An In-Silico Pipeline for Rapid Screening of DNA Aptamers against Mycotoxins: The Case-Study of Fumonisin B1, Aflatoxin B1 and Ochratoxin A
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Apparatus
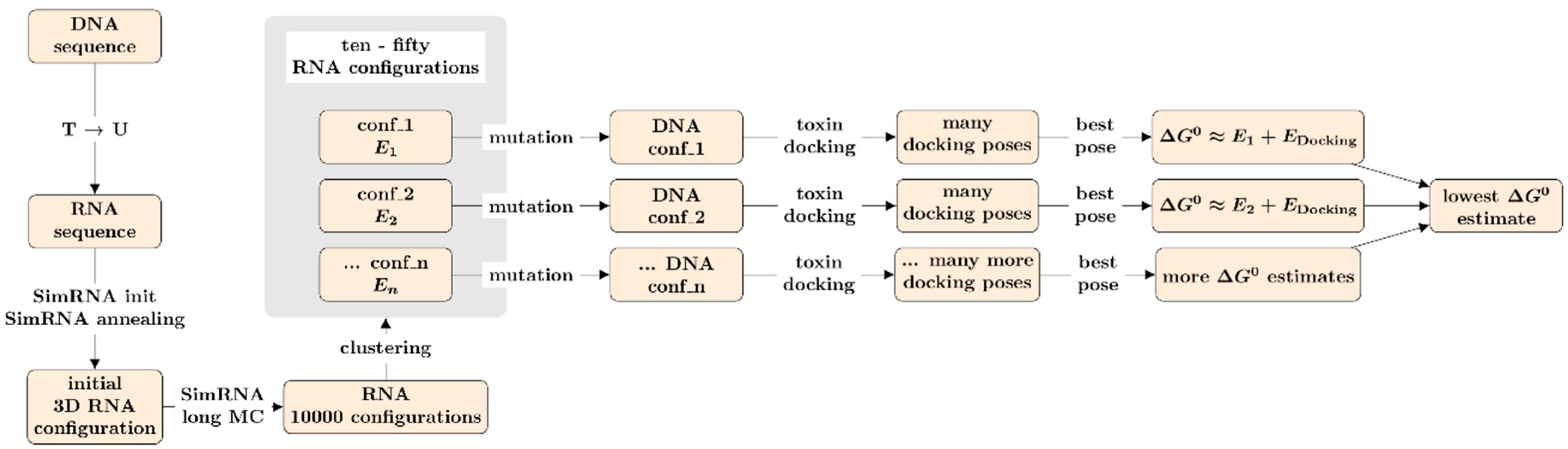
2.2. In-Silico Pipeline for Aptamer Screening
2.2.1. Hardware and Software
2.2.2. Prediction of the 3-D DNA-Aptamer Structures
2.2.3. The Docking Approach
2.2.4. Docking Calculations and Relation to Experimental Data
2.3. Experimental Assays
2.3.1. Fluorescent Microscale Thermophoresis (MST)
2.3.2. Magnetic Beads (MBs) Assay
3. Results and Discussion
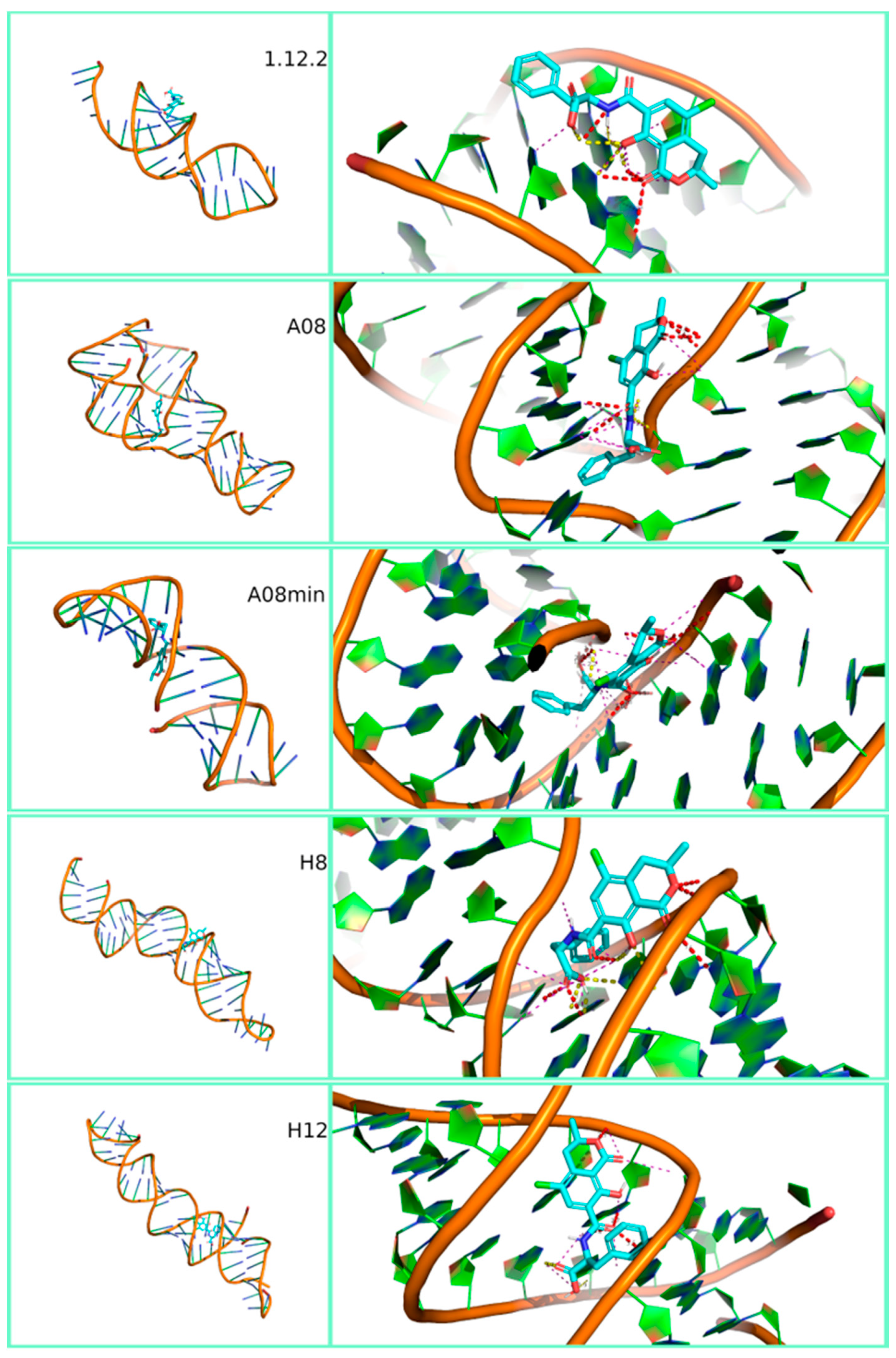
3.1. In-Silico Binding Affinity of Mycotoxins to Aptamers
3.2. Experimental Binding Affinity of Aptamers to Mycotoxins
3.3. Comparison of In-Silico and Experimental Binding Affinity of Aptamers to Mycotoxins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage t4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; Smith, M.; Goudreau, D.N.; DeRosa, M.C. Current status and future prospects for aptamer-based mycotoxin detection. J. AOAC Int. 2016, 99, 865–877. [Google Scholar] [CrossRef]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-based biosensors for antibiotic detection: A review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, F.R.W.; Valério, A.; de Oliveira, D.; Hotza, D. An overview and future prospects on aptamers for food safety. Appl. Micr. Biotech. 2020, 104, 6929–6939. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y. (Ed.) Aptamers for Analytical Applications: Affinity Optimization and Method Design; Wiley-VCH Verlag GmbH: New York, NY, USA, 2019; p. 414. [Google Scholar]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajslova, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 2019, 60, 2773–2789. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.R.N.; Salleh, B.; Saad, B.; Abbas, H.K.; Abel, C.A.; Shier, W.T. An overview of mycotoxin contamination in foods and its implications for human health. Toxin Rev. 2010, 29, 3–26. [Google Scholar] [CrossRef]
- Commission Regulation (EU) No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Union 2006, L 364, 5–24.
- Commission Recommendation (EC) No 165/2013 of 27 March 2013 on the presence of T-2 and HT-2 toxin in cereals and cereal products. Off. J. Eur. Union 2013, L 91, 12–15.
- Commission Recommendation (EC) No 576/2006 of 17 August 2006 on the presence of deoxynivalenol, zearalenone, ochratoxin A, T-2 and HT-2 and fumonisins in products intended for animal feeding. Off. J. Eur. Union 2006, L 229, 7–9.
- Commission Recommendation (EU) No 1319/2016 of 29 July 2016 amending Recommendation 2006/576/EC as regards deoxynivalenol, zearalenone and ochratoxin A in pet food. Off. J. Eur. Union 2016, L 208, 58–60.
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Velu, R.; Hill, K.; Bardoczy, V.; Meszaros, T.; DeRosa, M.C. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 2014, 6, 2435–2452. [Google Scholar] [CrossRef] [PubMed]
- Barthelmebs, L.; Jonca, J.; Hayat, A.; Prieto-Simon, B.; Marty, J.-L. Enzyme-linked aptamer assays (ELAAs), based on a competition format for a rapid and sensitive detection of ochratoxin A in wine. Food Contr. 2011, 22, 737–743. [Google Scholar] [CrossRef]
- Rhouati, A.; Yang, C.; Hayat, A.; Marty, J.-L. Aptamers: A promising tool for ochratoxin A detection in food analysis. Toxins 2013, 5, 1988–2008. [Google Scholar] [CrossRef] [PubMed]
- Le, L.C.; Cruz-Aguado, J.A.; Penner, G.A. DNA Ligands for Aflatoxin and Zearalenone. U.S. Patent PCT/CA2010/001292, 6 September 2011. NeoVentures Biotechnology. [Google Scholar]
- Ma, X.; Wang, W.; Chen, X.; Xia, Y.; Wu, S.; Duan, N.; Wang, Z. Selection, identification, and application of Aflatoxin B1 aptamer. Eur. Food Res. Technol. 2014, 238, 919–925. [Google Scholar] [CrossRef]
- Malhotra, S.; Pandey, A.K.; Rajput, Y.S.; Sharma, R. Selection of aptamers for aflatoxin M1 and their characterization. J. Mol. Recognit. 2014, 27, 493–500. [Google Scholar] [CrossRef]
- Setlem, K.; Mondal, B.; Ramlal, S.; Kingston, J. Immuno affinity SELEX for simple, rapid, and cost-effective aptamer enrichment and identification against aflatoxin B1. Front. Microbiol. 2016, 7, 1909. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.Y.V.V.; Renuka, R.M.; Achuth, J.; Mudili, V.; Poda, S. Development of a FRET-based fluorescence aptasensor for the detection of aflatoxin B1 in contaminated food grain samples. RSC Adv. 2018, 8, 10465–10473. [Google Scholar] [CrossRef] [Green Version]
- Danesh, N.M.; Bostan, H.B.; Abnous, K.; Ramezani, M.; Youssefi, K.; Taghdisi, S.M.; Karimi, G. Ultrasensitive detection of aflatoxin B1 and its major metabolite aflatoxin M1 using aptasensors: A review TrAC Trends Anal. Chem. 2018, 99, 117–128. [Google Scholar]
- Wang, Q.; Yang, Q.; Wu, W. Progress on structured biosensors for monitoring aflatoxin B1 from biofilms: A review. Front. Microb. 2020, 11, 408. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Bradley, C.R.; De Girolamo, A.; Visconti, A.; Miller, J.D.; DeRosa, M.C. Screening and initial binding assessment of fumonisin B1 aptamers. Int. J. Mol. Sci. 2010, 11, 4864–4881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frost, N.R.; McKeague, M.; Falcioni, D.; DeRosa, M.C. An in-solution assay for interrogation of affinity and rational minimer design for small molecule-binding aptamers. Analyst 2015, 140, 6643–6651. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Xia, Y.; Ma, X.; Zhu, C.; Jiang, Y.; Ding, Z.; Wang, Z. Selection and characterization of single stranded DNA aptamers recognizing fumonisin B1. Microchim. Acta 2014, 181, 1317–1324. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, S. Oligonucleotide Aptamer Special for Distinguishing Fumonisin B1. Patent No. CN 103013999 B, 28 May 2014. [Google Scholar]
- Guo, X.; Wen, F.; Zheng, N.; Saive, M.; Fauconnier, M.L.; Wang, J. Aptamer-based biosensor for detection of mycotoxins. Front. Chem. 2020, 8, 195. [Google Scholar] [CrossRef] [Green Version]
- Goud, K.Y.; Reddy, K.K.; Satyanarayana, M.; Kummari, S.; Gobi, K.V. A review on recent developments in optical and electrochemical aptamer-based assays for mycotoxins using advanced nanomaterials. Microchim. Acta 2020, 187, 29. [Google Scholar] [CrossRef]
- McKeague, M.; De Girolamo, A.; Valenzano, S.; Pascale, M.; Ruscito, A.; Velu, R.; Frost, N.R.; Hill, K.; Smith, M.; McConnell, E.M.; et al. Comprehensive analytical comparison of strategies used for small molecule aptamer evaluation. Anal Chem. 2015, 87, 8608–8612. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Liu, B.; Cui, X.; Li, Y.; Tang, J.; Wang, H.; Zhang, D.; Li, Z. Recent advances in aptasensors for mycotoxin detection: On the surface and in the colloid. Talanta 2021, 223, 121729. [Google Scholar] [CrossRef]
- McKeague, M.; DeRosa, M.C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 2012, 748913. [Google Scholar] [CrossRef]
- De Girolamo, A.; McKeague, M.; Pascale, M.; Cortese, M.; DeRosa, M.C. Immobilization of aptamers on substrates. In Aptamers for Analytical Applications: Affinity Optimization and Method Design; Dong, Y., Ed.; Wiley-VCH Verlag GmbH: New York, NY, USA, 2018; pp. 85–126. [Google Scholar]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Sabri, M.Z.; Hamid, A.A.A.; Hitam, S.M.S.; Rahim, M.Z.A. In-silico selection of aptamer: A review on the revolutionary approach to understand the aptamer design and interaction through computational chemistry. Mater. Today Proc. 2019, 19, 1572–1581. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, T.; Wang, Y.; Diao, C.; Zhou, Y.; Zhao, L.; Chen, H. Selection of a DNA aptamer against zearalenone and docking analysis for highly sensitive rapid visual detection with a label-free aptasensor. J. Agric. Food Chem. 2018, 66, 12102–12110. [Google Scholar] [CrossRef] [PubMed]
- Mousivand, M.; Anfossi, L.; Bagherzadeh, K.; Barbero, N.; Mirzadi-Gohari, A.; Javan-nikkhah, M. In silico maturation of affinity and selectivity of DNA aptamers against aflatoxin B1 for biosensor development. Anal. Chim. Acta 2020, 1105, 178–186. [Google Scholar] [CrossRef]
- McKeague, M.; Velu, R.; De Girolamo, A.; Valenzano, S.; Pascale, M.; Smith, M.; DeRosa, M.C. Comparison of in-solution biorecognition properties of aptamers against ochratoxin A. Toxins 2016, 8, 336. [Google Scholar] [CrossRef] [Green Version]
- Boniecki, M.J.; Lach, G.; Wayne, K.; Dawson, W.K.; Tomala, K.; Lukasz, P.; Soltysinski, T.; Rother, K.M.; Bujnicki, J.M. SimRNA (version 3.20) User Manual. 2014. Available online: http://genesilico.pl/DOWNLOAD/SimRNA_UserManual_v3_20_20141002.pdf (accessed on 13 December 2020).
- Boniecki, M.J.; Lach, G.; Wayne, K.; Dawson, W.K.; Tomala, K.; Lukasz, P.; Soltysinski, T.; Rother, K.M.; Bujnicki, J.M. SimRNA: A coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016, 44, e63. [Google Scholar] [CrossRef]
- Morley, S.D.; Afshar, M. Validation of an empirical RNA-ligand scoring function for fast flexible docking using RiboDock®. J. Comput. Aided Mol. Des. 2004, 18, 189–208. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, J.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Carmona, S.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A.B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R.E.; Morley, S.D. rDock: A fast, versatile and open-source program for docking ligands to proteins and nucleic acids. PLoS Comput. Biol. 2014, 10, e1003571. [Google Scholar] [CrossRef] [Green Version]
- Baaske, P.; Wienken, C.J.; Reineck, P.; Duhr, S.; Braun, D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed. 2010, 49, 2238–2241. [Google Scholar] [CrossRef] [PubMed]
- De Girolamo, A.; Pascale, M.; Visconti, A. Comparison of methods and optimization of the analysis of fumonisins B1 and B2 in masa flour, an alkaline cooked corn product. Food Add. Contam. Part A 2010, 28, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Duan, N.; Li, X.; Tan, G.; Ma, X.; Xia, Y.; Wang, Z.; Wang, H. Homogenous detection of fumonisin B(1) with a molecular beacon based on fluorescence resonance energy transfer between NaYF4: Yb, Ho upconversion nanoparticles and gold nanoparticles. Talanta 2013, 116, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, Y.; Marty, J.-L.; Yang, X. Aptamer-based colorimetric biosensing of ochratoxin A using unmodified gold nanoparticles indicator. Biosens. Bioelectron. 2011, 26, 2724–2727. [Google Scholar] [CrossRef] [PubMed]
- Entzian, C.; Schubert, T. Studying small molecule-aptamer interactions using MicroScale Thermophoresis (MST). Methods 2016, 97, 27–34. [Google Scholar] [CrossRef] [PubMed]




| Target Mycotoxin | Aptamer Name | ssDNA Sequence (5′-3′) | Buffer Composition | Reference |
|---|---|---|---|---|
| FB1 | FB1_39 | ATACCAGCTTATTCAATTAATCGCATTACCTTATACCAGCTTATTCAATTACGTCTGCACATACCAGCTTATTCAATTAGATAGTAAGTGCAATCT | 100 mM NaCl, 20 mM Tris, 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2, pH 7.6 | [25] |
| FB1_39t3 | ATACCAGCTTATTCAATTAATCGCATTACCTTATACCAGCTTATTCAATTACGTCTGCACATACCAGCTTATTCAATT | Same as FB1_39 | [26] | |
| FB1_10 | AGCAGCACAGAGGTCAGATGCGATCTGGATATTATTTTTGATACCCCTTTGGGGAGACATCCTATGCGTGCTACCGTGAA | 100 mM NaCl, 20 mM Tris–HCl, 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2, 0.02 % Tween 20 (pH 7.4) | [27] | |
| AFB1 | AF_AB3 | ATCCGTCACACCTGCTCTATTCCTCTGTTGAAGAACCACTTCCGGAAATAAGAGTGGTGTTGGCTCCCGTAT | 100 mM NaCl, 20 mM Tris–HCl (pH 7.6), 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2 | [20] |
| AF_APT1 | AGCAGCACAGAGGTCAGATGGTGCTATCATGCGCTCAATGGGAGACTTTAGCTGCCCCCACCTATGCGTGCTACCGTGAA | 100 mM NaCl, 20 mM Tris–HCl (pH 7.6), 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2, 0.02% Tween 20 (pH 7) | [19] | |
| OTA | 1.12.2 | GATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACA | 10 mM HEPES, 120 mM NaCl, 5 mM KCl, 10mM CaCl2, pH 7.0 | [14] |
| A08 | AGCCTCGTCTGTTCTCCCGGCAGTGTGGGCGAATCTATGCGTACCGTTCGATATCGTGGGGAAGACAAGCAGACGT | 10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4 | [15] | |
| A08min | GGCAGTGTGGGCGAATCTAT GCGTACCGTTCGATATCGTG | Same as A08 | [15] | |
| H8 | GGGAGGACGAAGCGGAACTGGGTGTGGGGTGATCAAGGGAGTAGACTACAGAAGACACGCCCGACA | 10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, pH 7.4 | [16] | |
| H12 | GGGAGGACGAAGCGGAACCGGGTGTGGGTGCCTTGATCCAGGGAGTCTCAGAAGACACGCCCGACA | Same as H8 | [16] |
| Target Mycotoxin | Aptamer Name | Docking Score (kcal/mol) | Configurational Energy (kcal/mol) | Best Estimated ΔG0 (kcal/mol) |
|---|---|---|---|---|
| Fumonisin B1 (FB1) | FB1_39 | −5.50 6.70 −9.04 −9.25 | 0.0 0.1 0.7 1.6 | −8.34 |
| FB1_39t3 | −11.33 | 0.0 | −11.33 | |
| FB1_10 | 10.98 1.19 −5.58 | 0.0 4.89 9.76 | 4.18 | |
| Aflatoxin B1 (AFB1) | AF_AB3 | −9.20 | 0.0 | −9.20 |
| AF_APT1 | −10.66 −11.16 −11.72 | 0.0 0.8 1.9 | −10.66 | |
| Ochratoxin A (OTA) | 1.12.2 | −10.38 | 0.0 | −10.38 |
| A08 | −17.08 | 0.0 | −17.08 | |
| A08min | −12.05 | 0.0 | −12.05 | |
| H8 | −12.91 | 0.0 | −12.91 | |
| H12 | −9.98 | 0.0 | −9.98 |
| Target Mycotoxin | Aptamer Name | MST Assay | MBs Assay | ||
|---|---|---|---|---|---|
| KD (nmol/L) | ΔG0 Energy a (kcal/mol) | KD (nmol/L) | ΔG0 Energy (kcal/mol) | ||
| Fumonisin B1 (FB1) | FB1_39 | 31 ± 22 | −10.24 ± 0.42 | 1.53 ± 0.67 | −12.03 ± 0.26 |
| FB1_39t3 | 2200 ± 1100 | −7.72 ± 0.30 | NB b | - c | |
| FB1_10 | 162 ± 137 | −9.26 ± 0.50 | NB | - | |
| Aflatoxin B1 (AFB1) | AF_AB3 | 178 ± 50 | −9.21 ± 0.17 | NB | - |
| AF_APT1 | NB | - | 1.40 ± 0.51 | −12.08 ± 0.22 | |
| Ochratoxin A (OTA) | 1.12.2 | 525 ± 147 d | −8.57 ± 0.17 | 374 ± 255 e | −8.77 ± 0.40 |
| A08 | 233 ± 81 d | −9.05 ± 0.21 | 286 ± 149 e | −8.93 ± 0.31 | |
| A08min | 97 ± 33 d | −9.57 ± 0.20 | 406 ± 166 e | −8.72 ± 0.24 | |
| H8 | 416 ± 119 d | −8.70 ± 0.17 | 14 ± 7 e | −10.71 ± 0.30 | |
| H12 | 630 ± 80 d | −8.46 ± 0.08 | 40 ± 14 e | −10.09 ± 0.21 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciriaco, F.; De Leo, V.; Catucci, L.; Pascale, M.; Logrieco, A.F.; DeRosa, M.C.; De Girolamo, A. An In-Silico Pipeline for Rapid Screening of DNA Aptamers against Mycotoxins: The Case-Study of Fumonisin B1, Aflatoxin B1 and Ochratoxin A. Polymers 2020, 12, 2983. https://doi.org/10.3390/polym12122983
Ciriaco F, De Leo V, Catucci L, Pascale M, Logrieco AF, DeRosa MC, De Girolamo A. An In-Silico Pipeline for Rapid Screening of DNA Aptamers against Mycotoxins: The Case-Study of Fumonisin B1, Aflatoxin B1 and Ochratoxin A. Polymers. 2020; 12(12):2983. https://doi.org/10.3390/polym12122983
Chicago/Turabian StyleCiriaco, Fulvio, Vincenzo De Leo, Lucia Catucci, Michelangelo Pascale, Antonio F. Logrieco, Maria C. DeRosa, and Annalisa De Girolamo. 2020. "An In-Silico Pipeline for Rapid Screening of DNA Aptamers against Mycotoxins: The Case-Study of Fumonisin B1, Aflatoxin B1 and Ochratoxin A" Polymers 12, no. 12: 2983. https://doi.org/10.3390/polym12122983
APA StyleCiriaco, F., De Leo, V., Catucci, L., Pascale, M., Logrieco, A. F., DeRosa, M. C., & De Girolamo, A. (2020). An In-Silico Pipeline for Rapid Screening of DNA Aptamers against Mycotoxins: The Case-Study of Fumonisin B1, Aflatoxin B1 and Ochratoxin A. Polymers, 12(12), 2983. https://doi.org/10.3390/polym12122983











