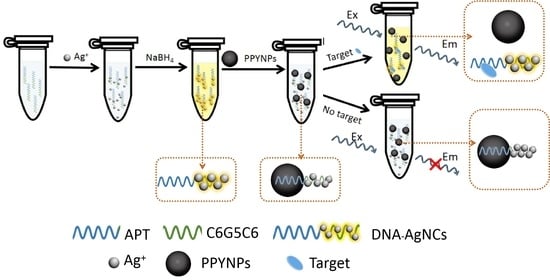
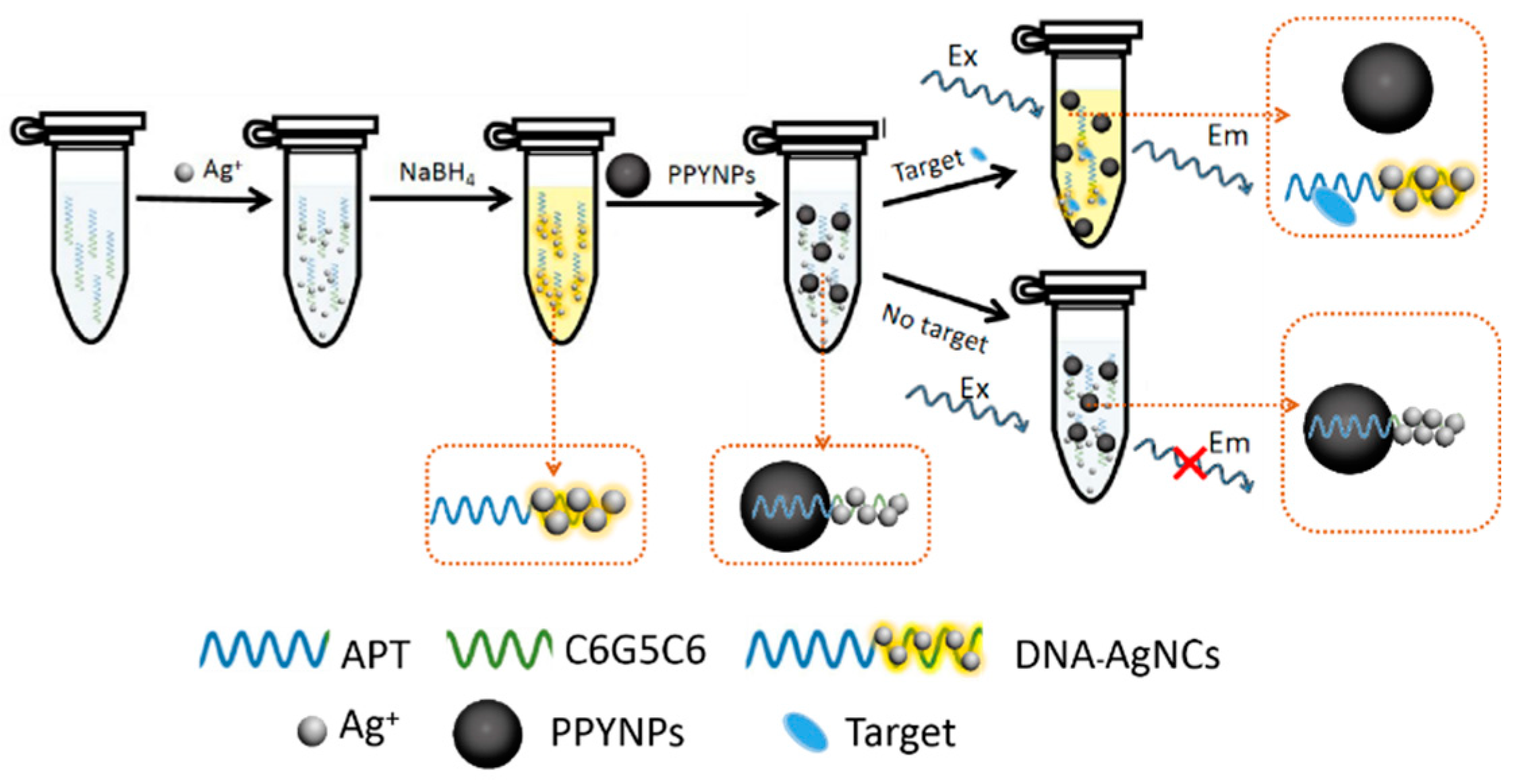
A Label-Free Fluorescent Aptasensor for Detection of Staphylococcal Enterotoxin A Based on Aptamer-Functionalized Silver Nanoclusters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. DNA-AgNC/PPyNP Characterization
2.3. Preparation of DNA-AgNCs
2.4. Preparation of PPyNPs
2.5. Fluorescence Detection of SEA Toxins
2.6. Sample Preparation and Measurement
3. Results
3.1. Principle of the Detection Method
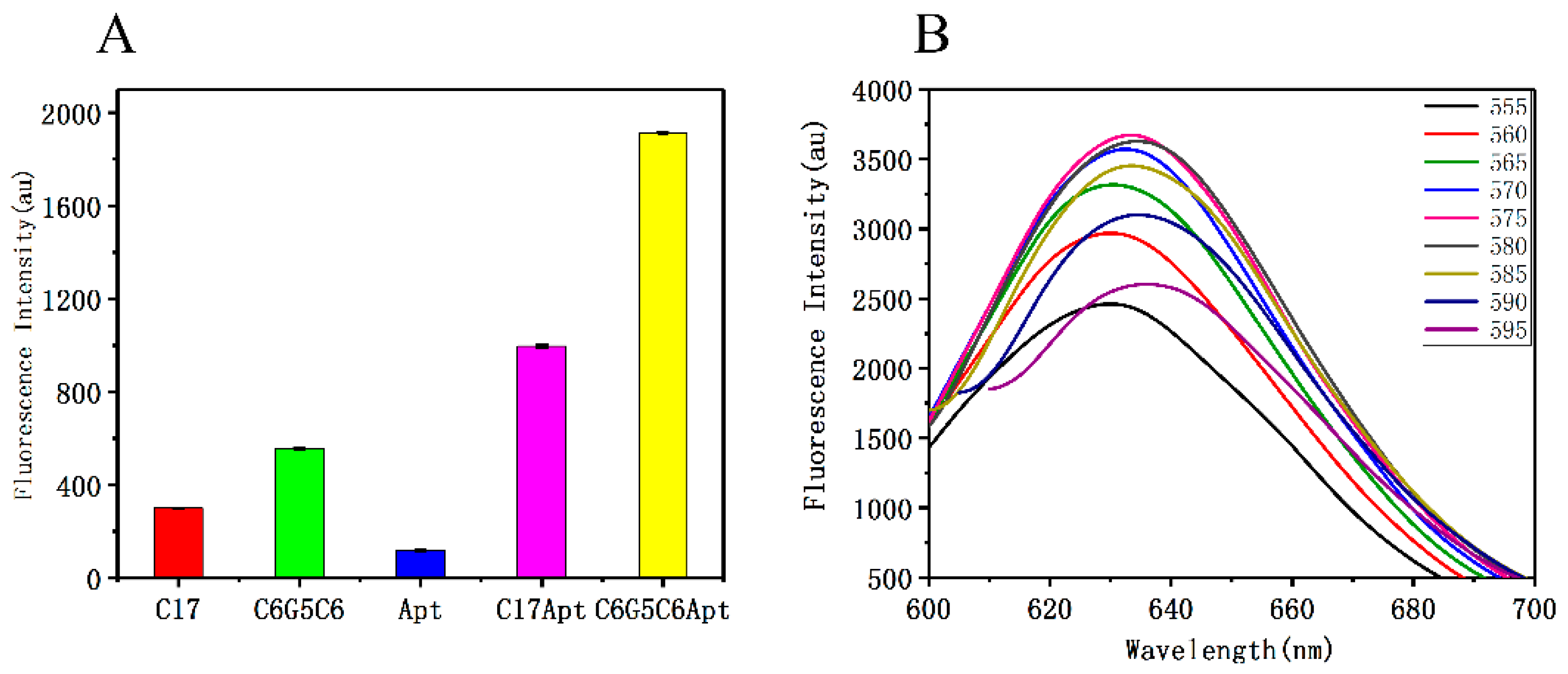
3.2. Optimization of DNA-AgNC Conditions
3.3. Characterization of AgNCs and PPyNPs
3.4. Optimized Detection Conditions
3.5. Feasibility Analysis
3.6. Analytical Performance
3.7. Detection of SEA in Milk Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rasooly, R.; Do, P.; He, X.; Hernlem, B. T cell Receptor V beta 9 in Method for Rapidly Quantifying Active Staphylococcal Enterotoxin Type-A without Live Animals. Toxins 2019, 11, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sospedra, I.; Soriano, J.M.; Mañes, J. Enterotoxinomics: The omic sciences in the study of staphylococcal toxins analyzed in food matrices. Food Res. Int. 2013, 54, 1052–1060. [Google Scholar] [CrossRef]
- Zeaki, N.; Johler, S.; Skandamis, P.N.; Schelin, J. The Role of Regulatory Mechanisms and Environmental Parameters in Staphylococcal Food Poisoning and Resulting Challenges to Risk Assessment. Front. Microbiol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desouza, I.A.; Camargo, E.A.; Mariano, N.S.; Optiz-Neto, J.B.; Resende, J.S.; Mello, G.C.; Costa, S.K.P.; De Nucci, G.; Antunes, E. Role of sensory innervation in the rat pulmonary neutrophil recruitment induced by staphylococcal enterotoxins type A and B. Eur. J. Pharmacol. 2009, 613, 128–134. [Google Scholar] [CrossRef]
- Evenson, M.L.; Hinds, M.W.; Bernstein, R.S.; Bergdoll, M.S. Estimation of human dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal food poisoning involving chocolate milk. Int. J. Food Microbiol. 1988, 7, 311–316. [Google Scholar] [CrossRef]
- Asao, T.; Kumeda, Y.; Kawai, T.; Shibata, T.; Oda, H.; Haruki, K.; Nakazawa, H.; Kozaki, S. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: Estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol. Infect. 2003, 130, 33–40. [Google Scholar] [CrossRef]
- Omwenga, I.; Aboge, G.O.; Mitema, E.S.; Obiero, G.; Ngaywa, C.; Ngwili, N.; Wamwere, G.; Wainaina, M.; Bett, B. Staphylococcus aureus enterotoxin genes detected in milk from various livestock species in northern pastoral region of Kenya. Food Control 2019, 103, 126–132. [Google Scholar] [CrossRef]
- Drougka, E.; Foka, A.; Giormezis, N.; Sergelidis, D.; Militsopoulou, M.; Jelastopulu, E.; Komodromos, D.; Sarrou, S.; Anastassiou, E.D.; Petinaki, E.; et al. Multidrug-resistant enterotoxigenic Staphylococcus aureus lineages isolated from animals, their carcasses, the personnel, and the environment of an abattoir in Greece. J. Food Process. Preserv. 2019, 43, e13961. [Google Scholar] [CrossRef]
- Lin, L.; Hu, J.Y.; Wu, Y.; Chen, M.; Ou, J.; Yan, W.L. Assessment of the inhibitory effects of sodium nitrite, nisin, potassium sorbate, and sodium lactate on Staphylococcus aureus growth and staphylococcal enterotoxin A production in cooked pork sausage using a predictive growth model. Food Sci. Hum. Wellness 2018, 7, 83–90. [Google Scholar] [CrossRef]
- Mahfoozi, A.; Shirzad-Aski, H.; Kaboosi, H.; Ghaemi, E.A. Identification of the classical enterotoxin genes of Staphylococcus aureus in various foods by multiplex PCR assay. Iran. J. Vet. Res. 2019, 20, 209–212. [Google Scholar]
- Gilquin, B.; Jaquinod, M.; Louwagie, M.; Kieffer-Jaquinod, S.; Kraut, A.; Ferro, M.; Becher, F.; Brun, V. A proteomics assay to detect eight CBRN-relevant toxins in food. Proteomics 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Elahi, S.; Fujikawa, H. Comprehensive Study of the Boundaries of Enterotoxin A Production and Growth of Staphylococcus aureus at Various Temperatures and Salt Concentrations. J. Food Sci. 2019, 84, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Schwan, W.R. Staphylococcus aureus Toxins: Armaments for a Significant Pathogen. Toxins 2019, 11, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmer, A.; Abobarin-Adeagbo, A.; Posa, A.; Jordan, B.; Delank, K.-S.; Staege, M.S.; Surov, A.; Zierz, S.; Kornhuber, M.E. Myositis in Lewis rats induced by the superantigen Staphylococcal enterotoxin A. Mol. Biol. Rep. 2019, 46, 4085–4094. [Google Scholar] [CrossRef]
- Aragao, B.B.; Trajano, S.C.; Silva, J.G.; Silva, B.P.; Oliveira, R.P.; Pinheiro Junior, J.W.; Peixoto, R.M.; Mota, R.A. Short communication: High frequency of beta-lactam-resistant Staphylococcus aureus in artisanal coalho cheese made from goat milk produced in northeastern Brazil. J. Dairy Sci. 2019, 102, 6923–6927. [Google Scholar] [CrossRef]
- Necidova, L.; Bursova, S.; Harustiakova, D.; Bogdanovicova, K.; Lacanin, I. Effect of heat treatment on activity of staphylococcal enterotoxins of type A, B, and C in milk. J. Dairy Sci. 2019, 102, 3924–3932. [Google Scholar] [CrossRef]
- Muratovic, A.Z.; Hagstrom, T.; Rosen, J.; Granelli, K.; Hellenas, K.-E. Quantitative Analysis of Staphylococcal Enterotoxins A and B in Food Matrices Using Ultra High-Performance Liquid Chromatography Tandem Mass Spectrometry (UPLC-MS/MS). Toxins 2015, 7, 3637–3656. [Google Scholar] [CrossRef] [Green Version]
- Koike, H.; Kanda, M.; Hayashi, H.; Matsushima, Y.; Ohba, Y.; Nakagawa, Y.; Nagano, C.; Sekimura, K.; Hirai, A.; Shindo, T.; et al. Quantification of staphylococcal enterotoxin type A in cow milk by using a stable isotope-labelled peptide via liquid chromatography-tandem mass spectrometry. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2019, 36, 1098–1108. [Google Scholar] [CrossRef]
- Andjelkovic, M.; Tsilia, V.; Rajkovic, A.; De Cremer, K.; Van Loco, J. Application of LC-MS/MS MRM to Determine Staphylococcal Enterotoxins (SEB and SEA) in Milk. Toxins 2016, 8, 118. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Li, X.; Ma, X.; Ou, G.; Gao, Z. Rapid and multiple detections of staphylococcal enterotoxins by two-dimensional molecularly imprinted film-coated QCM sensor. Sens. Actuators B Chem. 2014, 191, 326–331. [Google Scholar] [CrossRef]
- Wu, L.; Gao, B.; Zhang, F.; Sun, X.; Zhang, Y.; Li, Z. A novel electrochemical immunosensor based on magnetosomes for detection of staphylococcal enterotoxin B in milk. Talanta 2013, 106, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Pimenta-Martins, M.G.R.; Furtado, R.F.; Heneine, L.G.D.; Dias, R.S.; Borges, M.d.F.; Alves, C.R. Development of an amperometric immunosensor for detection of staphylococcal enterotoxin type A in cheese. J. Microbiol. Methods 2012, 91, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Yamada, K.; Ikami, M.; Kaji, N.; Tokeshi, M.; Atsumi, Y.; Mizutani, M.; Murai, A.; Okamoto, A.; Namikawa, T.; et al. Application of IgY to sandwich enzyme-linked immunosorbent assays, lateral flow devices, and immunopillar chips for detecting staphylococcal enterotoxins in milk and dairy products. J. Microbiol. Methods 2013, 92, 323–331. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Z.; Li, Y.; Li, Q.; Song, C.; Xu, Z.; Zhang, Y.; Zhang, Y.; Ma, Y.; Sun, Y.; et al. High sensitivity chemiluminescence enzyme immunoassay for detecting staphylococcal enterotoxin A in multi-matrices. Anal. Chim. Acta 2013, 796, 14–19. [Google Scholar] [CrossRef]
- Tsai, W.-C.; Li, I.-C. SPR-based immunosensor for determining staphylococcal enterotoxin A. Sens. Actuators B: Chem. 2009, 136, 8–12. [Google Scholar] [CrossRef]
- Lam, M.T.; Wan, Q.H.; Boulet, C.A.; Le, X.C. Competitive immunoassay for staphylococcal enterotoxin A using capillary electrophoresis with laser-induced fluorescence detection. J. Chromatogr. A 1999, 853, 545–553. [Google Scholar] [CrossRef]
- Sospedra, I.; Soler, C.; Manes, J.; Miguel Soriano, J. Rapid whole protein quantitation of staphylococcal enterotoxins A and B by liquid chromatography/mass spectrometry. J. Chromatogr. A 2012, 1238, 54–59. [Google Scholar] [CrossRef]
- Ben Haddada, M.; Salmain, M.; Boujday, S. Gold colloid-nanostructured surfaces for enhanced piezoelectric immunosensing of staphylococcal enterotoxin A. Sens. Actuators B Chem. 2018, 255, 1604–1613. [Google Scholar] [CrossRef] [Green Version]
- van Grinsven, B.; Eersels, K.; Peeters, M.; Losada-Pérez, P.; Vandenryt, T.; Cleij, T.J.; Wagner, P. The Heat-Transfer Method: A Versatile Low-Cost, Label-Free, Fast, and User-Friendly Readout Platform for Biosensor Applications. Acs. Appl. Mater. Interfaces 2014, 6, 13309–13318. [Google Scholar] [CrossRef] [Green Version]
- Peeters, M. Real-time monitoring of aptamer functionalization and detection of Ara H1 by electrochemical impedance spectroscopy and dissipation-mode quartz crystal microbalance. Biosens. Bioelectron. 2014, 5. [Google Scholar]
- Zhuo, Z.J.; Yu, Y.Y.; Wang, M.L.; Li, J.; Zhang, Z.K.; Liu, J.; Wu, X.H.; Lu, A.P.; Zhang, G.; Zhang, B.T. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wei, C. The aptamer DNA-templated fluorescence silver nanoclusters: ATP detection and preliminary mechanism investigation. Biosens. Bioelectron. 2017, 87, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Gang, J. The detection of a mismatched DNA by using hairpin DNA-templated silver nanoclusters. Anal. Biochem. 2018, 549, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Huili, T.; Nuo, D.; Shijia, W.; Zhouping, W. Selection and application of ssDNA aptamers against spermine based on Capture-SELEX. Anal. Chim. Acta 2019, 1081, 168–175. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, H.; Chen, X.; Wang, X.; Duan, N.; Wu, S.; Xu, B.; Wang, Z. A multicolor time-resolved fluorescence aptasensor for the simultaneous detection of multiplex Staphylococcus aureus enterotoxins in the milk. Biosens. Bioelectron. 2015, 74, 170–176. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, H.; Li, W.; Dong, Y.; Chi, Y. A novel hybrid platform of g-C3N4 nanosheets/nucleic-acid-stabilized silver nanoclusters for sensing protein. Anal. Chim. Acta 2019, 1091, 112–118. [Google Scholar] [CrossRef]
- Shen, F.; Cheng, Y.; Xie, Y.; Yu, H.; Yao, W.; Li, H.-W.; Guo, Y.; Qian, H. DNA-silver nanocluster probe for norovirus RNA detection based on changes in secondary structure of nucleic acids. Anal. Biochem. 2019, 583, 113365. [Google Scholar] [CrossRef]
- Liu, W.; Lai, H.; Huang, R.; Zhao, C.; Wang, Y.; Weng, X.; Zhou, X. DNA methyltransferase activity detection based on fluorescent silver nanocluster hairpin-shaped DNA probe with 5′-C-rich/G-rich-3′ tails. Biosens. Bioelectron. 2015, 68, 736–740. [Google Scholar] [CrossRef]
- Richards, C.I.; Choi, S.; Hsiang, J.-C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y.-L.; Dickson, R.M. Oligonucleotide-stabilized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039. [Google Scholar] [CrossRef] [Green Version]
- Diez, I.; Ras, R.H.A. Fluorescent silver nanoclusters. Nanoscale 2011, 3, 1963–1970. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Dickson, R.M.; Yu, J. Developing luminescent silver nanodots for biological applications. Chem. Soc. Rev. 2012, 41, 1867–1891. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhu, Y.; Chen, L.; Wang, L.; Zhang, X. Target-induced quenching for highly sensitive detection of nucleic acids based on label-free luminescent supersandwich DNA/silver nanoclusters. Analyst 2014, 139, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, J.; Yang, G.; Ma, S.; Zhang, M.; Yang, J. Cysteine-rich protein-templated silver nanoclusters as a fluorometric probe for mercury(ii) detection. Anal. Methods 2019, 11, 733–738. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, W.; Han, Y.; Ouyang, J.; Chen, M.; Hu, S.; Deng, L.; Liu, Y.-N. A label-free sensitive method for membrane protein detection based on aptamer and AgNCs transfer. Talanta 2017, 175, 470–476. [Google Scholar] [CrossRef]
- Lu, W.; Luo, Y.; Chang, G.; Qin, X.; Liao, F.; Sun, X. Polypyrrole colloidal nanospheres as an effective fluorescent sensing platform for DNA detection. Synth. Met. 2011, 161, 1766–1770. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.; Lu, Q.; Li, X.; Weng, C.; Yan, X.; Hong, J.; Zhou, X. A versatile fluorometric aptasensing scheme based on the use of a hybrid material composed of polypyrrole nanoparticles and DNA-silver nanoclusters: Application to the determination of adenosine, thrombin, or interferon-gamma. Microchim. Acta 2019, 186, 356. [Google Scholar] [CrossRef]
- Ke, K.; Lin, L.; Liang, H.; Chen, X.; Han, C.; Li, J.; Yang, H.-H. Polypyrrole nanoprobes with low non-specific protein adsorption for intracellular mRNA detection and photothermal therapy. Chem. Commun. 2015, 51, 6800–6803. [Google Scholar] [CrossRef]
- Pande, R.; Ruben, G.C.; Lim, J.O.; Tripathy, S.; Marx, K.A. DNA bound to polypyrrole films: High-resolution imaging, DNA binding kinetics and internal migration. Biomaterials 1998, 19, 1657–1667. [Google Scholar] [CrossRef]
- Khan, I.M.; Zhao, S.; Niazi, S.; Mohsin, A.; Shoaib, M.; Duan, N.; Wu, S.; Wang, Z. Silver nanoclusters based FRET aptasensor for sensitive and selective fluorescent detection of T-2 toxin. Sens. Actuators B-Chem. 2018, 277, 328–335. [Google Scholar] [CrossRef]
- Guo, B.; Zhao, J.; Wu, C.; Zheng, Y.; Ye, C.; Huang, M.; Wang, S. One-pot synthesis of polypyrrole nanoparticles with tunable photothermal conversion and drug loading capacity. Colloids Surf. B-Biointerfaces 2019, 177, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wei, C. Highly sensitive and selective fluorescence detection of Hg2+ based on turn-on aptamer DNA silver nanoclusters. RSC Adv. 2017, 7, 56289–56295. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Zhang, Y.; Pei, R.; Cheng, Y.; Xie, Y.; Yu, H.; Yao, W.; Li, H.-W.; Qian, H. Detecting the adulteration of antihypertensive health food using G-insertion enhanced fluorescent DNA-AgNCs. Sens. Actuators B Chem. 2019, 281, 493–498. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Teng, Y.; Lou, B.; Jia, X.; Gu, X.; Wang, E. G-quadruplex enhanced fluorescence of DNA–silver nanoclusters and their application in bioimaging. Nanoscale 2015, 7, 13224–13229. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.P.; Guo, W.W.; Wang, E.K. Oligonucleotide stabilized silver nanoclusters as fluorescence probe for drug-DNA interaction investigation. Anal. Chim. Acta 2011, 706, 338–342. [Google Scholar] [CrossRef]
- Wang, J.; Guo, X.; Liu, R.; Guo, J.; Zhang, Y.; Zhang, W.; Sang, S. Detection of carcinoembryonic antigen using a magnetoelastic nano-biosensor amplified with DNA-templated silver nanoclusters. Nanotechnology 2020, 31. [Google Scholar] [CrossRef]
- Zhang, B.; Wei, C. Highly sensitive and selective detection of Pb2+ using a turn-on fluorescent aptamer DNA silver nanoclusters sensor. Talanta 2018, 182, 125–130. [Google Scholar] [CrossRef]





| Sample | Added (ng/mL) | Detection (ng/mL) (Mean ± SD, n = 6) | Recovery Ratio (%) |
|---|---|---|---|
| Sample 1 | 50 | 47.3291 ± 3.0103 | 94.65% |
| Sample 2 | 100 | 87.7010 ± 5.6440 | 87.70% |
| Sample 3 | 150 | 138.4932 ± 5.2269 | 92.33% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Khan, I.M.; Ji, H.; Wang, Z.; Tian, H.; Cao, W.; Mi, W. A Label-Free Fluorescent Aptasensor for Detection of Staphylococcal Enterotoxin A Based on Aptamer-Functionalized Silver Nanoclusters. Polymers 2020, 12, 152. https://doi.org/10.3390/polym12010152
Zhang X, Khan IM, Ji H, Wang Z, Tian H, Cao W, Mi W. A Label-Free Fluorescent Aptasensor for Detection of Staphylococcal Enterotoxin A Based on Aptamer-Functionalized Silver Nanoclusters. Polymers. 2020; 12(1):152. https://doi.org/10.3390/polym12010152
Chicago/Turabian StyleZhang, Xueyan, Imran Mahmood Khan, Hua Ji, Zhouping Wang, Huili Tian, Wenbo Cao, and Weiyu Mi. 2020. "A Label-Free Fluorescent Aptasensor for Detection of Staphylococcal Enterotoxin A Based on Aptamer-Functionalized Silver Nanoclusters" Polymers 12, no. 1: 152. https://doi.org/10.3390/polym12010152