, L.H. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 1998, 27, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, R.; Wurleitner, E.; Wacenovsky, C.; Aro, N.; Stricker, A.R.; Zeilinger, S.; Kubicek, C.P.; Penttila, M.; Mach, R.L. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell 2006, 5, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Gielkens, M.M.; Dekkers, E.; Visser, J.; de Graaff, L.H. Two cellobiohydrolase-encoding genes from Aspergillus niger require D-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 1999, 65, 4340–4345. [Google Scholar] [CrossRef] [PubMed]
- Stricker, A.R.; Grosstessner-Hain, K.; Wurleitner, E.; Mach, R.L. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 2006, 5, 2128–2137. [Google Scholar] [CrossRef] [PubMed]
- Hasper, A.A.; Trindade, L.M.; van der Veen, D.; van Ooyen, A.J.J.; de Graaff, L.H. Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology 2004, 150, 1367–1375. [Google Scholar] [CrossRef]
- Battaglia, E.; Zhou, M.; de Vries, R.P. The transcriptional activators AraR and XlnR from Aspergillus niger regulate expression of pentose catabolic and pentose phosphate pathway genes. Res. Microbiol. 2014, 165, 531–540. [Google Scholar] [CrossRef]
- Ishikawa, K.; Kunitake, E.; Kawase, T.; Atsumi, M.; Noguchi, Y.; Ishikawa, S.; Ogawa, M.; Koyama, Y.; Kimura, M.; Kanamaru, K.; et al. Comparison of the paralogous transcription factors AraR and XlnR in Aspergillus oryzae. Curr. Genet. 2018, 64, 1245–1260. [Google Scholar] [CrossRef]
- Xiao, W.J.; Li, H.N.; Xia, W.C.; Yang, Y.X.; Hu, P.; Zhou, S.N.; Hu, Y.M.; Liu, X.P.; Dai, Y.J.; Jiang, Z.B. Co-expression of cellulase and xylanase genes in Sacchromyces cerevisiae toward enhanced bioethanol production from corn stover. Bioengineered 2019, 10, 513–521. [Google Scholar] [CrossRef]
- Nitta, M.; Furukawa, T.; Shida, Y.; Mori, K.; Kuhara, S.; Morikawa, Y.; Ogasawara, W. A new Zn(II)(2)Cys(6)-type transcription factor BglR regulates beta-glucosidase expression in Trichoderma reesei. Fungal Genet. Biol. 2012, 49, 388–397. [Google Scholar] [CrossRef]
- Kunitake, E.; Tani, S.; Sumitani, J.; Kawaguchi, T. A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulose-responsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus. Appl. Microbiol. Biotechnol. 2013, 97, 2017–2028. [Google Scholar] [CrossRef] [PubMed]
- Huberman, L.B.; Coradetti, S.T.; Glass, N.L. Network of nutrient-sensing pathways and a conserved kinase cascade integrate osmolarity and carbon sensing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2017, 114, E8665–E8674. [Google Scholar] [CrossRef] [PubMed]
- Coradetti, S.T.; Craig, J.P.; Xiong, Y.; Shock, T.; Tian, C.; Glass, N.L. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 7397–7402. [Google Scholar] [CrossRef] [PubMed]
- Coradetti, S.T.; Xiong, Y.; Glass, N.L. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2013, 2, 595–609. [Google Scholar] [CrossRef]
- Craig, J.P.; Coradetti, S.T.; Starr, T.L.; Glass, N.L. Direct Target Network of the Neurospora crassa Plant Cell Wall Deconstruction Regulators CLR-1, CLR-2, and XLR-1. Mbio 2015, 6, e01452-15. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism-From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Dowzer, C.E.; Kelly, J.M. Cloning of the creA gene from Aspergillus nidulans: A gene involved in carbon catabolite repression. Curr. Genet. 1989, 15, 457–459. [Google Scholar] [CrossRef]
- Dowzer, C.E.; Kelly, J.M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell Biol. 1991, 11, 5701–5709. [Google Scholar] [CrossRef]
- de Vries, R.P.; Visser, J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 2001, 65, 497–522. [Google Scholar] [CrossRef]
- Takashima, S.; Iikura, H.; Nakamura, A.; Masaki, H.; Uozumi, T. Analysis of Cre1 binding sites in the Trichoderma reesei cbh1 upstream region. FEMS Microbiol. Lett. 1996, 145, 361–366. [Google Scholar] [CrossRef]
- Roy, P.; Lockington, R.A.; Kelly, J.M. CreA-mediated repression in Aspergillus nidulans does not require transcriptional auto-regulation, regulated intracellular localisation or degradation of CreA. Fungal Genet. Biol. 2008, 45, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Flipphi, M.J.; Visser, J.; van der Veen, P.; de Graaff, L.H. Arabinase gene expression in Aspergillus niger: Indications for coordinated regulation. Microbiology 1994, 140, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.; Mach, R.L.; Zeilinger, S.; Hartler, G.; Stoffler, G.; Wolschek, M.; Kubicek, C.P. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 1995, 376, 103–107. [Google Scholar] [CrossRef]
- Cziferszky, A.; Mach, R.L.; Kubicek, C.P. Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 2002, 277, 14688–14694. [Google Scholar] [CrossRef] [PubMed]
- Ilmen, M.; Thrane, C.; Penttila, M. The glucose repressor gene cre1 of Trichoderma: Isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 1996, 251, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Ilmen, M.; Onnela, M.L.; Klemsdal, S.; Keranen, S.; Penttila, M. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol. Gen. Genet. 1996, 253, 303–314. [Google Scholar] [CrossRef]
- Zeilinger, S.; Mach, R.L.; Kubicek, C.P. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 1998, 273, 34463–34471. [Google Scholar] [CrossRef]
- Wurleitner, E.; Pera, L.; Wacenovsky, C.; Cziferszky, A.; Zeilinger, S.; Kubicek, C.P.; Mach, R.L. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot. Cell 2003, 2, 150–158. [Google Scholar] [CrossRef][Green Version]
- Denton, J.A.; Kelly, J.M. Disruption of Trichoderma reesei cre2, encoding an ubiquitin C-terminal hydrolase, results in increased cellulase activity. BMC Biotechnol. 2011, 11, 103. [Google Scholar] [CrossRef]
- Lockington, R.A.; Kelly, J.M. The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol. Microbiol. 2002, 43, 1173–1182. [Google Scholar] [CrossRef]
- Boase, N.A.; Kelly, J.M. A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol. Microbiol. 2004, 53, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Aro, N.; Ilmen, M.; Saloheimo, A.; Penttila, M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 2003, 69, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Chilton, I.J.; Delaney, C.E.; Barham-Morris, J.; Fincham, D.A.; Hooley, P.; Whitehead, M.P. The Aspergillus nidulans stress response transcription factor StzA is ascomycete-specific and shows species-specific polymorphisms in the C-terminal region. Mycol. Res. 2008, 112, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Han, J.; Li, Y.Y.; Liu, J.; Gan, L.H.; Long, M.N. Promoting cellulase and hemicellulase production from Trichoderma orientalis EU7-22 by overexpression of transcription factors Xyr1 and Ace3. Bioresour. Technol. 2020, 296, 122355. [Google Scholar] [CrossRef] [PubMed]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Penalva, M.A.; Arst, H.N., Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar] [CrossRef]
- He, R.L.; Ma, L.J.; Li, C.; Jia, W.D.; Li, D.M.; Zhang, D.Y.; Chen, S.L. Trpac1, a pH response transcription regulator, is involved in cellulase gene expression in Trichoderma reesei. Enzym. Microb. Technol. 2014, 67, 17–26. [Google Scholar] [CrossRef]
- Hakkinen, M.; Sivasiddarthan, D.; Aro, N.; Saloheimo, M.; Pakula, T.M. The effects of extracellular pH and of the transcriptional regulator PACI on the transcriptome of Trichoderma reesei. Microb. Cell Fact. 2015, 14, 63. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, Q.; Wang, J.X.; Liao, X.Z.; Guo, H.; Li, C.X.; Zhang, F.F.; Liao, L.S.; Luo, X.M.; Feng, J.X. Differential transcriptomic profiling of filamentous fungus during solid-state and submerged fermentation and identification of an essential regulatory gene PoxMBF1 that directly regulated cellulase and xylanase gene expression. Biotechnol. Biofuels 2019, 12, 103. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Andrianopoulos, A.; Kato, M.; Steidl, S.; Davis, M.A.; Tsukagoshi, N.; Hynes, M.J. HAP-Like CCAAT-binding complexes in filamentous fungi: Implications for biotechnology. Fungal. Genet. Biol. 1999, 27, 243–252. [Google Scholar] [CrossRef]
- Zeilinger, S.; Ebner, A.; Marosits, T.; Mach, R.; Kubicek, C.P. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol. Genet. Genom. 2001, 266, 56–63. [Google Scholar] [CrossRef]
- Wirsel, S.; Lachmund, A.; Wildhardt, G.; Ruttkowski, E. Three alpha-amylase genes of Aspergillus oryzae exhibit identical intron-exon organization. Mol. Microbiol. 1989, 3, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Tsukagoshi, N.; Furukawa, M.; Nagaba, H.; Kirita, N.; Tsuboi, A.; Udaka, S. Isolation of a cDNA encoding Aspergillus oryzae Taka-amylase A: Evidence for multiple related genes. Gene 1989, 84, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, T.; Maruyama, J.; Kitamoto, K. Contribution ratios of amyA, amyB, amyC genes to high-level alpha-amylase expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2012, 76, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Hata, Y.; Kitamoto, K.; Gomi, K.; Kumagai, C.; Tamura, G.; Hara, S. The glucoamylase cDNA from Aspergillus oryzae: Its cloning, nucleotide sequence, and expression in Saccharomyces cerevisiae. Agric Biol. Chem. 1991, 55, 941–949. [Google Scholar]
- Hata, Y.; Tsuchiya, K.; Kitamoto, K.; Gomi, K.; Kumagai, C.; Tamura, G.; Hara, S. Nucleotide sequence and expression of the glucoamylase-encoding gene (glaA) from Aspergillus oryzae. Gene 1991, 108, 145–150. [Google Scholar] [CrossRef]
- Minetoki, T.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Nucleotide sequence and expression of alpha-glucosidase-encoding gene (agdA) from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1995, 59, 1516–1521. [Google Scholar] [CrossRef]
- Hata, Y.; Ishida, H.; Ichikawa, E.; Kawato, A.; Suginami, K.; Imayasu, S. Nucleotide sequence of an alternative glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Gene 1998, 207, 127–134. [Google Scholar] [CrossRef]
- Tada, S.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G.; Hara, S. Identification of the promoter region of the Taka-amylase A gene required for starch induction. Agric. Biol. Chem. 1991, 55, 1939–1941. [Google Scholar]
- Tsuchiya, K.; Tada, S.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Deletion analysis of the Taka-amylase A gene promoter using a homologous transformation system in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 1992, 56, 1849–1853. [Google Scholar] [CrossRef]
- Kanemori, Y.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Insertion analysis of putative functional elements in the promoter region of the Aspergillus oryzae Taka-amylase A gene (amyB) using a heterologous Aspergillus nidulans amdS-lacZ fusion gene system. Biosci. Biotechnol. Biochem. 1999, 63, 180–183. [Google Scholar] [CrossRef]
- Minetoki, T.; Kumagai, C.; Gomi, K.; Kitamoto, K.; Takahashi, K. Improvement of promoter activity by the introduction of multiple copies of the conserved region III sequence, involved in the efficient expression of Aspergillus oryzae amylase-encoding genes. Appl. Microbiol. Biotechnol. 1998, 50, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Gomi, K.; Akeno, T.; Minetoki, T.; Ozeki, K.; Kumagai, C.; Okazaki, N.; Iimura, Y. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2000, 64, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.L.; Lehmbeck, J.; Christensen, T. A new transcriptional activator for amylase genes in Aspergillus. Mol. Gen. Genet. 1999, 262, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Tanaka, H.; Mogi, Y.; Yamazaki, T.; Suzuki, K.; Watanabe, T.; Yamada, O.; Akita, O. Loss of Aspergillus oryzae amyR function indirectly affects hemicellulolytic and cellulolytic enzyme production. J. Biosci. Bioeng. 2011, 111, 408–413. [Google Scholar] [CrossRef]
- Tanaka, M.; Yoshimura, M.; Ogawa, M.; Koyama, Y.; Shintani, T.; Gomi, K. The C2H2-type transcription factor, FlbC, is involved in the transcriptional regulation of Aspergillus oryzae glucoamylase and protease genes specifically expressed in solid-state culture. Appl. Microbiol. Biotechnol. 2016, 100, 5859–5868. [Google Scholar] [CrossRef]
- Kwon, N.J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 2010, 77, 1203–1219. [Google Scholar] [CrossRef]
- Ogawa, M.; Tokuoka, M.; Jin, F.J.; Takahashi, T.; Koyama, Y. Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 10–18. [Google Scholar] [CrossRef]
- Zhuang, M.; Zhang, Z.M.; Jin, L.; Wang, B.T.; Koyama, Y.; Jin, F.L. The Basic-Region Helix-Loop-Helix Transcription Factor DevR Significantly Affects Polysaccharide Metabolism in Aspergillus oryzae. Appl. Environ. Microbiol. 2019, 85, e00089-19. [Google Scholar] [CrossRef]
- Hasegawa, S.; Takizawa, M.; Suyama, H.; Shintani, T.; Gomi, K. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 1–9. [Google Scholar] [CrossRef]
- Needleman, R.B.; Kaback, D.B.; Dubin, R.A.; Perkins, E.L.; Rosenberg, N.G.; Sutherland, K.A.; Forrest, D.B.; Michels, C.A. MAL6 of Saccharomyces: A complex genetic locus containing three genes required for maltose fermentation. Proc. Natl. Acad. Sci. USA 1984, 81, 2811–2815. [Google Scholar] [CrossRef]
- Hiramoto, T.; Tanaka, M.; Ichikawa, T.; Matsuura, Y.; Hasegawa-Shiro, S.; Shintani, T.; Gomi, K. Endocytosis of a maltose permease is induced when amylolytic enzyme production is repressed in Aspergillus oryzae. Fungal Genet. Biol. 2015, 82, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, S.; Tanaka, M.; Shintani, T.; Gomi, K. Improved alpha-amylase production by Aspergillus oryzae after a double deletion of genes involved in carbon catabolite repression. Appl. Microbiol. Biotechnol. 2014, 98, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Florencio, C.; Cunha, F.M.; Badino, A.C.; Farinas, C.S.; Ximenes, E.; Ladisch, M.R. Secretome analysis of Trichoderma reesei and Aspergillus niger cultivated by submerged and sequential fermentation processes: Enzyme production for sugarcane bagasse hydrolysis. Enzym. Microb. Technol. 2016, 90, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zhang, H.; Liu, S.; Zhang, L.; Gao, P.; Chen, G.; Wang, L. Comparative Secretome Analysis of Aspergillus niger, Trichoderma reesei, and Penicillium oxalicum During Solid-State Fermentation. Appl. Biochem. Biotechnol. 2015, 177, 1252–1271. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Moller, L.L.H.; Larsen, T.O.; Kumar, R.; Arnau, J. Safety of the fungal workhorses of industrial biotechnology: Update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 2018, 102, 9481–9515. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.-T.; Hu, S.; Yu, X.-Y.; Jin, L.; Zhu, Y.-J.; Jin, F.-J. Studies of Cellulose and Starch Utilization and the Regulatory Mechanisms of Related Enzymes in Fungi. Polymers 2020, 12, 530. https://doi.org/10.3390/polym12030530
Wang B-T, Hu S, Yu X-Y, Jin L, Zhu Y-J, Jin F-J. Studies of Cellulose and Starch Utilization and the Regulatory Mechanisms of Related Enzymes in Fungi. Polymers. 2020; 12(3):530. https://doi.org/10.3390/polym12030530
Chicago/Turabian StyleWang, Bao-Teng, Shuang Hu, Xing-Ye Yu, Long Jin, Yun-Jia Zhu, and Feng-Jie Jin. 2020. "Studies of Cellulose and Starch Utilization and the Regulatory Mechanisms of Related Enzymes in Fungi" Polymers 12, no. 3: 530. https://doi.org/10.3390/polym12030530
APA StyleWang, B.-T., Hu, S., Yu, X.-Y., Jin, L., Zhu, Y.-J., & Jin, F.-J. (2020). Studies of Cellulose and Starch Utilization and the Regulatory Mechanisms of Related Enzymes in Fungi. Polymers, 12(3), 530. https://doi.org/10.3390/polym12030530




