Identifications of False Positives Amongst Sodium(I) Cations in Protein Three-Dimensional Structures—A Validation Approach Extendible to Any Alkali or Alkaline Earth Cation and to Any Monoatomic Anion
Abstract
1. Introduction
1.1. Overview of the Validation Tools
1.2. Aberrant and Inaccurate Structures
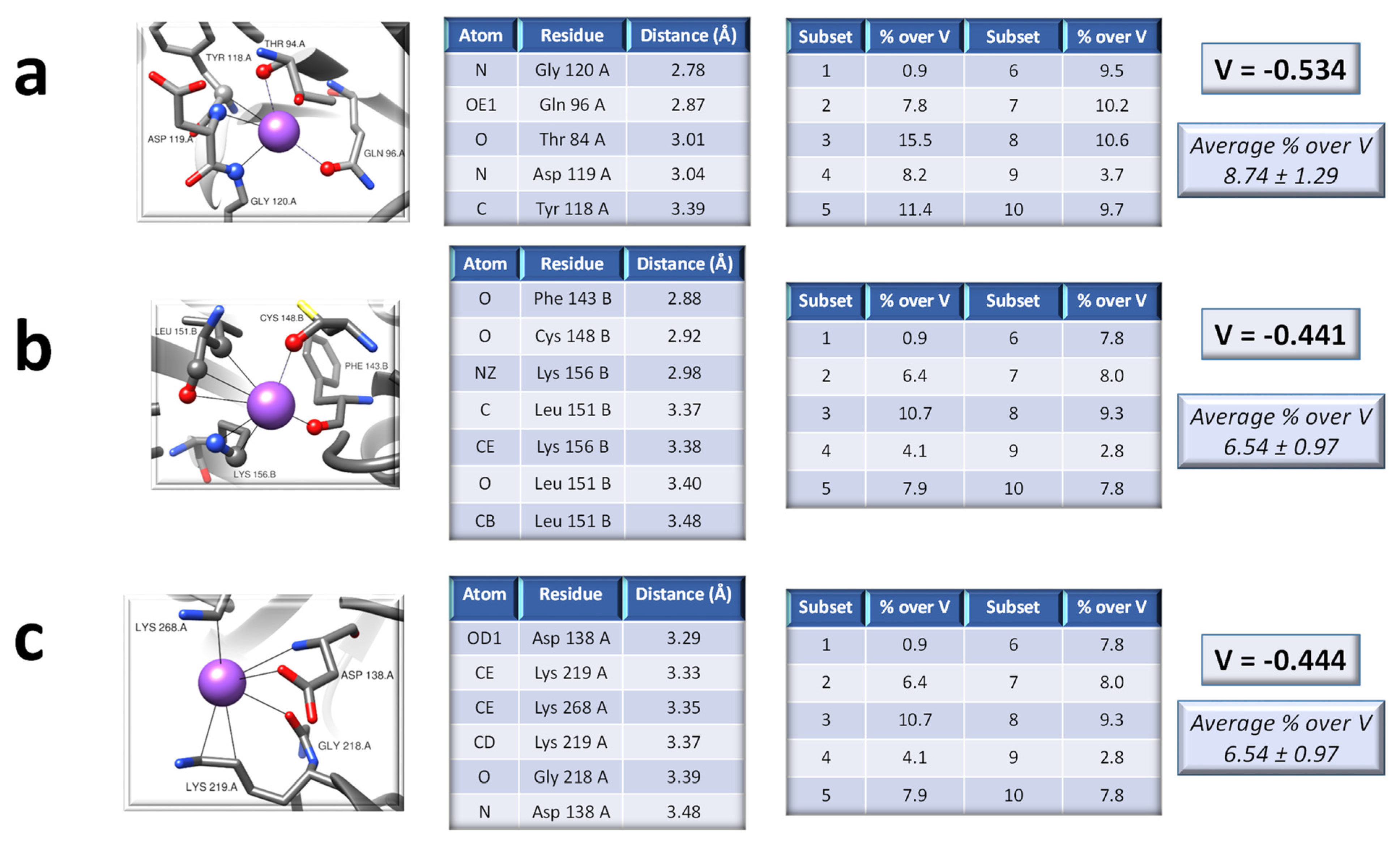
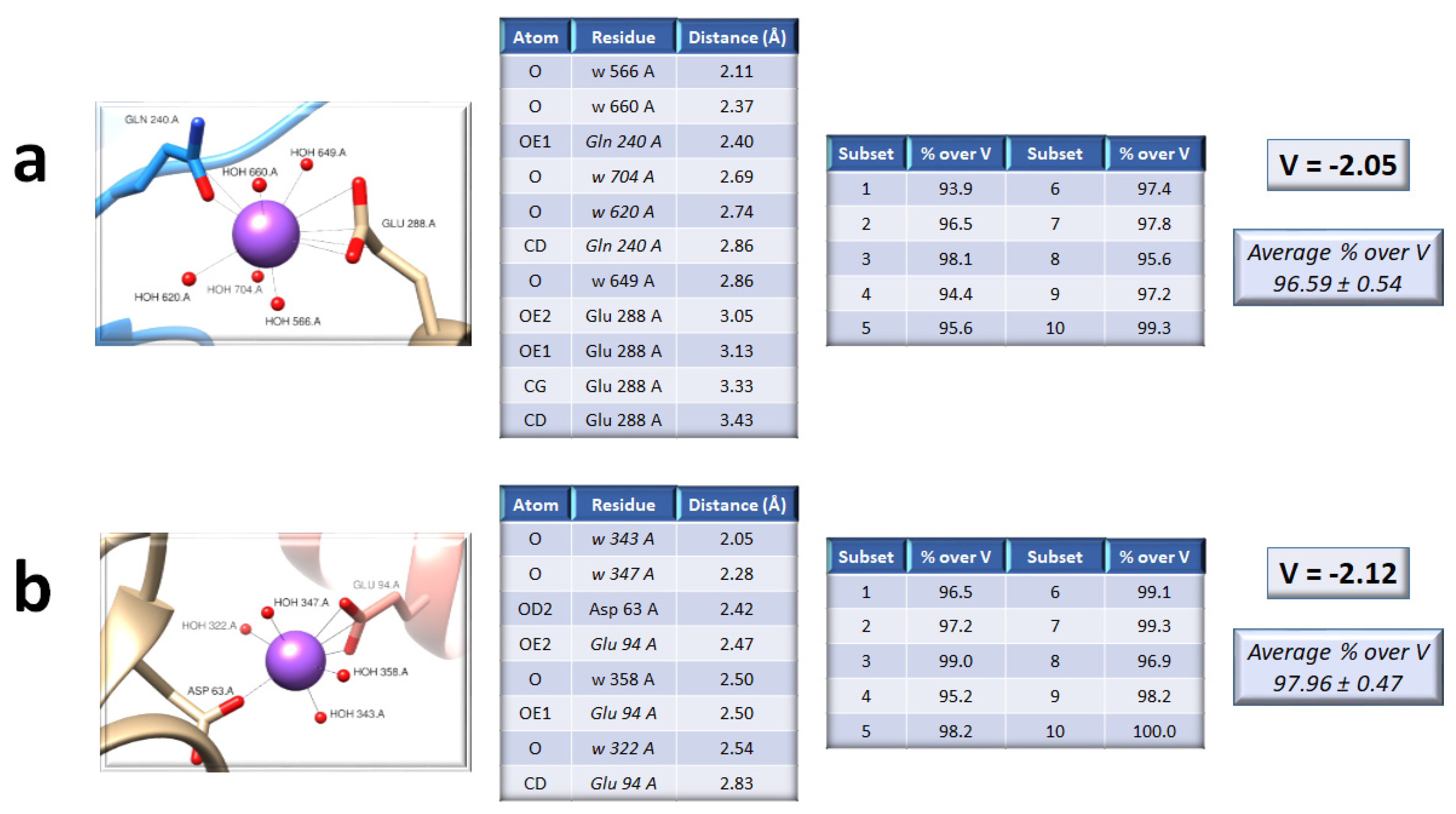
2. Results
3. Discussion
4. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popper, K. Logik der Forschung; Verlag von Julius Springer: Heidelberg, Germany, 1934. [Google Scholar]
- Carugo, O.; Djinovic-Carugo, K. Half a century of Ramachandran plots. Acta Crystallogr. 2013, D69, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain conformations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, W.M. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010, D66, 12–21. [Google Scholar] [CrossRef]
- Davis, I.W.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004, 32, W615–W619. [Google Scholar] [CrossRef]
- Hintze, B.J.; Lewis, S.M.; Richardson, J.S.; Richardson, D.C. Molprobity’s ultimate rotamer-library distributions for model validation. Proteins 2016, 84, 1177–1189. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Hooft, R.W.; Sander, C.; Vriend, G. Objectively judging the quality of a protein structure from a Ramachandran plot. Comput. Appl. Biosci. 1997, 13, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, O.V.; Afonine, P.V.; Moriarty, N.W.; Hekkelman, M.L.; Joosten, R.P.; Perrakis, A.; Adams, P.D. A Global Ramachandran Score Identifies Protein Structures with Unlikely Stereochemistry. Structure 2020, 28, 1249–1258.e1242. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Sobolev, O.V.; Moriarty, N.W.; Terwilliger, T.C.; Adams, P.D. Overall protein structure quality assessment using hydrogen-bonding parameters. Acta Cryst. 2023, D79, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Vaguine, A.A.; Richelle, J.; Wodak, S.J. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. 1999, D55, 191–205. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Benkert, P.; Tosatto, S.C.; Schomburg, D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins 2008, 71, 261–277. [Google Scholar] [CrossRef]
- Praznikar, J.; Tomic, M.; Turk, D. Validation and quality assessment of macromolecular structures using complex network analysis. Sci. Rep. 2019, 9, 1678. [Google Scholar] [CrossRef]
- Studer, G.; Biasini, M.; Schwede, T. Assessing the local structural quality of transmembrane protein models using statistical potentials (QMEANBrane). Bioinformatics 2014, 30, i505–i511. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Shao, C.; Liu, Z.; Yang, H.; Wang, S.; Burley, S.K. Outlier analyses of the Protein Data Bank archive using a probability-density-ranking approach. Sci. Data 2018, 5, 180293. [Google Scholar] [CrossRef]
- Shao, C.; Bittrich, S.; Wang, S.; Burley, S.K. Assessing PDB macromolecular crystal structure confidence at the individual amino acid residue level. Structure 2022, 30, 1385–1394. [Google Scholar] [CrossRef]
- Pereira, J.; Lamzin, V.S. A distance geometry-based description and validation of protein main-chain conformation. IUCrJ 2017, 4, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Trewhella, J.; Hendrickson, W.A.; Kleywegt, G.J.; Sali, A.; Sato, M.; Schwede, T.; Svergun, D.I.; Tainer, J.A.; Westbrook, J.; Berman, H.M. Report of the wwPDB Small-Angle Scattering Task Force: Data requirements for biomolecular modeling and the PDB. Structure 2013, 21, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Barad, B.A.; Echols, N.; Wang, R.Y.; Cheng, Y.; DiMaio, F.; Adams, P.D.; Fraser, J.S. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 2015, 12, 943–946. [Google Scholar] [CrossRef]
- Gore, S.; Sanz Garcia, E.; Hendrickx, P.M.S.; Gutmanas, A.; Westbrook, J.D.; Yang, H.; Feng, Z.; Baskaran, K.; Berrisford, J.M.; Hudson, B.P.; et al. Validation of Structures in the Protein Data Bank. Structure 2017, 25, 1916–1927. [Google Scholar] [CrossRef]
- Smart, O.S.; Horský, V.; Gore, S.; Svobodová Vařeková, R.; Bendová, V.; Kleywegt, G.J.; Velankar, S. Worldwide Protein Data Bank validation information: Usage and trends. Acta Cryst. 2018, 74, 237–244. [Google Scholar] [CrossRef]
- Dauter, Z.; Wlodawer, A.; Minor, W.; Jaskolski, M.; Rupp, B. Avoidable errors in deposited macromolecular structures: An impediment to efficient data mining. IUCrJ 2014, 1, 179–193. [Google Scholar] [CrossRef]
- Shabalin, I.; Dauter, Z.; Jaskolski, M.; Minor, W.; Wlodawer, A. Crystallography and chemistry should always go together: A cautionary tale of protein complexes with cisplatin and carboplatin. Acta Cryst. 2015, D71, 1965–1979. [Google Scholar] [CrossRef]
- Raczynska, J.E.; Wlodawer, A.; Jaskolski, M. Prior knowledge or freedom of interpretation? A critical look at a recently published classification of “novel” Zn binding sites. Proteins 2016, 84, 700–776. [Google Scholar] [CrossRef]
- Raczynska, J.; Shabalin, I.G.; Minor, W.; Wlodawer, A.; Jaskolski, M. A close look onto structural models and primary ligands of metallo-β-lactamases. Drug Resist. Updat. 2018, 40, 1–12. [Google Scholar] [CrossRef]
- Brezinski, D.; Kowiel, M.; Cooper, D.R.; Cymborowski, M.; Grabowski, M.; Wlodawer, A.; Dauter, Z.; Shabalin, I.G.; Gilski, M.; Rupp, B.; et al. Covid-19.bioreproducibility.org: A web resource for SARS-CoV-2-related structural models. Protein Sci. 2021, 30, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Jaskolski, M.; Dauter, Z.; Shabalin, I.G.; Gilski, M.; Brzezinski, D.; Kowiel, M.; Rupp, B.; Wlodawer, A. Crystallographic models of SARS-CoV-2 3CLpro: In-depth assessment of structure quality and validation. IUCrJ 2021, 8, 238–256. [Google Scholar] [CrossRef] [PubMed]
- Wlodawer, A.; Dauter, Z.; Shabalin, I.; Gilski, M.; Brezinski, D.; Kowiel, M.; Minor, W.; Rupp, B.; Jaskolski, M. Ligand-centered assessment of SARS-CoV-2 drug target models in the Protein Data Bank. FEBS J. 2020, 287, 3703–3718. [Google Scholar] [CrossRef]
- Wlodawer, A.; Dauter, Z.; Lubkowski, J.; Loch, J.; Brezinski, D.; Gilski, M. Towards a dependable dataset of structures for L-asparaginase research. Acta Cryst. 2024, D80, 506–527. [Google Scholar]
- Jaskolski, M.; Wlodawer, A.; Dauter, Z.; Minor, W.; Rupp, B. Group deposition to the Protein Data Bank need adequate presentation and different archiving protocol. Protein Sci. 2022, 31, 784–786. [Google Scholar] [CrossRef]
- Domagalski, M.J.; Zheng, H.; Zimmerman, M.D.; Dauter, Z.; Wlodawer, A.; Minor, W. The quality and validation of structures from structural genomics. Meth. Mol. Biol. 2014, 2091, 297–314. [Google Scholar]
- Djinovic-Carugo, K.; Carugo, O. Naked Metal Cations Swimming in Protein Crystals. Crystals 2019, 9, 581. [Google Scholar] [CrossRef]
- Rupp, B.; Wlodawer, A.; Minor, W.; Helliwell, J.R.; Jaskolski, M. Correcting the record of structural publications requires joint effort of the community and journal editors. FEBS J. 2016, 283, 4452–4457. [Google Scholar] [CrossRef]
- Wlodawer, A.; Dauter, Z.; Minor, W.; Stanfield, R.; Porebski, P.; Jaskolski, M.; Pozjarski, E.; Weichenberger, C.X.; Rupp, B. Detect, Correct, Retract: How to manage incorrect structural models. FEBS J. 2018, 285, 444–466. [Google Scholar] [CrossRef]
- Brown, I.D.; Wu, K.K. Empirical Parameters for Calculating Cation-Oxygen Bond Valences. Acta Cryst. 1975, B32, 1957–1959. [Google Scholar] [CrossRef]
- Carugo, O. Buried chloride stereochemistry in the protein data bank. BMC Struct. Biol. 2014, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Gucwa, M.; Bijak, V.; Zheng, H.; Murzyn, K.; Minor, W. CheckMyMetal (CMM): Validating metal-binding sites in X-ray and cryo-EM data. IUCrJ 2024, 11, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.M. The geometry of metal-ligand interactions relevant to proteins. Acta Cryst. 1999, D55, 1432–1443. [Google Scholar] [CrossRef]
- Harding, M.M. The geometry of metal-ligand interactions relevant to proteins. II. Angles at the metal atom, additional weak metal-donor interactions. Acta Cryst. 2000, D56, 857–867. [Google Scholar] [CrossRef]
- Harding, M.M. Geometry of metal-ligand interactions in proteins. Acta Cryst. 2001, D57, 401–411. [Google Scholar] [CrossRef]
- Harding, M.M. The architecture of metal coordination groups in proteins. Acta Cryst. 2004, D60, 849–859. [Google Scholar] [CrossRef]
- Harding, M.M. Small revisions to predicted distances around metal sites in proteins. Acta Cryst. 2006, D62, 678–682. [Google Scholar] [CrossRef]
- Harding, M.M.; Nowicki, M.W.; Walkinshaw, M.D. Metals in protein structures: A review of their principal features. Cryst. Rev. 2010, 16, 247–302. [Google Scholar] [CrossRef]
- Hsin, K.; Sheng, Y.; Harding, M.M.; Taylor, P.; Walkinshaw, M.D. MESPEUS: A database of the geometry of metal sites in proteins. J. Appl. Cryst. 2008, 41, 963–968. [Google Scholar] [CrossRef]
- Lin, G.-Y.; Su, Y.-C.; Huang, Y.L.; Hsin, K.-Y. MESPEUS: A database of metal coordination groups in proteins. Nucl. Acids Res. 2024, 52, D483–D493. [Google Scholar] [CrossRef]
- Mueller, P.; Koepke, S.; Sheldrick, G.M. Is the bond-valence method able to identify metal atoms in protein structures? Acta Cryst. 2003, D59, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Nayal, M.; Di Cera, E. Valence Screening of Water in Protein Crystals Reveals Potential Na+ Binding Sites. J. Mol. Biol. 1996, 256, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Bazayeva, M.; Andreini, C.; Rosato, A. A adatabase overview of meta-coordination distances in metalloproteins. Acta Cryst. 2024, D80, 362–376. [Google Scholar]
- Minor, W.; Dauter, Z.; Helliwell, J.R.; Jaskolski, M.; Wlodawer, A. Safeguarding Structural Data Repositories against Bad Apples. Structure 2016, 24, 216–220. [Google Scholar] [CrossRef][Green Version]
- Levi, P. If This Is a Man and the Truce; Abacus: London, UK, 2003. [Google Scholar]
- Levi, P. Se Questo e’ un Uomo; Einaudi: Turin, Italy, 2014. [Google Scholar]
- Carugo, O. Random sampling of the Protein Data Bank: RaSPDB. Sci. Rep. 2021, 11, 24178. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Anjanappa, R.; Garcia-Alai, M.; Kopicki, J.D.; Lockhauserbaumer, J.; Aboelmagd, M.; Hinrichs, J.; Nemtanu, I.M.; Uetrecht, C.; Zacharias, M.; Springer, S.; et al. Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection. Nat. Commun. 2020, 11, 1314. [Google Scholar] [CrossRef]
- Thomas, M.E.; Grinshpon, R.; Swartz, P.; Clark, A.C. Modifications to a common phosphorylation network provide individualized control in caspases. J. Biol. Chem. 2018, 293, 5447–5461. [Google Scholar] [CrossRef]
- Podvalnaya, N.; Bronkhorst, A.W.; Lichtenberger, R.; Hellmann, S.; Nischwitz, E.; Falk, T.; Karaulanov, E.; Butter, F.; Falk, S.; Ketting, R.F. piRNA processing by a trimeric Schlafen-domain nuclease. Nature 2023, 622, 402–409. [Google Scholar] [CrossRef]
- Rupp, B. Biomolecular Crystallography: Principles, Practice, and Application to Structural Biology; Garland Science: New York, NY, USA, 2010. [Google Scholar]
- Wlodawer, A.; Minor, W.; Dauter, Z.; Jaskolski, M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008, 275, 1–21. [Google Scholar] [CrossRef]
- Connick, W.B.; Henling, W.M.; Marsh, R.E. Revision of structure of (bipyridyl-N,N’)disyanoplatinum(II). Acta Cryst. 1996, B52, 817–822. [Google Scholar] [CrossRef]
- Marsh, R.E. P1 or P-1? Or something else? Acta Cryst. 1999, B55, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Marsh, R.E. Space groups P1 and Cc: How are they doing? Acta Cryst. 2009, B65, 782–783. [Google Scholar] [CrossRef]
- Meng, A.Q.; Diment, L.A.; Abdi, A.; Hubbs, V.J.; Jeffreys, E.A.; O’Dell, M.; Ou, X.; Park, K.A.; Quillin, B.T.; Dickie, D.A. Using data from the Cambridge Structural Database to practice crystallographic skills and revise erroneous structures. Cryst. Growth Des. 2024, 24, 4690–4696. [Google Scholar] [CrossRef]
- Thompson, A.J.; Whittaker, J.J.; Brock, A.J.; Baanoon, H.A.; Sankalpa, A.-F.K.; Arachichage, A.; Pfunder, M.C.; McMurtrie, J.C. Is a crystal structure enough? Reflecting on the reliability of SCXRD in a age of automation. Cryst. Growth Des. 2024, 24, 5349–5354. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carugo, O. Identifications of False Positives Amongst Sodium(I) Cations in Protein Three-Dimensional Structures—A Validation Approach Extendible to Any Alkali or Alkaline Earth Cation and to Any Monoatomic Anion. Crystals 2024, 14, 918. https://doi.org/10.3390/cryst14110918
Carugo O. Identifications of False Positives Amongst Sodium(I) Cations in Protein Three-Dimensional Structures—A Validation Approach Extendible to Any Alkali or Alkaline Earth Cation and to Any Monoatomic Anion. Crystals. 2024; 14(11):918. https://doi.org/10.3390/cryst14110918
Chicago/Turabian StyleCarugo, Oliviero. 2024. "Identifications of False Positives Amongst Sodium(I) Cations in Protein Three-Dimensional Structures—A Validation Approach Extendible to Any Alkali or Alkaline Earth Cation and to Any Monoatomic Anion" Crystals 14, no. 11: 918. https://doi.org/10.3390/cryst14110918
APA StyleCarugo, O. (2024). Identifications of False Positives Amongst Sodium(I) Cations in Protein Three-Dimensional Structures—A Validation Approach Extendible to Any Alkali or Alkaline Earth Cation and to Any Monoatomic Anion. Crystals, 14(11), 918. https://doi.org/10.3390/cryst14110918





