Abstract
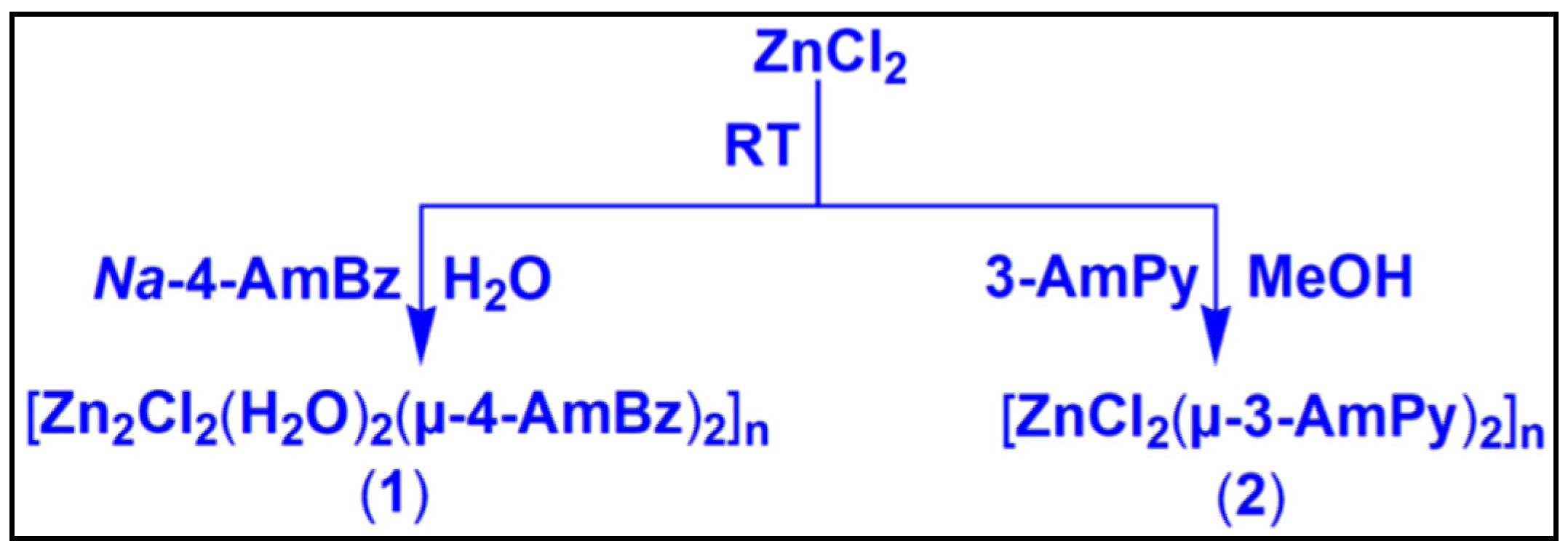
Two Zn(II) coordination polymers, viz., [Zn2Cl2(H2O)2(µ-4-AmBz)2]n (1) and [ZnCl2(µ-3-AmPy)2]n (2) (4-AmBz = 4-aminobenzoate, 3-AmPy = 3-aminopyridine) have been prepared at room temperature and characterized using elemental analysis, FT-IR, electronic spectroscopy, TGA (thermogravimetric analysis) and single crystal XRD. Crystal structure analyses of the polymers unfold the presence of non-covalent anion–π, π-stacking and unusual NH2(amino)⋯π interactions which provide rigidity to the crystal structures. Unconventional Type I Cl⋯Cl interactions also play a pivotal role in the stability of compound 1. Molecular electrostatic potential (MEP) surface analysis reveals that the MEP values over the center of the aromatic rings of coordinated 4-AmBz and 3-AmPy moieties are positive on one side and negative on the other side which confirms the dual non-covalent donor-acceptor topologies of the aromatic rings and explains the concurrent formation of unusual non-covalent NH2···π and anion–π interactions. DFT (density functional theory) calculations, QTAIM (quantum theory of atoms in molecules) and NCI plot (non-covalent index) index analyses reveal that among various non-covalent contacts involved in the crystal packing of the compounds, H-bonds in compound 1 and π-interactions (NH2···π, π-π, anion–π) in compound 2 are energetically significant. We have explored in vitro cytotoxic potential of the compounds in Dalton’s lymphoma (DL) cancer cells using trypan blue and apoptosis assays. The studies show that compounds 1 and 2 can significantly exhibit cytotoxicity in DL cells with minimum cytotoxicity in healthy PBMC cells. Molecular docking studies reveal that the compounds effectively bind with the antiapoptotic target proteins; thereby establishing a structure activity relationship of the compounds.
1. Introduction
Coordination compounds with organic bridging ligands have gained appreciable research interest not only due to their intriguing structural topologies but also because of their myriad potential applications in biological systems, luminescence, catalysis, drug development, non-linear optics, magnetism, etc. [1,2,3,4,5]. Looking at the potential applications of poly-nuclear compounds, various coordination compounds with one-dimensional polymeric structures have been developed [6,7]. However, it is still a difficult task for the researchers to design the architectures with desired structural topologies as the self-assembly of molecular entities mainly depends on a variety of experimental conditions, viz., metal-to-ligand ratio, pH of the medium, solvents, temperature, and coordination geometry of the metal centers, among others [8].
Studies on non-covalent interactions such as hydrogen bonding, electrostatic and charge-transfer interactions, π-stacking and metal ion coordination, etc. have attracted researchers from the crystal engineering viewpoint [9,10,11]. Hydrogen bonding—one of the commonly observed non-covalent interactions—can effectively control the secondary coordination sphere of metal ions [12,13]. Supramolecular non-covalent contacts involving the aromatic π-systems have also played important roles in the stability of the crystal structures [14]. It has been well established that metal-coordinated organic moieties with aromatic rings can be either electron-rich (donor) or electron-deficient (acceptor) to direct molecular self-assemblies with diverse structural topologies [15,16]. However, dual non-covalent donor-acceptor topologies of aromatic rings of metal-coordinated organic ligands are still only scarcely explored in the literature. Cl⋯Cl interaction has also emerged as a potential non-covalent interaction which can also be termed a donor-acceptor interaction [17]. Moreover, unusual NH2⋯π type non-covalent interactions have also gained interest from researchers in the field of supramolecular chemistry [18]. Experimental analysis aided by computational studies is important to visualize such unusual non-covalent interactions which can be used for engineering crystal structures with desired physical and chemical properties.
Aromatic organic donors with carboxyl groups can be used as potential candidates to develop coordination compounds with multi-dimensional assemblies because of the flexible coordination motifs [19,20]. Pyridine-based donors have also prompted immense research interest in coordination chemistry due to their potential applications in various fields [21,22]. Coordination compounds of aminopyridines have shown a wide range of biological applications [23]. Zinc is the second most abundant human body trace element which can protect against tissue damage [24,25]. Zinc plays a key role in cell proliferation as well as in DNA/RNA synthesis; hence, a deficiency of zinc can cause depression in the immune system [26]. Coordination compounds of zinc involving benzoate and substituted benzoate derivatives have been reported to possess interesting structural topologies [27,28]. Zinc compounds have also been reported to show significant pharmacological properties such as anticancer [29], antidiabetic [30], anti-inflammatory [31], antimicrobial [32,33,34,35], antioxidant [36], and in Alzheimer’s disease [37]. There are several reports of anticancer activities of coordination compounds against DL cancer cells [38,39]. Jayendran and Kurup have reported a Zn(II) coordination compound involving NNO donor Schiff base ligand and explored its significant in vitro anticancer activity in the DL cancer cell line [40].
To explore the self-assembly, unconventional structural topologies, and anticancer properties of metal-organic polymers; in the present study, we have reported the synthesis and crystal structures of two Zn(II) coordination polymers, viz., [Zn2Cl2(H2O)2(µ-4-AmBz)2]n (1) and [ZnCl2(µ-3-AmPy)2]n (2). The compounds have been further characterized using spectroscopic techniques (FT-IR and electronic), elemental analysis, and TGA. Crystal structure analysis of compound 1 reveals the presence of Type I Cl⋯Cl and unusual NH2(amino)⋯π interactions which stabilize the crystal structure. Similarly, NH2(amino)⋯π, anion⋯π, and π-π interactions stabilize the 2D assembly of compound 2. We have explored the energetic features of the unusual supramolecular assemblies observed in compounds 1 and 2 using DFT studies. The interactions have been further characterized by using several computational tools such as molecular electrostatic potential (MEP) surface, non-covalent index (NCI) plot, and quantum theory of atoms in molecules (QTAIM). The cytotoxic potential of the compounds has been investigated using Trypan blue and apoptosis assays in the DL cancer cell line. Molecular docking studies of the compounds were performed to explore the possible interactions of the compounds with the target proteins which are related to cancer growth and progression.
2. Experimental Section
2.1. Materials and Methods
The chemicals required for the synthesis of the compounds, viz., anhydrous zinc(II) chloride, 4-aminobenzoic acid, and 3-aminopyridine were bought from Sigma Aldrich and used as received. The single crystal X-ray data of the compounds were recorded using D8 Venture diffractometer, having a Photon III 14 detector and Incoatec high brilliance IμS DIAMOND Cu tube equipped with an Incoatec Helios MX multilayer optics. The data collection of the compounds was carried out at 100 K. The elemental analysis of the compounds was performed using Perkin Elmer 2400 Series II CHNS/O analyzer. We have recorded the KBr phase FT-IR spectra of the compounds using Bruker alpha (II) infrared spectrophotometer in the frequency range 4000–500 cm−1. Electronic spectra of the compounds were recorded using Shimadzu UV-2600 spectrophotometer. BaSO4 powder was used as reference to record the UV-Vis-NIR spectra. 1H-NMR spectra of the compounds were recorded using Avance III HD 400 NMR spectrometer with DMSO-d6 as solvent and tetramethylsilane (TMS) as the chemical shift reference. Thermal studies of the compounds were carried out using Mettler Toledo TGA/DSC1 STARe system with the flow of N2 gas at the heating rate of 10 °C min−1.
2.2. Syntheses
2.2.1. Synthesis of [Zn2Cl2(H2O)2(µ-4-AmBz)2]n (1)
Anhydrous ZnCl2 (0.136 g, 1 mmol) and sodium salt of 4-aminobenzoic acid (0.159 g, 1 mmol) were mixed in 10 mL of de-ionized water and mechanically stirred at room temperature for two hours (Scheme 1). After two hours, the colourless resulting solution was kept unperturbed in cooling conditions (2–4 °C) for crystallization. After a few days, block-shaped colorless crystals suitable for single-crystal XRD were obtained from the mother liquor. Yield: 0.444 g (87%). Anal. calcd. for C14H16N2O6Cl2Zn2: C, 32.97%; H, 3.16%; N, 5.49%; Found: C, 32.88%; H, 3.07%; N, 5.39%. FT-IR (KBr pellet, cm−1): 3448(br), 2077(w), 1621(s), 1428(w), 1388(s), 1356(sh), 1220(w), 1181(w), 1141(w), 1067(m), 862(w), 783(m), 699(w), 679(w) (s, strong; m, medium; w, weak; br, broad; sh, shoulder).
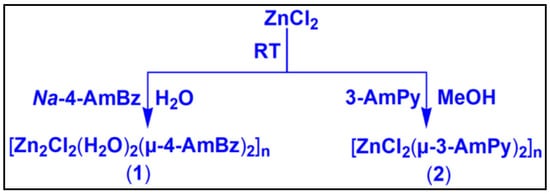
Scheme 1.
Synthesis of compounds 1 and 2.
2.2.2. Synthesis of [ZnCl2(µ-3-AmPy)2]n (2)
A mixture of anhydrous ZnCl2 (0.136 g, 1 mmol) and 3-aminopyridine (0.188 g, 2 mmol) was dissolved in methanol (10 mL) and mechanically stirred at room temperature for two hours (Scheme 1). The resulting colorless solution was then kept unperturbed in a refrigerator (below 4 °C) for crystallization. After several days, colorless single crystals were obtained from the mother liquor. Yield: 0.196 g (85%). Anal. calcd. for C5H6N2Cl2Zn: C, 26.06%; H, 2.62%; N, 12.16%; Found: C, 25.97%; H, 2.57%; N, 12.08%. FT-IR (KBr pellet, cm−1): 3448(br), 3258(w), 3129(w), 2108(w), 1639(s), 1555(sh), 1484(w), 1454(m), 1408(m), 1339(w), 1255(w), 1195(w), 1097(w), 1057(m), 1029(sh), 905(w), 816(w), 692(m), 651(m)
2.3. Crystallographic Data Collection and Refinement
The selected single crystals of 1 and 2 were covered with Parabar 10,320 (formally known as Paratone N) and mounted on a cryoloop on a D8 Venture diffractometer, with a Photon III 14 detector, using an Incoatec high brilliance IμS DIAMOND Cu tube equipped with an Incoatec Helios MX multilayer optics. The data collections of the compounds were carried out at 100 K. Data reduction and cell refinements were performed using the Bruker APEX3 program [41]. SADABS program was used for scaling and absorption corrections in all cases [41]. The crystal structures of the compounds were solved by direct method and refined by full-matrix least-squares techniques with SHELXL-2018/3 [42] using WinGX [43] software. All non-hydrogen atoms were refined with anisotropic thermal parameters by full-matrix least-squares calculations on F2. Hydrogen atoms were inserted at calculated positions and refined as riders. The structures were checked for higher symmetry with the help of the program PLATON [44]. The graphical materials have been prepared with the help of Mercury software [45]. Diamond 3.2 software is used to draw the molecular structures and the packing diagrams [46]. Collected data and refinement parameters for compounds 1 and 2 are summarized in Table 1.

Table 1.
Crystallographic data and structure refinement details for compounds 1 and 2.
CCDC 2123028 and 2123034 contain the supplementary crystallographic data for compounds 1 and 2, respectively. These data can be obtained free of charge at http://www.ccdc.cam.ac.uk (accessed on 22 February 2023) or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223-336-033; or E-mail: deposit@ccdc.cam.ac.uk.
2.4. Computational Methods
The theoretical study reported herein was performed using RI-BP86-D3/def2-TZVP [47,48] level of theory. For the calculations, the X-ray coordinates were used by means of the program Turbomole 7.2 [49] since we are interested in evaluating the noncovalent contacts as they stand in the solid state. The NCI plot [50] via reduced density gradient (RGD) isosurfaces and QTAIM [51] methods were used to characterize the non-covalent interactions at the same level of theory. The MULTIWFN program [52] was for the calculation of the NCIplot isosurfaces and critical points. They were represented using the VMD software [53].
2.5. Cell Line and Drug Preparation
To explore in vitro anticancer activities of the compounds, Dalton’s lymphoma (DL) cancer cells were used. DL is a malignant, transplantable T-cell lymphoma cell line; which has been effectively used in cancer research [54]. When choosing a cell line as the model system; the cell line’s genetic stability and heterogeneity, host animal immunogenicity, and biological endpoints are always considered. In this regard, Dalton’s lymphoma has evolved as an excellent model system where active components of a large number of natural plant products have been studied with biological endpoints [55]. It has been well established that peripheral blood mononuclear cells (PBMCs) are potential cell models that can be used to screen and investigate the effects of different synthesized compounds or drug molecules in the context of cancer research [56]. To explore the cytotoxicity of the compounds in normal cells; peripheral blood mononuclear cells (PBMC) were used. RPMI media was used to grow the DL cells which are used with a blood product, 10% FBS, streptomycin (100 g/mL), and penicillin (100 U/mL). The media was kept at 37 °C in CO2 incubator with 5% CO2. 50 mg of the compounds was dissolved in 1 mL of phosphate-buffered saline (PBS)/dimethyl sulfoxide (DMSO) (pH = 7.4) to prepare the stock solution of the compounds and then diluted to prepare the required solutions (0.01, 0.1, 0.5, 1, 5 and 10 μM).
2.6. Cytotoxicity and Apoptosis Assays
Trypan blue exclusion assay was performed in DL cancer cell line to evaluate the cytotoxic potential of the synthesized compounds [57]. Trypan blue dye is unable to penetrate the cell membrane of living cells but can go inside dead cells having damaged cell membranes [58]. Various concentrations (0.01, 0.1, 0.5, 1, 5, and 10 μM) of the compounds were used to explore the short-term (24 h) cytotoxic potential in 96 cell culture plates (Thermo Scientific, Cat. No: 265301). IC50 (concentration required for 50% cell death) values of the compounds in DL and PBMC cell lines were determined using the dose response-linear curve fit method. For DL and PBMC cell lines; 0.01–100 and 0.01–400 μM concentration ranges were used, respectively. The non-linear curve fit function that was used for the measurement of the IC50 is as follows:
y = A1 + (A2 − A1)/(1 + 10^((LOGx0-x)*p))
(where A1 = bottom asymptote, A2 = top asymptote, LOGx0 = centre and p = hill slope).
Acridine orange/ethidium bromide (AO/EB) fluorescence-based dual staining method was used to evaluate the apoptotic cell death induced by the compounds [59]. AO can go inside the intact cell membrane of a living cell and stain it green; whereas, EB can only color the apoptotic cells red/orange. After 24 h of treatment with the compounds, the cells were stained with equimolar solution of AO/EB (sigma, St. Louis, MO; 1 μL of the mixture containing 100 μg/mL of AO and EB each) and kept in dark for 3 min by covering with cover slip. Viable and apoptotic cells can be recognized as green and red/orange when observed under high-resolution microscope [60]. About 1000 cells were photographed under fluorescence microscope and the percentage of apoptosis has been determined.
2.7. Molecular Docking Simulation
Molecular docking is a theoretical approach that can be effectively used to predict the possible interactions of a target protein with molecules/drugs/compounds under investigation in order to dissect their possible mode of action [61]. In the present study, the molecular docking simulation was carried out for compounds 1 and 2 with the antiapoptotic cancer target proteins, viz., BCL-2 (PDB ID = 2O22) and BCL-XL (PDB ID = 2YXJ) using molegro virtual docker (MVD 2010.4.0) software [62]. The docking parameters were run using standard parameters [63]. After molecular docking simulation; the analysis of the protein compounds was performed using chimera (https://www.cgl.ucsf.edu/chimera/ (accessed on 22 February 2022)) and discovery studio visualization-BIOVIA software (https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/ (accessed on 22 February 2022)) [64].
3. Results
3.1. Syntheses and General Aspects
[Zn2Cl2(H2O)2(µ-4-AmBz)2]n(1) has been prepared by reacting zinc(II) chloride and sodium salt of 4-aminobenzoic acid in 1:1 molar ratio at room temperature in water. [ZnCl2(µ-3-AmPy)2]n (2) has been synthesized from the reaction between zinc chloride and 3-aminopyridine in a 1:2 molar ratio under similar conditions in methanol. Both compounds are fairly soluble in water and in common organic solvents. The crystal structure of compound 2 has been re-determined with slightly different unit cell parameters with an improved goodness of fit (S) value (Table S1) from those of the previously reported structure [65]. We have performed a thorough structural investigation of the compound which unfolds non-covalent anion–π, π-stacking, and unusual NH2(amino)⋯π interactions which stabilize the crystal packing (vide infra). We have also carried out computational studies to corroborate the presence of these supramolecular contacts in the crystal structure (vide infra).
3.2. Crystal Structure Analysis
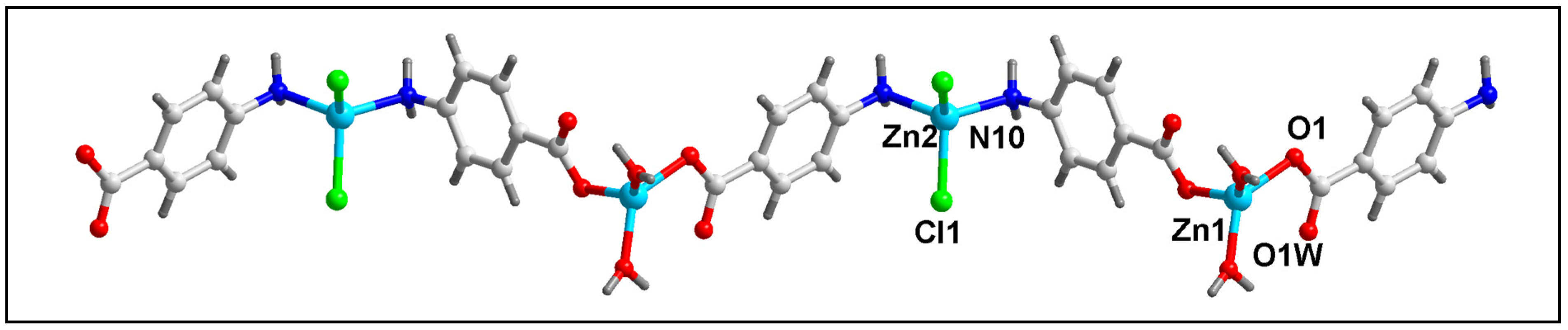
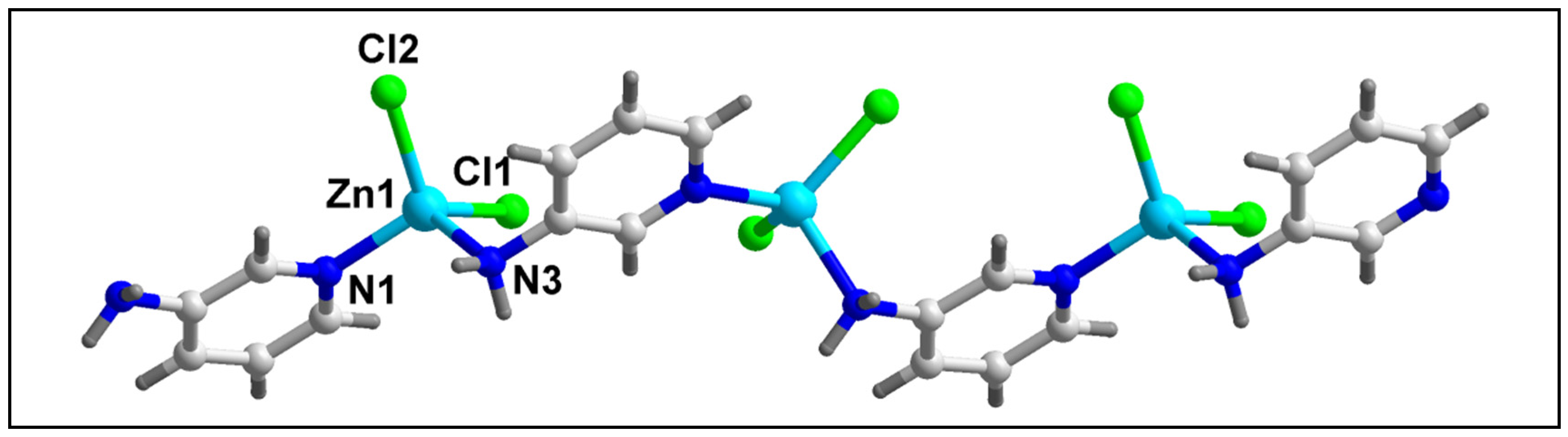
Figure 1 represents the molecular structure of compound 1. Table 2 contains the bond lengths and the bond angles around the central Zn(II) metal centers. Compound 1 crystallizes in the monoclinic P2/c space group. The polymeric chain of compound 1 contains two different Zn(II) centers (Zn1 and Zn2) with different coordination environments. The Zn1 center is tetra-coordinated with two coordinated water (O1W and O1W’) and two O-atoms (O1 and O1’) of the two bridging 4-AmBz ligands. Similarly, the Zn2 center is also tetra-coordinated with two Cl and two N-atoms (N10 and N10’) of –NH2 groups of the two bridging 4-AmBz moieties. Both Zn1 and Zn2 centers have nearly tetrahedral geometries with the corresponding bond angles ranging from 90.2 to 108.2°. The average metal-ligand bond lengths are comparable to the previously reported Zn(II) compounds [66]. The Zn1–Zn1, Zn1–Zn2, and Zn2–Zn2 distances in the molecular structure of compound 1 are found to be 16.323, 9.139, and 16.323 Å, respectively.
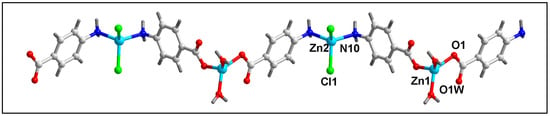
Figure 1.
Molecular structure of [Zn2Cl2(H2O)2(µ-4-AmBz)2]n (1).

Table 2.
Selected bond lengths (Å) and bond angles (deg.) of Zn(II) centers in 1 and 2.
Intramolecular O–H⋯O hydrogen bonding interactions are observed in the polymeric chain of compound 1 which stabilize the crystal structure (Figure S1). Uncoordinated O3 atoms of the bridging 4-AmBz and –OH (O1WH1WA) moiety of the coordinated water molecule are involved in O–H⋯O hydrogen bonding interactions having O1W–H1WA⋯O3 distance of 2.63 Å (Table 3).

Table 3.
Selected parameters for hydrogen bonding interactions in compounds 1 and 2.
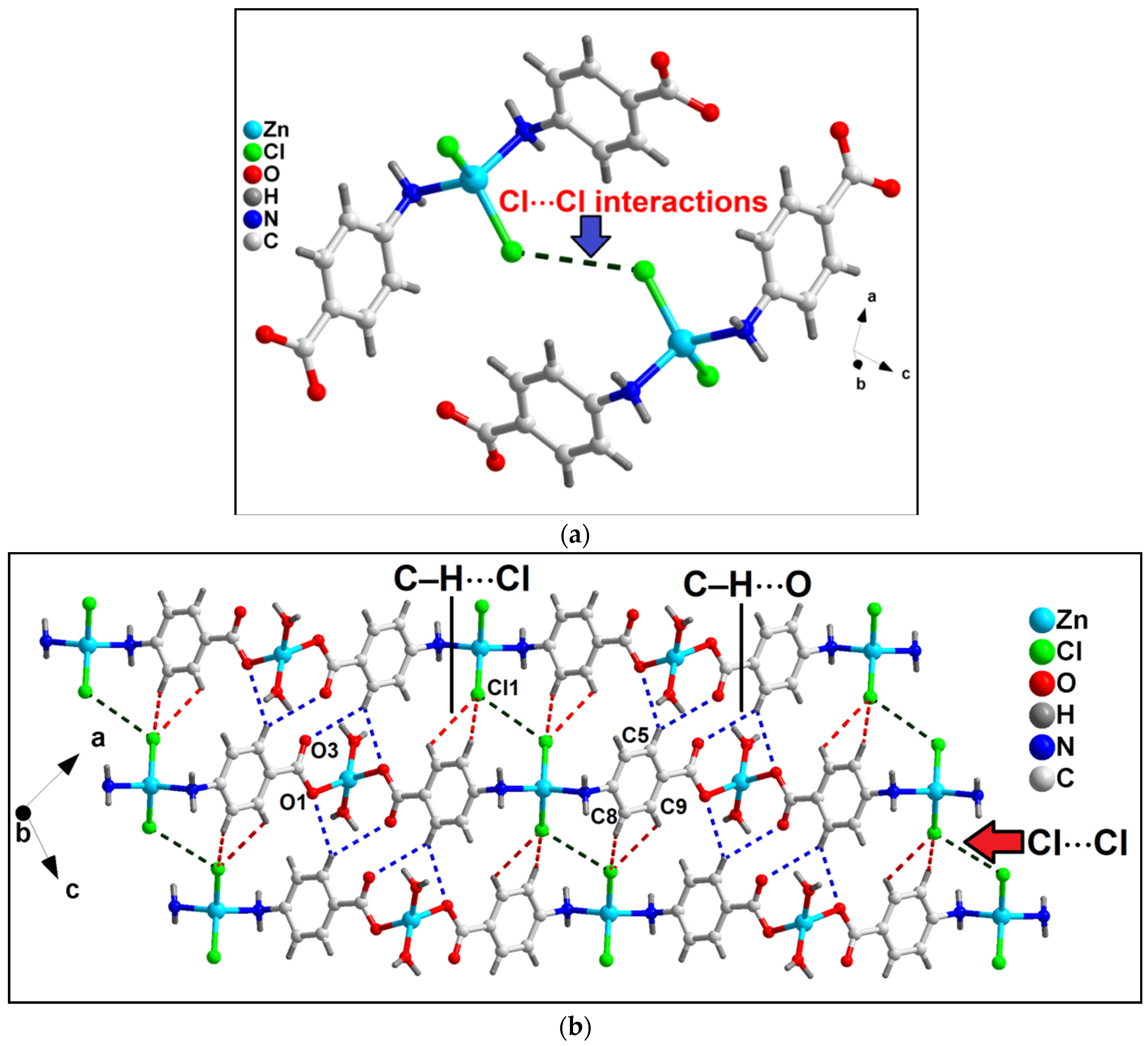
In the crystallographic ac plane, the neighboring 1D chains of compound 1 are interconnected through C–H⋯O and C–H⋯Cl hydrogen bonding and Type I Cl⋯Cl interactions to form the layered assembly [67] (Figure 2b). C–H⋯O hydrogen bonding interactions are observed involving the–CH moiety (–C5H5) of 4-AmBz and the coordinated O1 and uncoordinated O3 atoms of the bridging 4-AmBz having C5–H5⋯O1 and C5–H5⋯O3 distances of 2.73 and 2.84 Å, respectively. Cl1 ion and –CH moieties are involved in C–H⋯Cl interactions with the C8–H8⋯Cl1 and C9–H9⋯Cl1 distances of 3.05 and 3.02 Å, respectively.
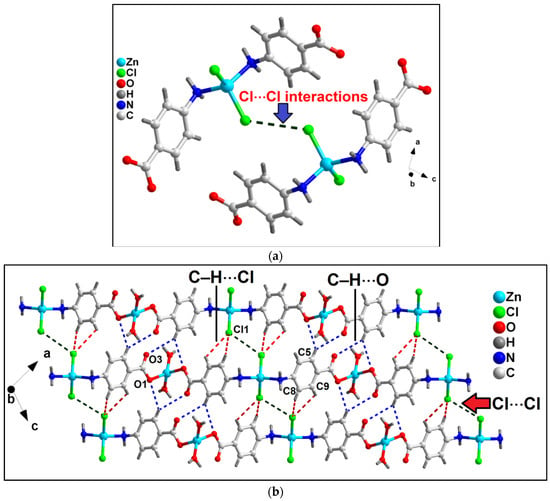
Figure 2.
(a) Unconventional Type I Cl⋯Cl interactions observed in compound 1; (b) 2D network architecture of compound 1 along the crystallographic ac plane aided by intermolecular C–H⋯O hydrogen bonding and Cl⋯Cl interactions.
Moreover, Cl⋯Cl interaction is also observed involving the Cl atoms (Cl1) having Cl1⋯Cl1 separation of 3.68 Å (Figure 2a). Usually, such intermolecular C–X1⋯X2–C contacts (X = F, Cl, Br, I) can be classified into two types depending on the corresponding angles, θ1 = ∠C–X1⋯X2 and θ2 = ∠X1⋯X2–C.
The interactions θ1 = θ2 are called Type I; whereas θ1 ≠ θ2 belongs to Type II contacts [68]. The Cl⋯Cl interactions observed in the crystal packing of 1 can be considered Type I with the corresponding angles of 127.1°. Li et al. have explored similar Cl⋯Cl interactions in a coordination polymer of Mn(II), viz., [Mn(tcpa)2(bipy)]n (where Htcpa = 3,5,6-trichloropyridine-2-oxyacetic acid and bipy = 2,2′-bipyridine) [69]. We have further used computational tools to support the presence of the interactions (vide infra).
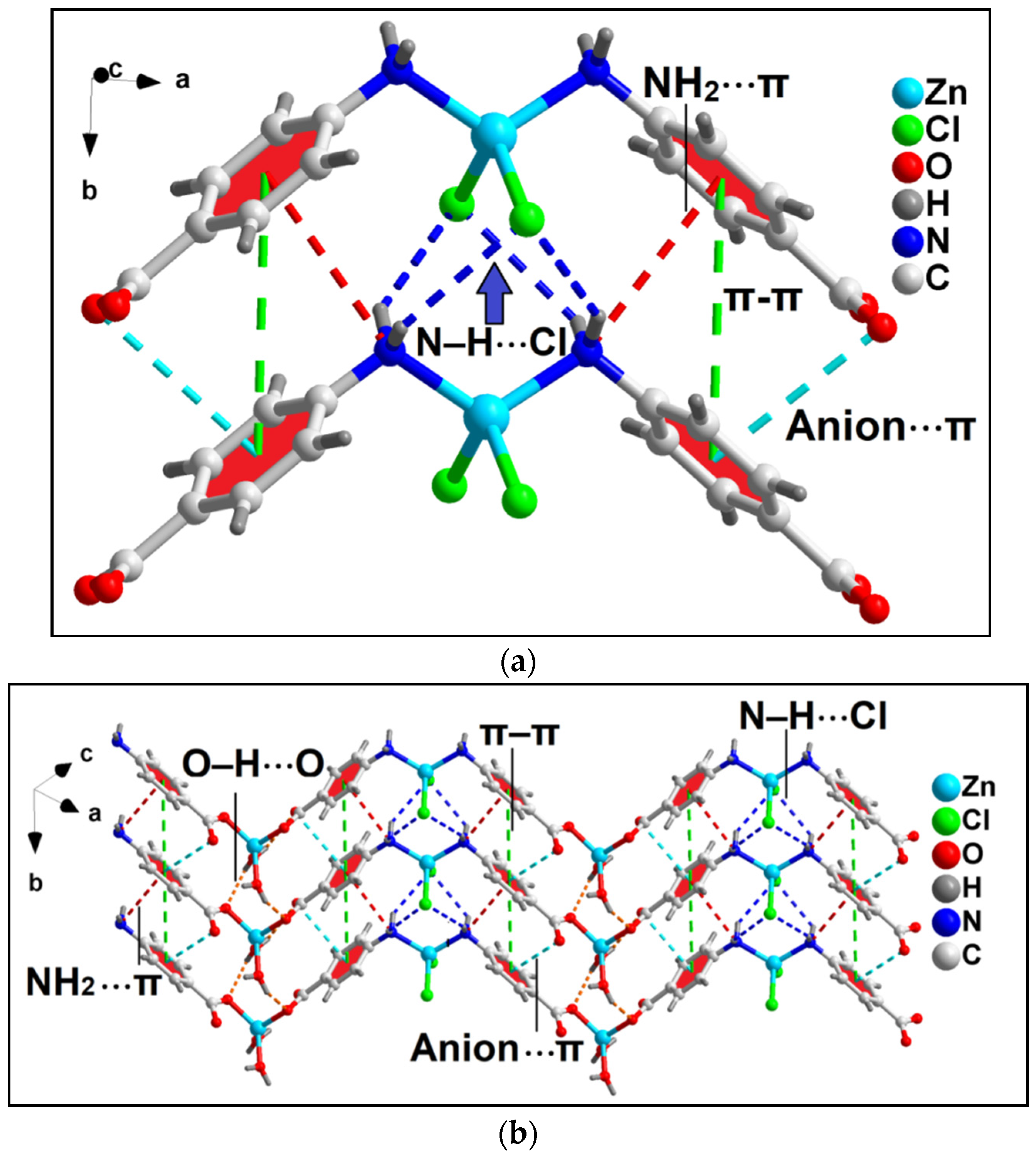
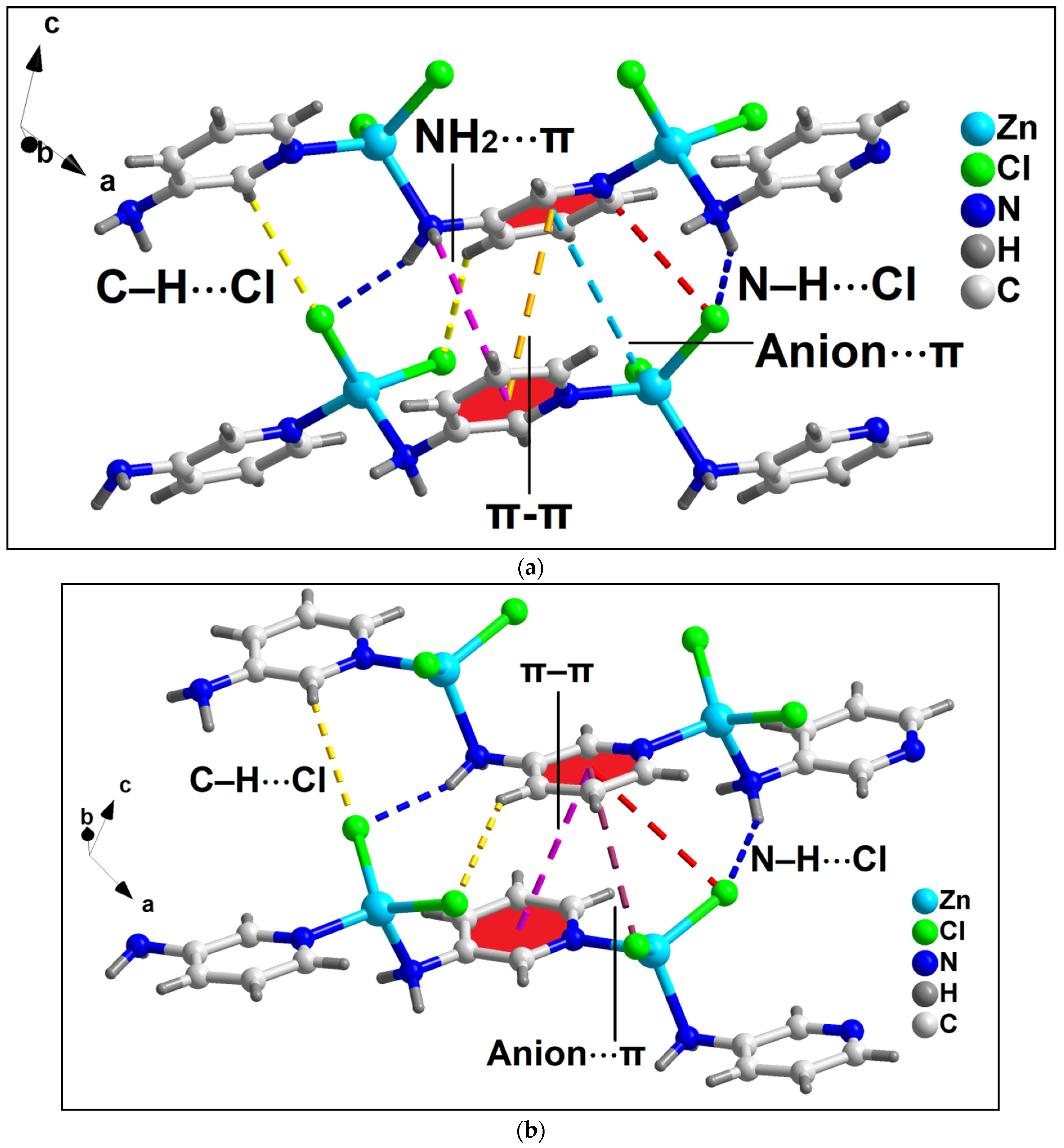
Unusual NH2(amino)⋯π, π–π and anion⋯π interactions involving the aromatic ring of coordinated 4-AmBz ligand also stabilize the crystal structure of compound 1 (Figure 3a). NH2(amino)⋯π interactions are observed having N⋯Cg (Cg is the ring centroid defined by the atoms C4-C9) distance of 3.67 Å. Anion⋯π contacts are observed involving the uncoordinated O3 atom of bridging 4-AmBzmoiety and the aromatic ring of 4-AmBz with the O3⋯Cg distance of 3.68 Å (where Cg is the ring centroid defined by the atoms C4-C9). The angle of 95.2° involving O3, Cg, and the aromatic plane reveals the strong nature of the interaction.
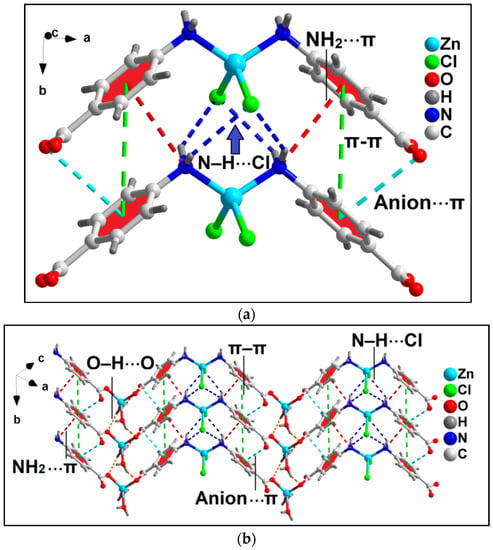
Figure 3.
(a) Model dimeric assembly in the crystal structure of the polymer 1 involving anion⋯π, NH2(amino)⋯π, π–π, and N–H⋯Cl interactions; (b) 2D network architecture of compound 1 along the crystallographic ab plane.
In addition, π–stacking contacts are also observed involving the neighboring aromatic 4-AmBz moieties with the neighboring C-C (C4–C7) separation of 3.58 Å. The dihedral angle between the aromatic rings is found to be 0.03°; whereas the slipped angle (angle between the ring normal and the vector joining the ring centroids) is found to be 17.3°. The perpendicular distance between the two aromatic rings is found to be 3.87 Å. These NH2(amino)⋯π, anion⋯π, and π-π interactions along with the O–H⋯O and N–H⋯Cl hydrogen bonding interactions stabilize the 2D architecture of the compounds along the crystallographic ab plane (Figure 3b). O–H⋯O interactions are observed involving the O1 atom of the bridging 4-AmBz and –OH moiety (–O1WH1WB) of the coordinated water molecule having O1W–H1WB⋯O1 distance of 2.11 Å. In addition, N–H⋯Cl interactions are also observed between the –NH fragments (–N10H10B and –N10H10A) and Cl1 moiety having N10–H10B⋯Cl1 and N10–H10A⋯Cl1 distances of 2.69 and 2.86 Å, respectively.
The molecular structure of compound 2 has been depicted in Figure 4. Table 2 contains the bond lengths and bond angles around the central Zn(II) center. Compound 2 crystallizes in the monoclinic Cc space group. Compound 2 contains one Zn(II) center, two bridging 3-AmPy ligands, and two Cl atoms. The Zn(II) center in polymer 2 has a nearly tetrahedral coordination geometry which is formed by two Cl atoms and two N atoms of 3-AmPy, having bond angles in the range of 102.01 to 119.8°. Zn–Cl bond lengths are in the range from 2.231 to 2.236 Å; whereas the Zn–N bond lengths are in the range of 2.025 to 2.086 Å (see Table 2) [70,71]. The adjacent Zn(II) centers of compound 2 are separated by a distance of 6.181 Å.
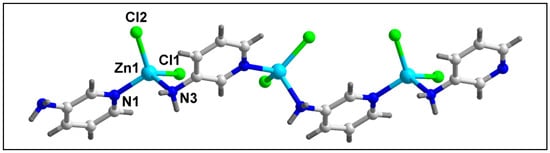
Figure 4.
Molecular structure of [ZnCl2(µ-3-AmPy)2]n (2).
The presence of carboxyl, pyridine N-atom, amino groups, and aromatic rings in compounds 1 and 2 may facilitate hydrogen bond donors, acceptors, and hydrophobic regions during the compound’s interaction with target proteins. H-bond donors and acceptors are the key pharmacophore features in anticancer active molecules/complexes as they are effectively involved in H-bonding interactions with the active sites of antiapoptotic proteins; thereby inhibiting the activities of antiapoptotic proteins [72]. Hydrophobic regions present in a molecule can efficiently inhibit the polymerization and repair of DNA of cancerous cells [73]. Therefore, we have performed molecular docking studies of the compounds (vide infra) to investigate the possible interactions of the compounds with antiapoptotic target proteins.
In compound 2, intramolecular C–H⋯Cl hydrogen bonding interactions are observed which stabilize the 1D polymeric chain along the crystallographic a axis (Figure S2). Intra-molecular C–H⋯Cl contact is observed between –CH (–C6H6) moiety of 3-AmPy and Cl atom (Cl1) having C6–H6⋯Cl1 distance of 2.99 Å.
Anion⋯π, NH2⋯π, and π–stacking interactions in addition to C–H⋯Cl and N–H⋯Cl contacts play decisive roles in the stability of the 2D architecture along the ac plane (Figure 5 and Figure 6). The details of the C–H⋯Cl and N–H⋯Cl interactions observed in the 2D assembly have been tabulated in Table 3. The coordinated Cl1 and Cl2 atoms are involved in anion⋯π interactions with C5 and C6 atoms of bridging 3-AmPy moiety having C5⋯Cl2 and C6⋯Cl1 distances of 3.67 and 3.53 Å, respectively. NH2(amino)⋯π interactions are observed in the layered assembly of the compound with NH2(amino)⋯C5 separation of 3.56 Å. In addition, π–π interactions are also observed between the aromatic rings of neighboring 3-AMpy with the nearest C-C (C3–C6) separation of 3.60 Å. For this π-stacking interaction, dihedral and slipped angles are found to be 0.04° and 19.1°, respectively. The perpendicular distance between the two aromatic rings is found to be 3.89 Å. These interactions (shown in the model dimeric assembly; Figure 5a) have been further studied theoretically (vide infra). Similar supramolecular contacts have been obtained in another model dimeric assembly (Figure 5b) having minor differences in separation distances (C4–H4⋯Cl2 = 2.86 Å; N3–H3A⋯Cl2 = 2.50 Å; N3–H3B⋯Cl1 = 2.69 Å; C5⋯Cl1 = 3.63 Å; C6⋯Cl2 distances of 3.62 Å; C3–C6 = 3.60 Å) (Figure 6).
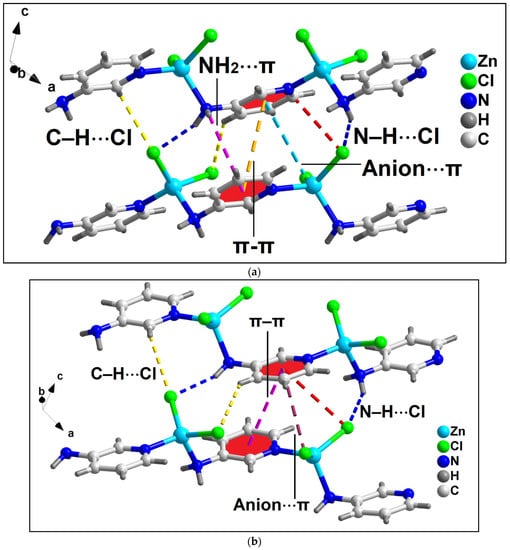
Figure 5.
Two partial views (a,b) of the layered assembly of compound 2 along the crystallographic ac plane aided by anion⋯π, NH2(amino)⋯π, π–π, N–H⋯Cl, and C–H⋯Cl interactions.
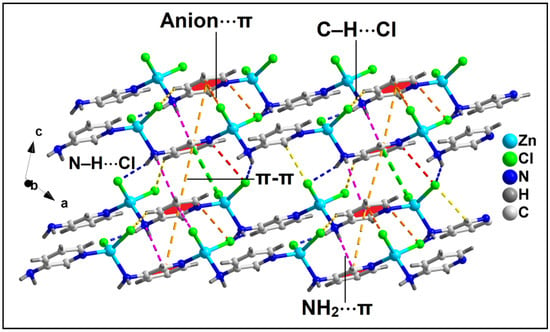
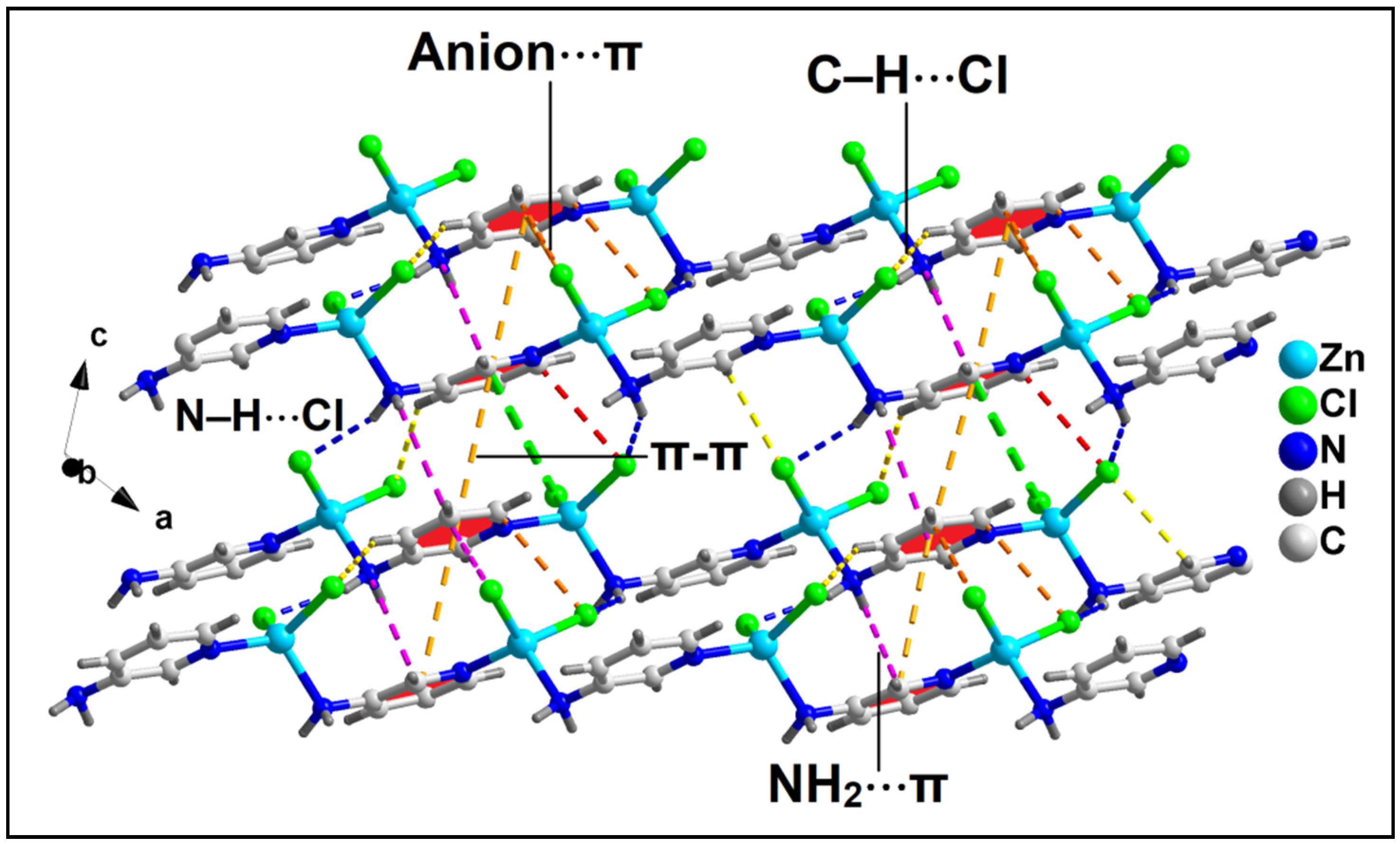
Figure 6.
Layered assembly of compound 2 along the crystallographic ac plane involving anion⋯π, NH2⋯π, and π–π interactions along with C–H⋯Cl and N–H⋯Cl hydrogen bonding interactions.
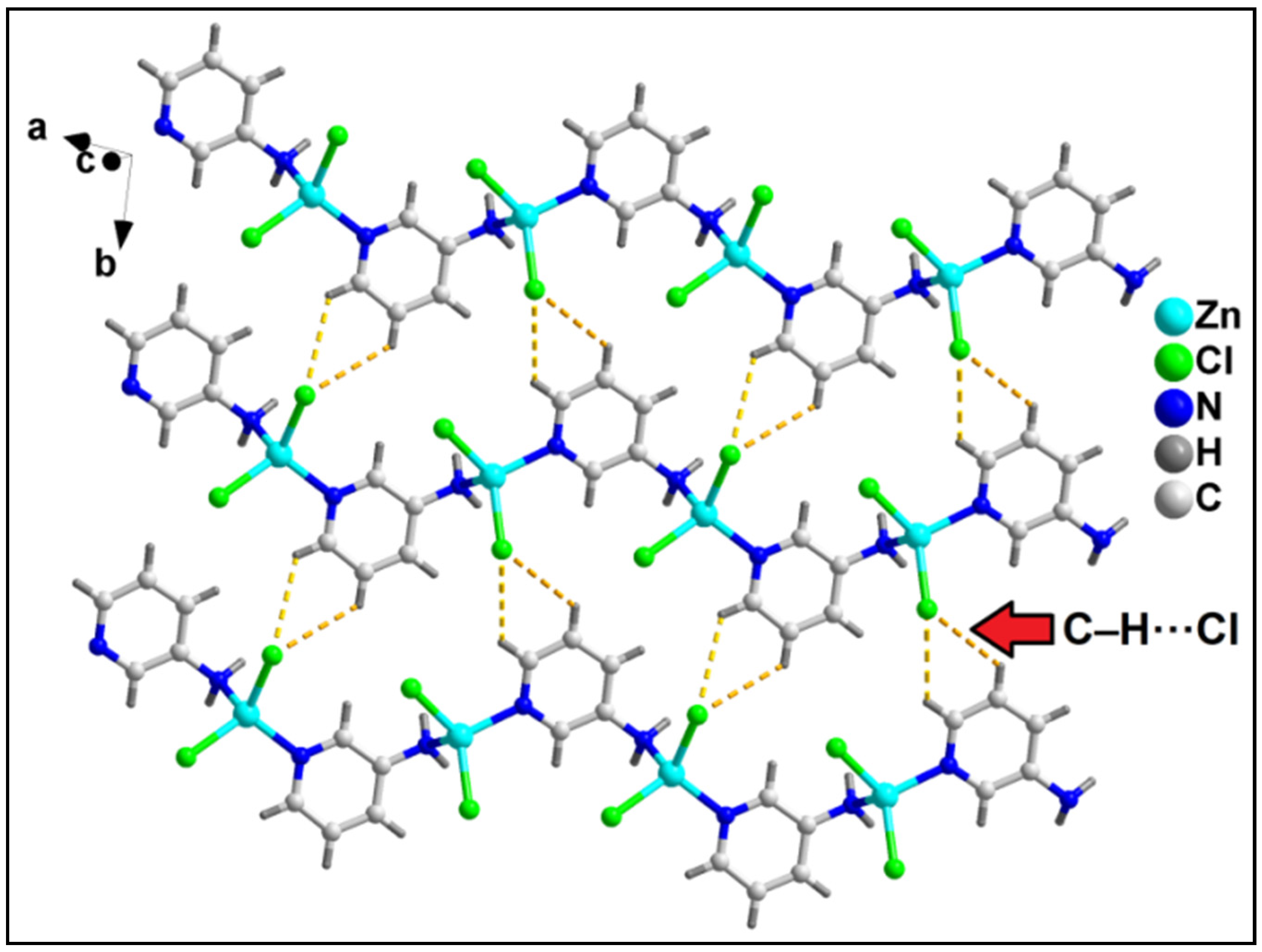
In the crystallographic ab plane, the 2D network is stabilized by C–H⋯Cl interactions (C6–H6⋯Cl2 = 3.04 Å) (Figure 7).
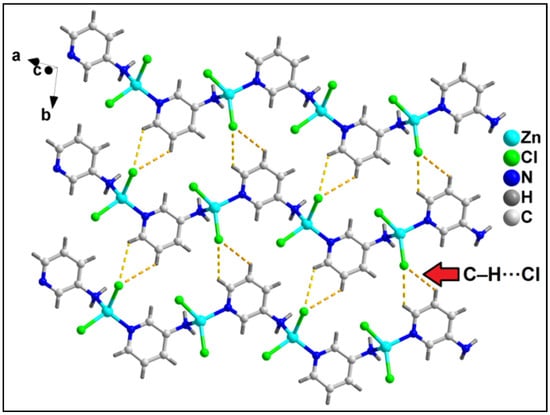
Figure 7.
Layered assembly of compound 2 along the crystallographic ab plane involving C–H⋯Cl hydrogen bonding interactions.
3.3. Spectral Studies
3.3.1. FT-IR Spectroscopy
The FT-IR spectra of compounds 1 and 2 have been recorded in the region 4000–500 cm−1 (Figure S3). The broad absorption peak in the 3410–3450 cm−1 region in the FT-IR spectrum for compound 1 can be attributed to the ν(O–H) stretching vibrations of coordinated water molecules [74,75,76]. The absorption peaks of [νas(OCO)] and [νs(OCO)] bands for the carboxylate moiety of 4-AmBz in 1, appear at 1621 and 1388 cm−1, respectively. The difference between the asymmetric and symmetric stretching vibrations is found to be greater than 200 cm−1 (233 cm−1) which supports the monodentate coordination of the carboxylate groups to Zn(II) centers in compound 1 [77]. However, the [νas(NH2)] and [νs(NH2)] bands of the amino groups appear at 3222, 3118, and 3107, 3228 cm−1, respectively, for compounds 1 and 2. These values are significantly lower than those of the free ligands which corroborates the coordination of the amino groups to the Zn(II) centers in the compounds [78].
3.3.2. Electronic Spectroscopy
Figures S4 and S5 depict the electronic spectra of compounds 1 and 2, respectively, in both the solid as well as in aqueous phases The diamagnetic Zn(II) coordination polymers do not show absorption bands in the visible region [79]. However, peaks due to the π-π* transitions of the aromatic ligands are observed in the UV region.
3.3.3. 1H-NMR Spectroscopy
Figures S6 and S7 depict the 1H-NMR spectra of compounds 1 and 2, respectively, in the DMSO-d6 solvent. For compound 1, the proton signals at 7.93 and 7.56 ppm can be attributed to C(2)H and C(3)H protons of the benzoate ring of 4-AmBz moiety [80]. In compound 2, the coordinated 3-AMpy gives proton signals at 7.68, 7.91, 7.02, 7.10 ppm due to the C(1)H, C(2)H, C(3)H, and C(4)H protons (Figure S7), respectively [81]. The absence of signals at >8 ppm in the spectrum of 1 supports the deprotonation of the carboxyl groups; consistent with the non-protonated 4-AmBz present in the crystal structure [82]. The presence of coordinated water molecules in 1 can be corroborated by the presence of a signal at 3.32 ppm [83]. The signals for the amine groups in aromatic ligands may have merged with the signal of DMSO-d6 in the NMR spectra of the compounds [84]. Therefore, the 1H-NMR data supports the stability of the polymers in the solution phase [85].
3.4. Thermogravimetric Analysis
The thermogravimetric analysis of compounds 1 and 2 have been carried out in the temperature range of 25–800 °C under the N2 atmosphere at the heating rate of 10 °C/min (Figure S8). For compound 1; in the temperature range of 70–170 °C, two coordinated water molecules undergo decomposition (obs. = 4.9%; calcd. = 6.2%) [86]. In the temperature range of 172–254 °C, one 4-AmBz moiety undergoes decomposition (obs. = 28.4%; calcd. = 26.69%) [87]. Two coordinated Cl moieties are decomposed at 255–410 °C with the observed weight loss of 14.2% (calcd. = 13.92%) [88]. The compound further decomposes in an unidentified manner beyond that temperature. Compound 2 undergoes single-step decomposition in the temperature range 350–515 °C with the loss of one coordinated 3-AmPy and two coordinated Cl moieties (obs. = 68.4%; calcd. = 70.2%) [89].
3.5. Theoretical Studies
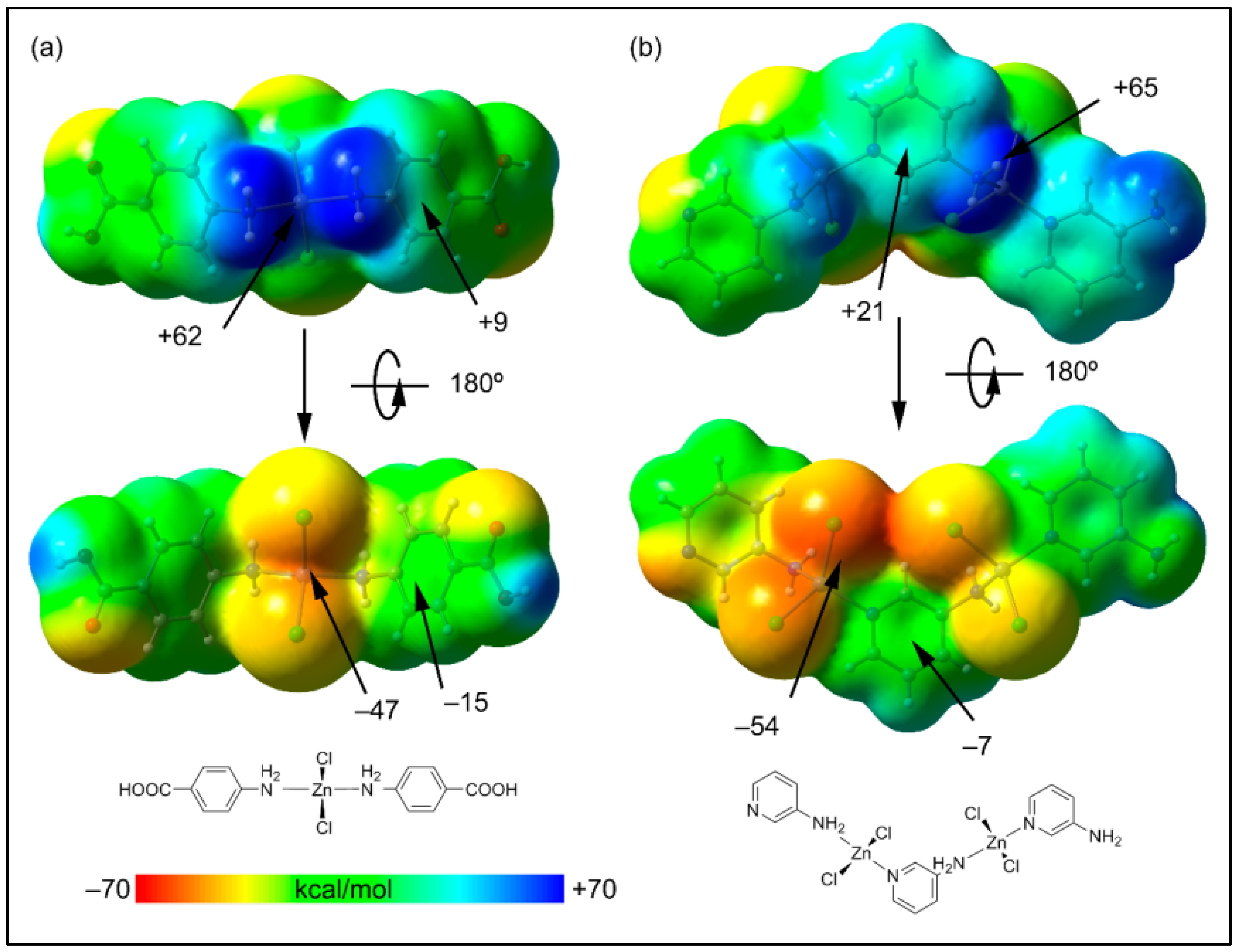
As commented above, among the myriad of non-covalent interactions, the aromatic rings participate in both anion-π (involving either the chlorido or carboxylato ligands) and NH2···π interactions involving the coordinated amino groups. Both interactions are opposite in nature since in the former, the aromatic ring is accepting charge and in the latter is donating charge. Therefore, the aromatic ring is acting as both an electron donor and acceptor simultaneously. To shed light on this unexpected behavior, we have first computed the molecular electrostatic potential surfaces of simplified models of polymers 1 and 2 (Figure 8). The molecular electrostatic potential maximum is located at the H-atoms of the amino groups (+62 kcal/mol in 1 and +65 kcal/mol in 2). These significant molecular electrostatic potential values are due to the enhanced acidity of the –NH2 protons due to the coordination of the amino group to the Zn2+ ion. The molecular electrostatic potential minimum is located at the chlorido ligands, as expected (−47 kcal/mol in 1 and −54 kcal/mol in 2). These results agree with the abundance of N–H···Cl interactions in the solid state of both compounds. In addition, more interesting are the MEP values over the center of the coordinated 4-AmBz and 3-AmPy ligands (Figure 8). The values are positive on one side (+9 and +21 kcal/mol in 1 and 2, respectively), and negative over the opposite side (−15 and −7 kcal/mol in 1 and 2, respectively), thus revealing the duality of these coordinated ligands and explaining the simultaneous formation of NH2···π and anion–π interactions.
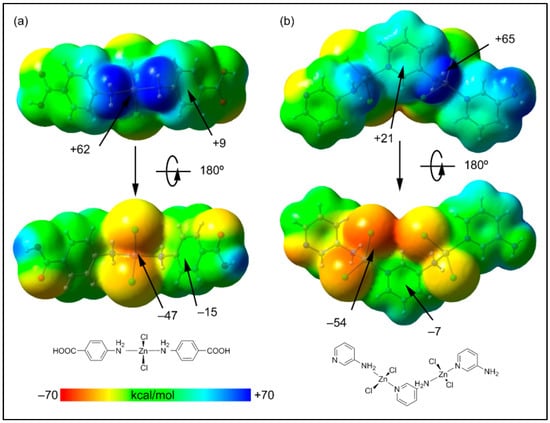
Figure 8.
Two views of the molecular electrostatic potential surfaces of the minimalistic models of the polymers 1 (a) and 2 (b) (isosurface 0.001 a.u.). The molecular electrostatic potential energies at some points of the surfaces are given in kcal/mol. The schematic drawings of the models are indicated in the lower part of the figure.
We have used the (QTAIM) and (NCI plot) index analyses in an extended dimeric model of the layered structure commented above (Figure 3b), that is stabilized by H-bonds, π–stacking, anion–π, and NH2···π interactions. The NCI plot index is an intuitive visualization index that facilitates the representation of non-covalent interactions in real space, showing which molecular regions interconnect.
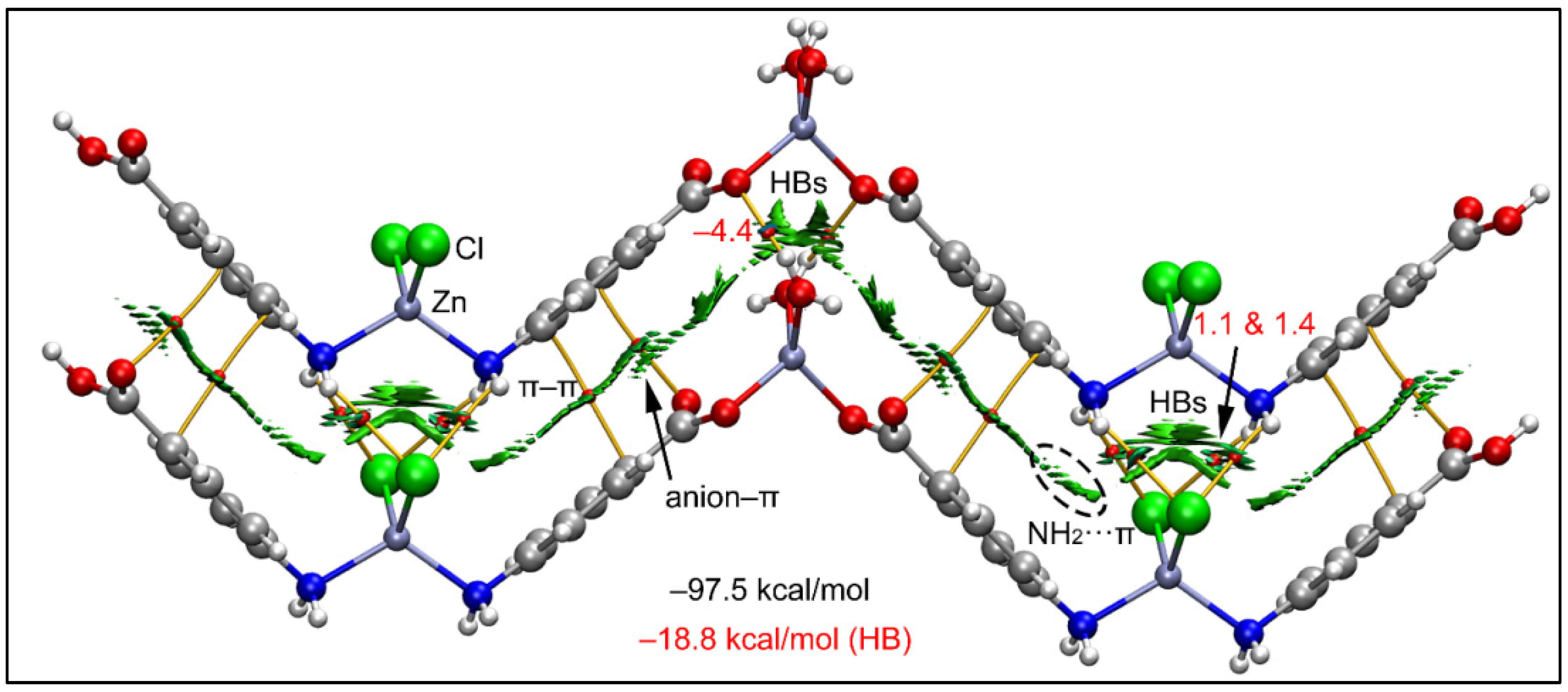
The analysis of the dimer of compound 1 reveals the presence of a green and extended isosurface located between both monomers, thus confirming the strong complementarity of the polymeric chains of the compound (Figure 9). For all HBs, a small reduced density gradient (RDG) isosurface appears coincident with the location of the bond CPs (critical points). We have assessed the formation energies of the H-bonds by using the value of Vr (potential energy density) at the bond CPs and the equation proposed in the literature (E = 0.5*Vr) [90]. The anion–π interaction is characterized by a bond critical point (CP, red sphere) and bond path (orange line) connecting the O-atom of the carboxylato group to one C-atom of the ring. Moreover, the NH2···π is also confirmed by the QTAIM analysis, revealing a bond CP and bond path connecting the H-atom of the amino group to one C-atom of the ring. Both contacts are characterized by green RDG (reduced density gradient) isosurfaces situated between the π-cloud of the ligand and both the amino and carboxylato groups.
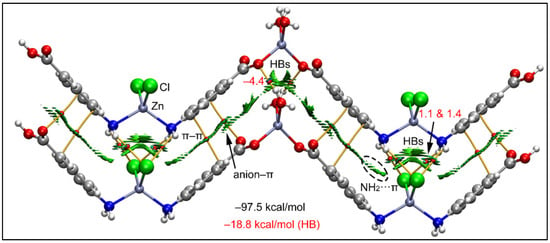
Figure 9.
QTAIM (bond paths in orange and bond CPs in red) and RDG surface analyses of the model of compound 1; the gradient cut-off is ρ = 0.04 a.u., s = 0.4, and the color scale is −0.035 a.u. < ρ < 0.035 a.u. The interaction energies are also indicated, computed at the RI-BP86-D3/def2-TZVP level of theory. Only intermolecular interactions are represented. The total contribution of the H-bonds is also indicated, estimated using the Vr energy predictor.
Interestingly, there is also a network of H-bonds that is energetically significant with a total H-bond interaction energy of −18.8 kcal/mol calculated using Vr values at the bond CPs. It can be observed that the chlorido ligands interact with the NH2 groups establishing four H-bonds, each one characterized by a bond CP and bond path interconnecting the Cl and H-atoms. Similarly, the coordinated water molecules of the other Zn(II) ion also establish two H-bonds (as donors) with the coordinated carboxylato groups. The formation energy of this dimeric assembly is very large (−97.5 kcal/mol) due to this intricate combination of interactions and evidencing the importance of these contacts in the crystal packing of 1. Using a minimalistic model of compound 1, we have also studied the Cl···Cl contact commented above (Figure 2).
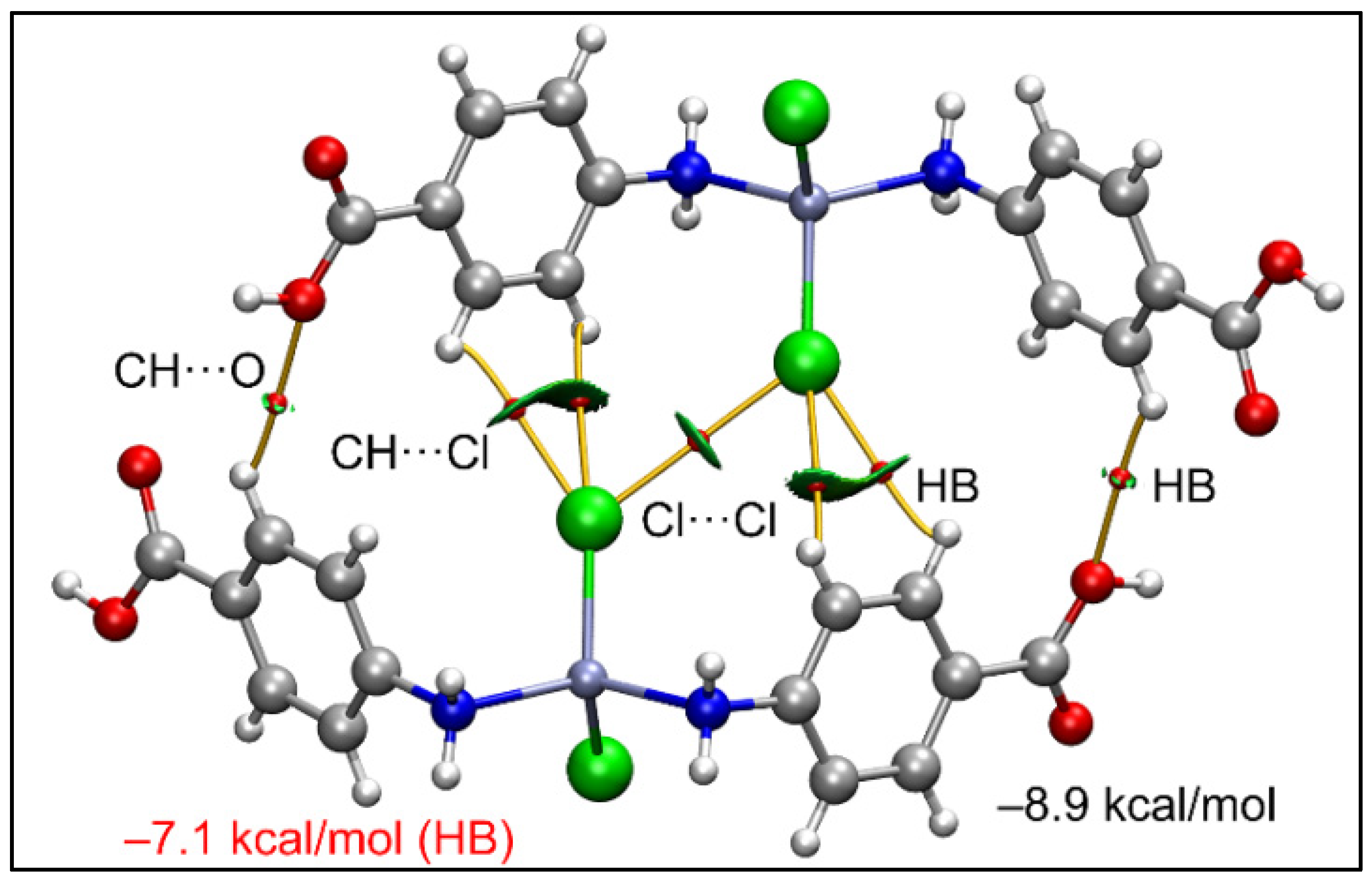
The joint QTAIM/NCI plot analysis is shown in Figure 10, where it can be observed that the existence of the Cl···Cl contact is corroborated by both methods (bond CP connecting the Cl-atoms and green isosurface). Moreover, the analysis also discloses the existence of other interactions, like C–H···O and C–H···Cl contacts that are also relevant for the formation of this assembly. The dimerization energy is moderately strong (−8.9 kcal/mol) where the most important contribution is the H-bonding (−7.1 kcal/mol), thus suggesting that the Cl···Cl contact is weak, as usual in Type I halogen···halogen contacts [91].
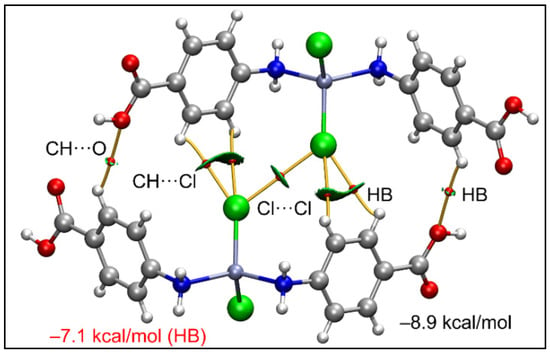
Figure 10.
QTAIM (bond paths in orange and bond CPs in red) and RDG surfaces of the model dimeric assembly of compound 1; the gradient cut-off is ρ = 0.04 a.u., s = 0.35, and the color scale is −0.04 a.u. < ρ < 0.04 a.u. Only intermolecular interactions are represented. The total contribution of the H-bonds is also indicated, estimated using the Vr energy predictor.
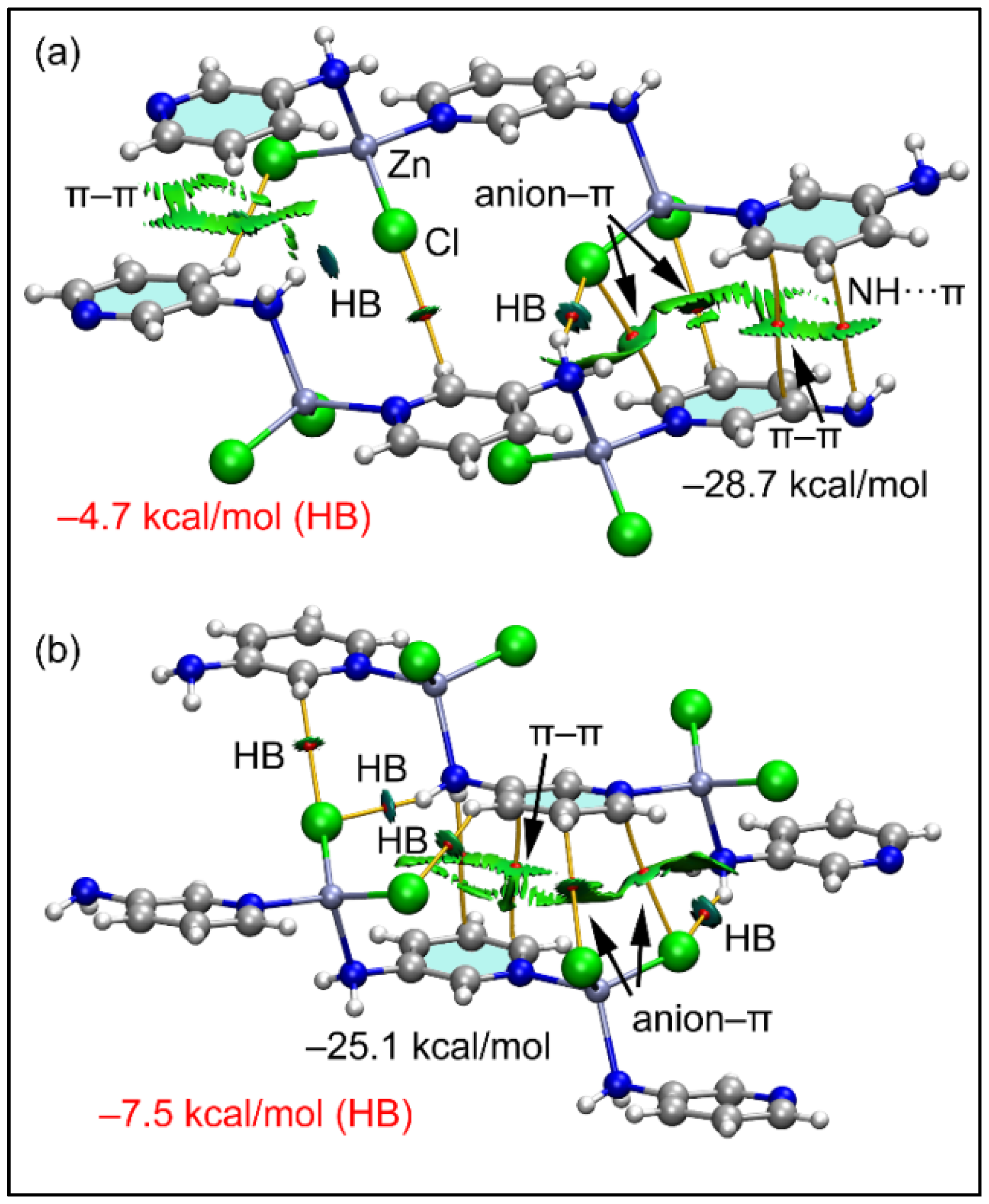
In compound 2, using the model depicted in the lower part of Figure 5b, we have analyzed two dimers retrieved from its solid state. In both dimers, the aromatic ring establishes both anion-π and NH2···π interactions, similar to compound 1. Both dimers are represented in Figure 11 exhibiting similar dimerization energies. The π-interactions (π–π, NH2···π and anion–π) are characterized by the corresponding bond CPs and bond paths interconnecting the atoms of the counterparts. All three interactions are also revealed by the NCI plot analysis and characterized by green RDG isosurfaces, confirming their attractive nature. The analysis also shows the presence of C–H···Cl and N–H···Cl H-bonding interactions that contribute −4.7 and −7.5 kcal/mol for the dimers in Figure 11a,b, respectively, thus evidencing that the π-interactions dominate the formation of the dimers. These results evidence that π-interactions are more relevant than the H-bonds in the crystal packing of compound 2.
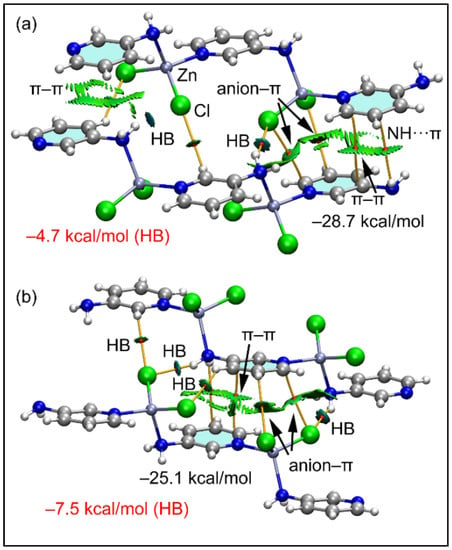
Figure 11.
(a,b) QTAIM (bond paths in orange and bond CPs in red) and RDG surfaces of two dimeric models of compound 2. The gradient cut-off is ρ = 0.04 a.u., s = 0.35, and the color scale is −0.04 a.u. < ρ < 0.04 a.u. Only intermolecular interactions are represented. The total contribution of the H-bonds is also indicated, estimated using the Vr energy predictor.
3.6. Cytotoxicity and Apoptosis Assays
Trypan blue is a polar dye that cannot penetrate through the intact cell membrane of normal cells; but can enter and color the non-viable cells due to damaged membrane integrity [92]. The percentage of dead cells can be calculated from the number of colored (stained) cells compared to the total population of cells [93]. The results of the trypan blue assay reveal that compounds 1 and 2 induced remarkable concentration-dependent cytotoxicity in DL cancer cells with negligible cytotoxicity in PBMC cells (Figure 12 and Figure 13). The cytotoxicity assay of the metal salt (ZnCl2) and the corresponding ligands (4-AmBz, 3-AmPy) under the same experimental conditions reveal that they exhibit negligible cytotoxicity against DL cancer cell after 24 h of treatment. The study reveals that the significant cytotoxicity exhibited by the compounds in a cancerous cell line is a combined effect of the metal center and the ligands [94]. Comparative analysis revealed that compound 1 induces better cytotoxicity in DL cells compared to that of compound 2. The higher positive charge of Zn(II) center in compound 1 than in compound 2; due to the smaller Zn-ligand bond length (2.06 Å in 1; 2.14 Å in 2); can be correlated to the higher cytotoxicity of compound 1 [95]. In addition, compound 1 interacts more efficiently with BCL-2 antiapoptotic protein than that of compound 2 (vide infra); which may also play a significant role in more cytotoxicity of 1.
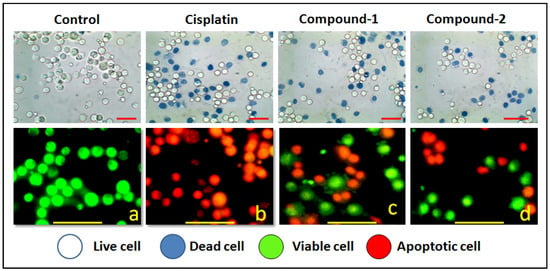
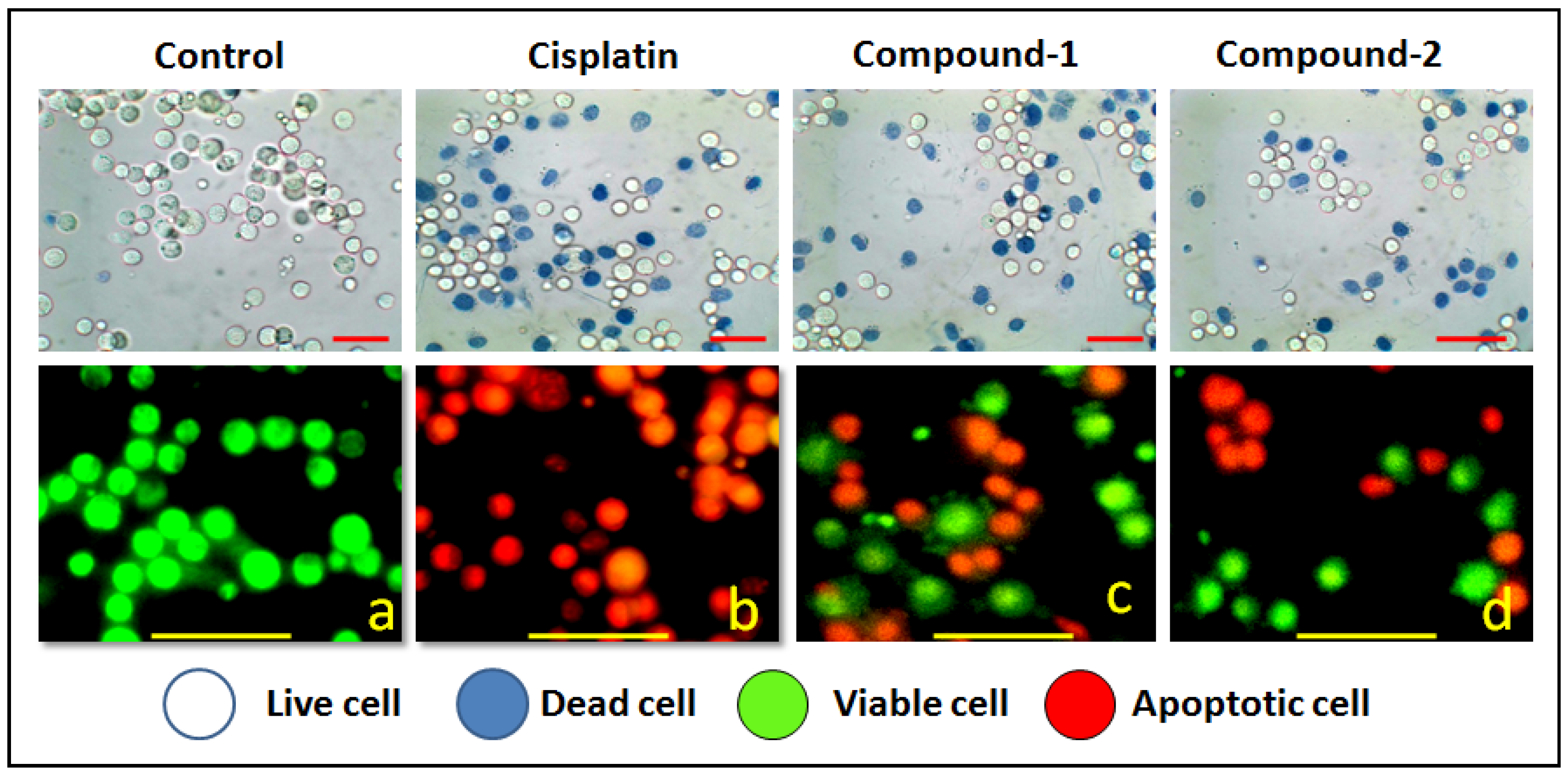
Figure 12.
Upper panel shows the results of trypan blue assay of the compound’s DL cells. Lower panel represents the morphological feature of living (green) and apoptotic (red) DL cancer cells observed under fluorescence microscope. Figure corresponds to apoptosis (450×) induced by the compounds at highest potent dose (10 μM).
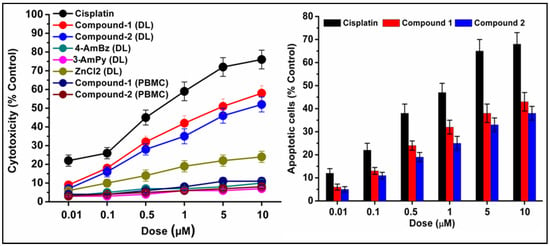
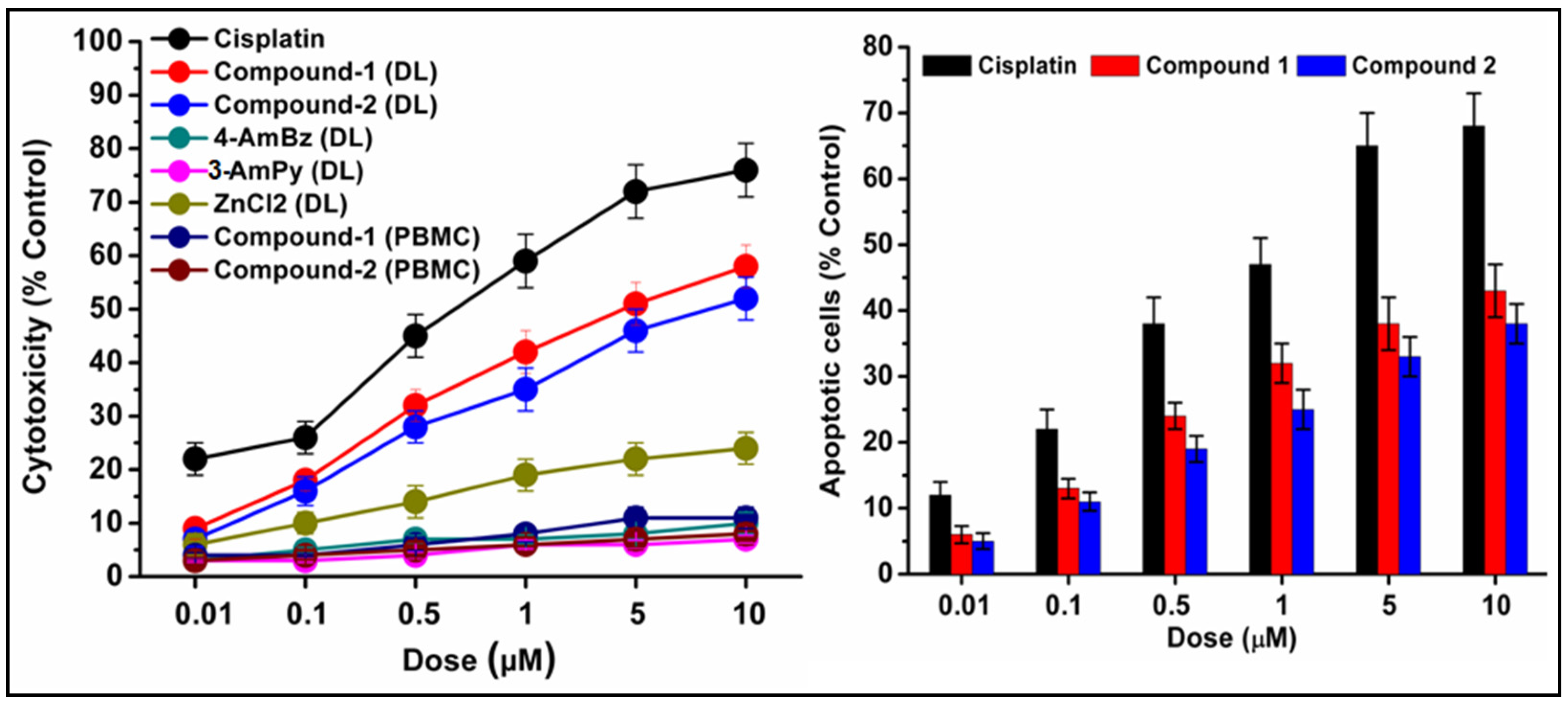
Figure 13.
Concentration-dependent cytotoxicity (left) induced by compounds 1 and 2 in DL cells. The results of the compounds were compared with cisplatin (reference drug). Percentage apoptotic cells (right) in DL cell line after exposure to compounds 1 and 2. Data are mean ± S.D., n = 3.
We have also carried out an AO/EB dual staining approach to investigate the apoptotic cell death induced by the compounds in DL cancer cells. AO is a vital dye that can penetrate the cell membrane of normal healthy cells and stain the nucleus as green; whereas, EB can only penetrate the damaged cell membrane of apoptotic cells and stain as red/orange [96]. The presence of red/orange fluorescence after exposure to compounds 1 and 2 indicates apoptosis-inducing abilities of the compounds (Figure 12 and Figure 13). A comparative study revealed that compound 1 induced higher apoptosis compared to compound 2 and the result is in line with that of the cytotoxicity assay (trypan blue exclusion assay). The results obtained for the cytotoxicity and apoptosis assays have been further compared with cisplatin (Sigma Aldrich), a reference drug. There are few reports of significant antiproliferative evaluation of coordination polymers involving bridging amine-based ligands in human cancerous cells [97,98]. Li and coworkers have reported the in vitro antiproliferative evaluation of a Co(II) coordination polymer with 4-AmBz derivatives against human liver cancer cells [99].
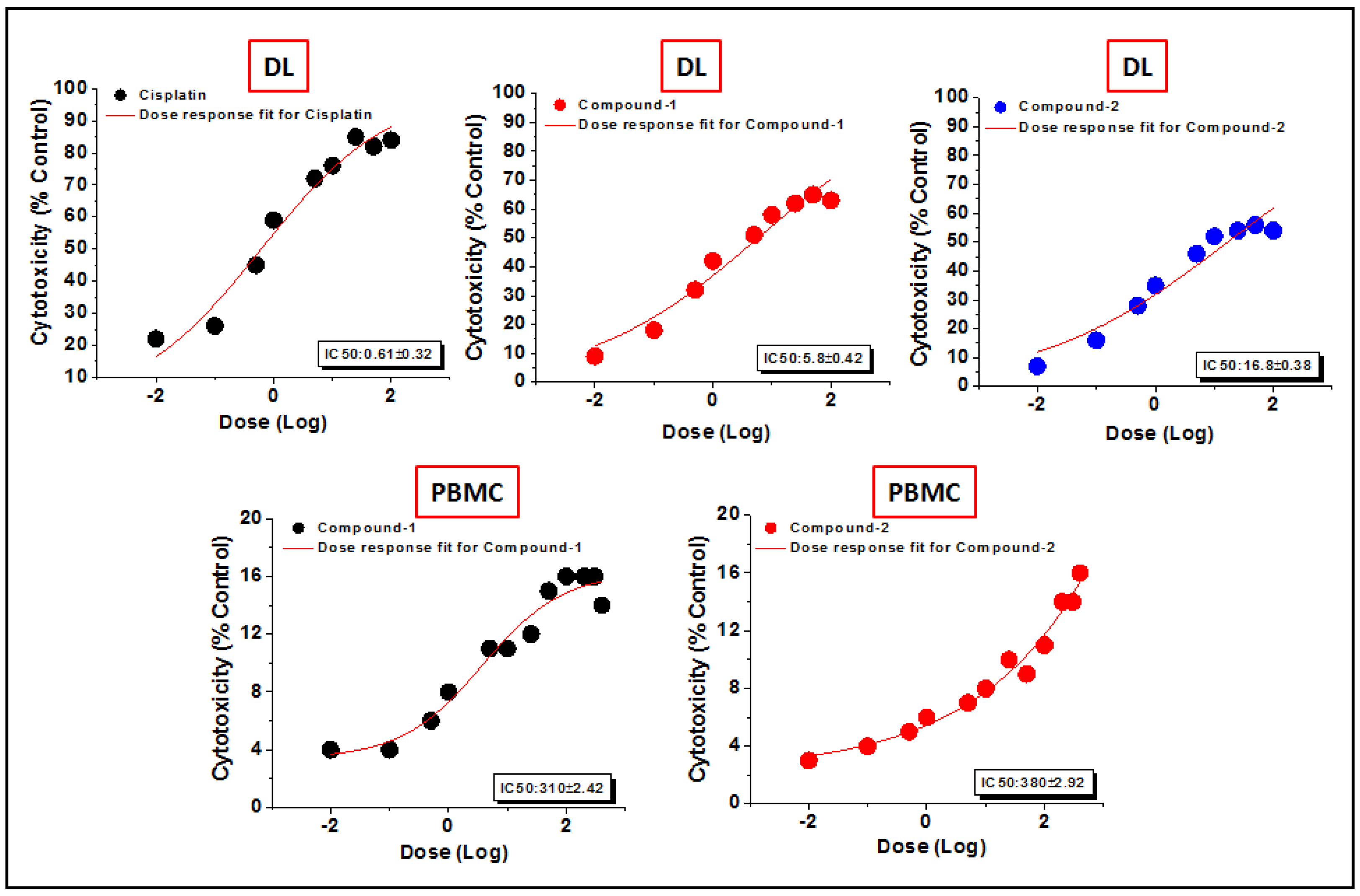
The preclinical assessment of newly synthesized drugs/compounds can be typically evaluated by the half maximum inhibitory concentration (IC50) which can be measured by the substance’s ability to inhibit 50% of biological processes [100]. The dose-response curves and the IC50 values of the compounds have been represented in Figure 14 and Table S2, respectively. Compound 1 exhibits a lower IC50 value (5.8 μM) than that of compound 2 (16.8 μM) in DL cells after 24 h of treatment (Figure 14). The IC50 value of cisplatin (reference drug) ranges from 0.5–0.6 μM under the same experimental conditions. Several research groups have reported the significant anticancer activities of transition metal compounds against DL cells [38,39,40]. However, they have not investigated the cytotoxicity of the compounds in normal cell lines. In our study, the promising outcome of the results of the cytotoxicity assay is that the compounds induced significant concentration-dependent cytotoxicity in DL cancer cells with negligible cytotoxicity in PBMC cells. Moreover, most of the previous reports do not perform the molecular docking simulation (vide infra) to establish the probable mechanism of the synthesized compounds in the cytotoxic assay.
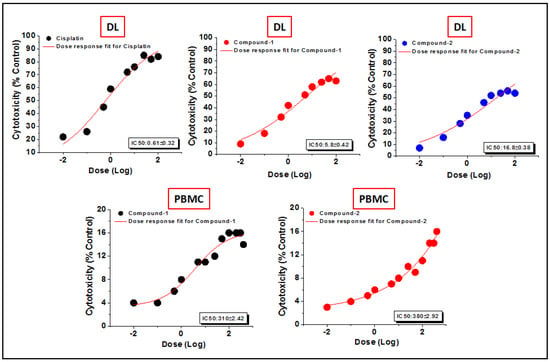
Figure 14.
The dosage response curves of compounds 1 and 2 to determine the IC50 values in DL and PBMC cell lines.
3.7. Molecular Docking Simulation
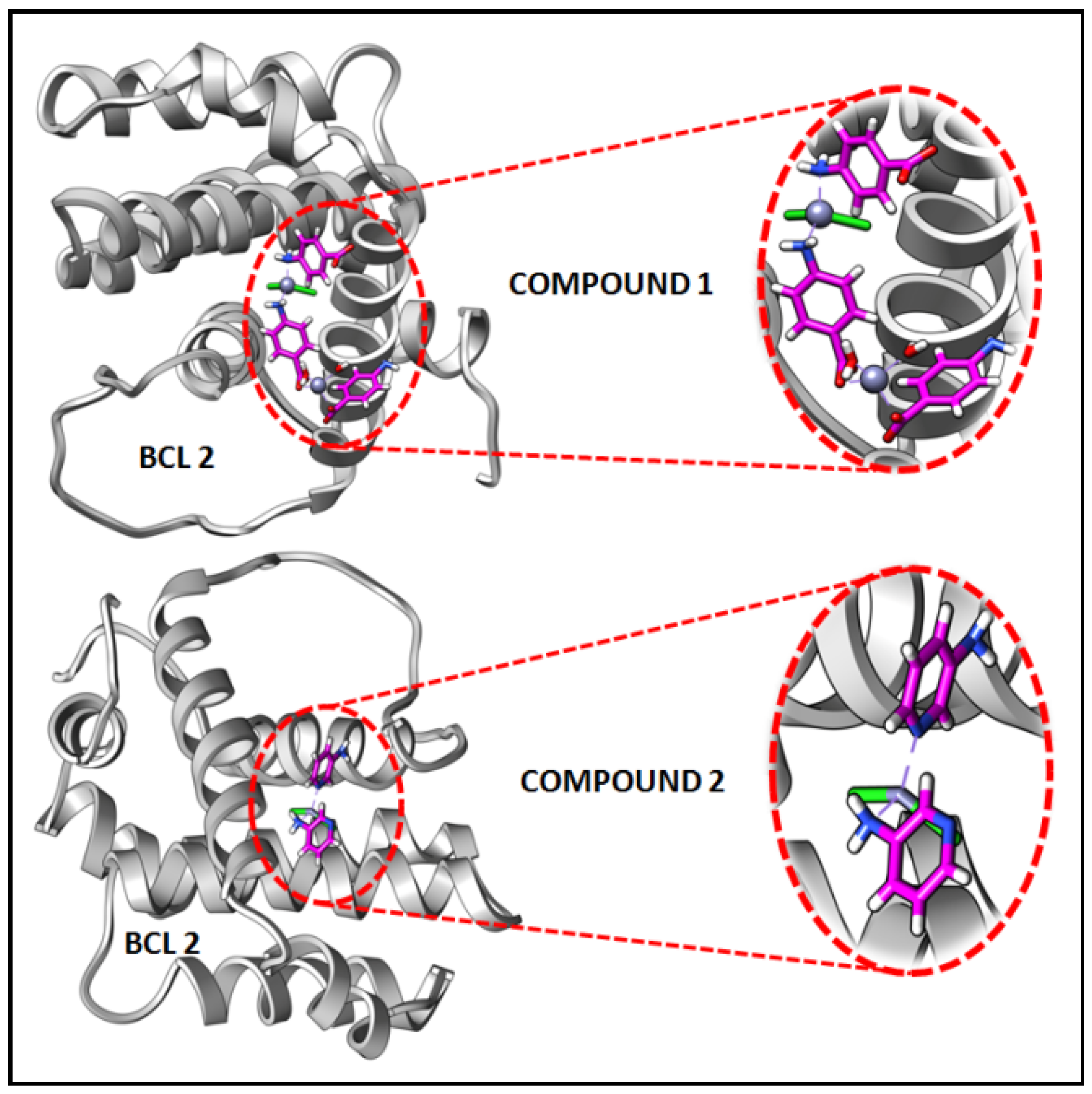
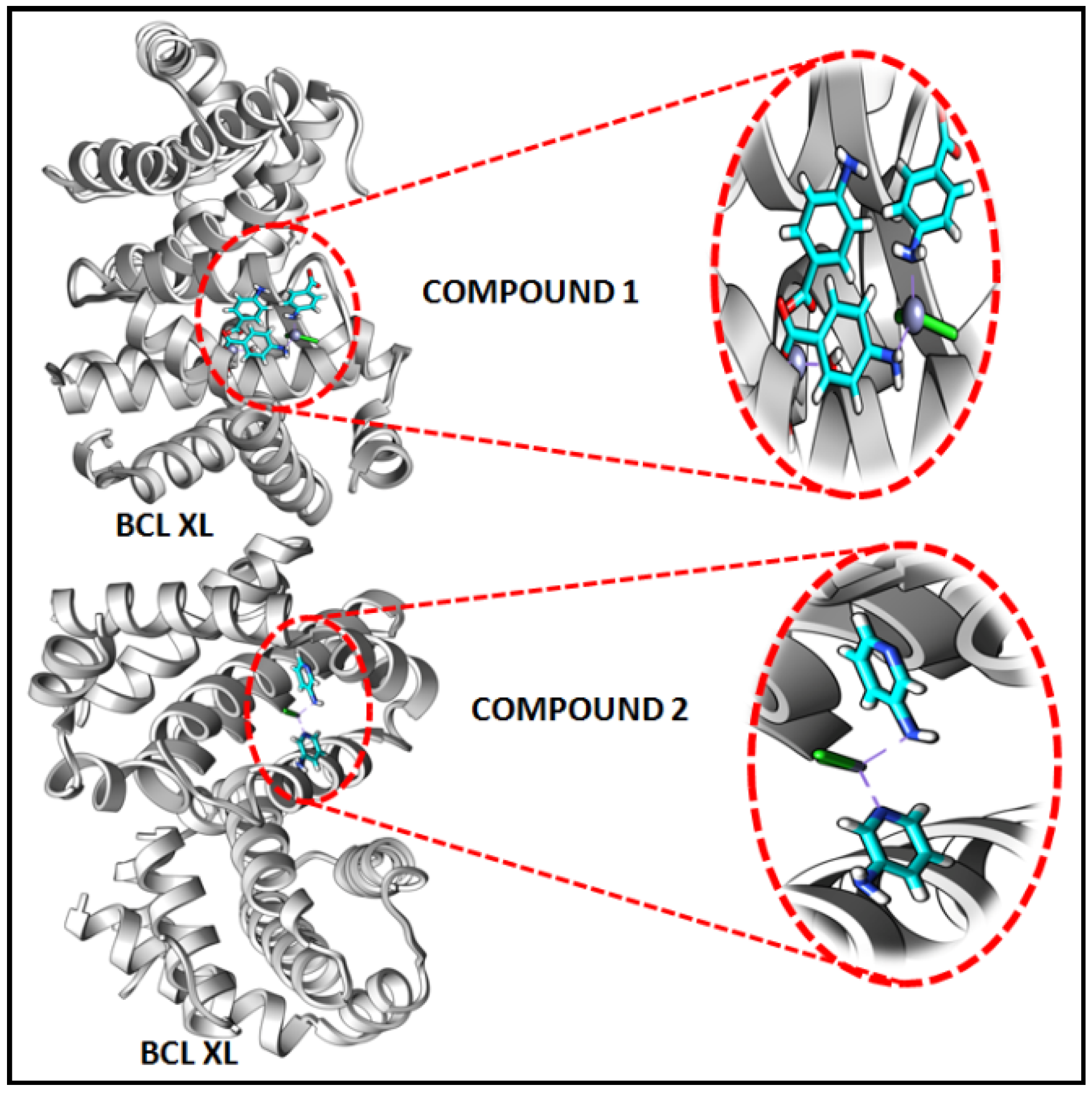
In the present study, in silico molecular docking simulations of the structures of the compounds have been performed to explore the probable interactions of the compounds with antiapoptotic BCL family proteins which are closely related to cancer progression and metastasis [101]. Molecular docking is a potential approach in computer-aided drug designing (CADD) for the virtual screening of drugs with the 3D structures of biological target proteins [102]. Molecular docking simulation was performed with the antiapoptotic target proteins [BCL-2 (PDB ID = 2O22) and BCL-XL (PDB ID = 2YXJ)] to elucidate the possible binding modes of the modeled fragments of the compounds with the active sites of the receptors. It has been well established that antiapoptotic BCL family proteins are actively involved in cancer cell progression and metastasis [103,104,105]. Studies on BCL family proteins have revealed their active involvement in tumor pathogenesis and therefore they can be effectively utilized as targets for developing more precisely tailored drugs/chemicals [62]. Figure 15 and Figure 16 represent the docking structures of the compounds (modeled fragment) with the antiapoptotic target proteins which reveal significant binding affinity of the modeled fragments of the compounds with the receptor proteins. Compound 1 possesses a stronger binding affinity with anti-apoptotic protein BCL-2 than that of compound 2, while interactions with BCL-XL protein revealed similar arrangements in both compounds. We have already mentioned in the cytotoxic assay that the comparatively higher positive charge of the Zn(II) center in compound 1 due to smaller Zn-ligand bond lengths than that of compound 2 can be correlated to the higher cytotoxicity of compound 1 [95]. The metal-ligand bond lengths and bond angles play crucial roles in the fitting of the compound in the receptor-binding pockets of the target proteins [106]. The smaller Zn-ligand bond lengths in 1 may play a crucial role in higher binding affinity with the anti-apoptotic protein BCL-2.
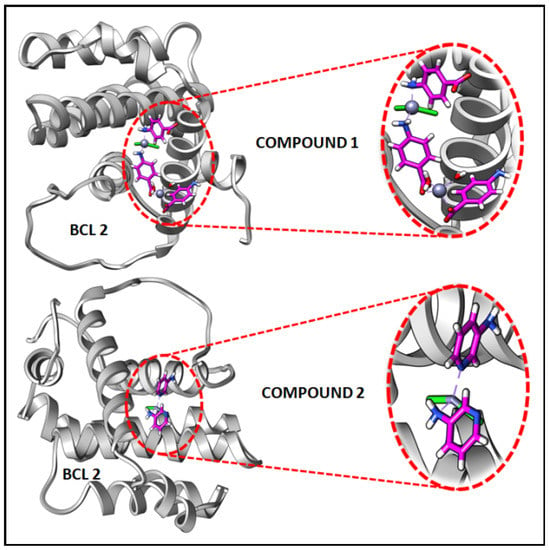
Figure 15.
Docking results of modelled fragments of the compounds 1 and 2 with BCL-2 receptor.
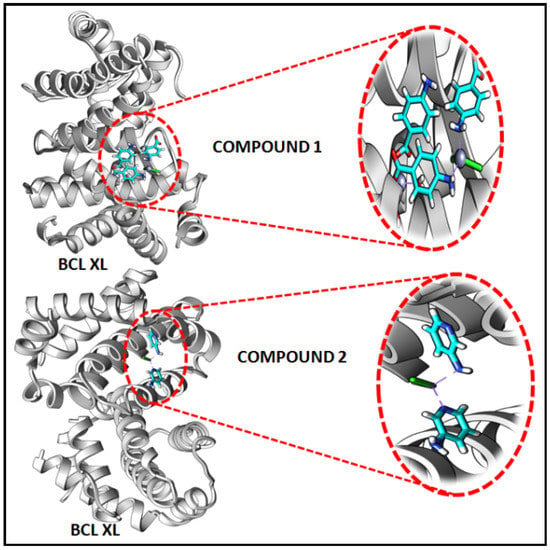
Figure 16.
Docking results of modelled fragments of the compounds 1 and 2 with BCL-XL receptor.
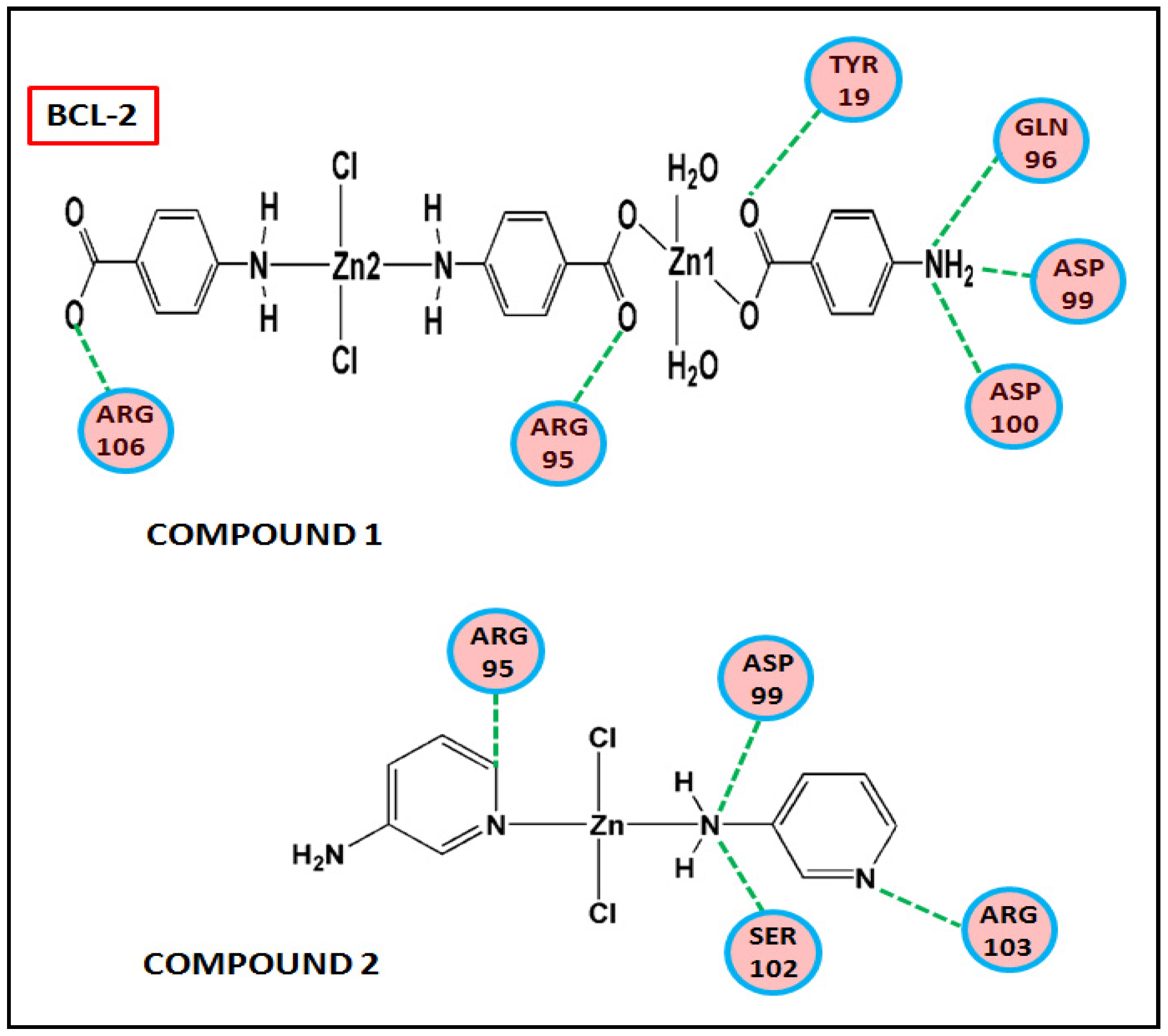
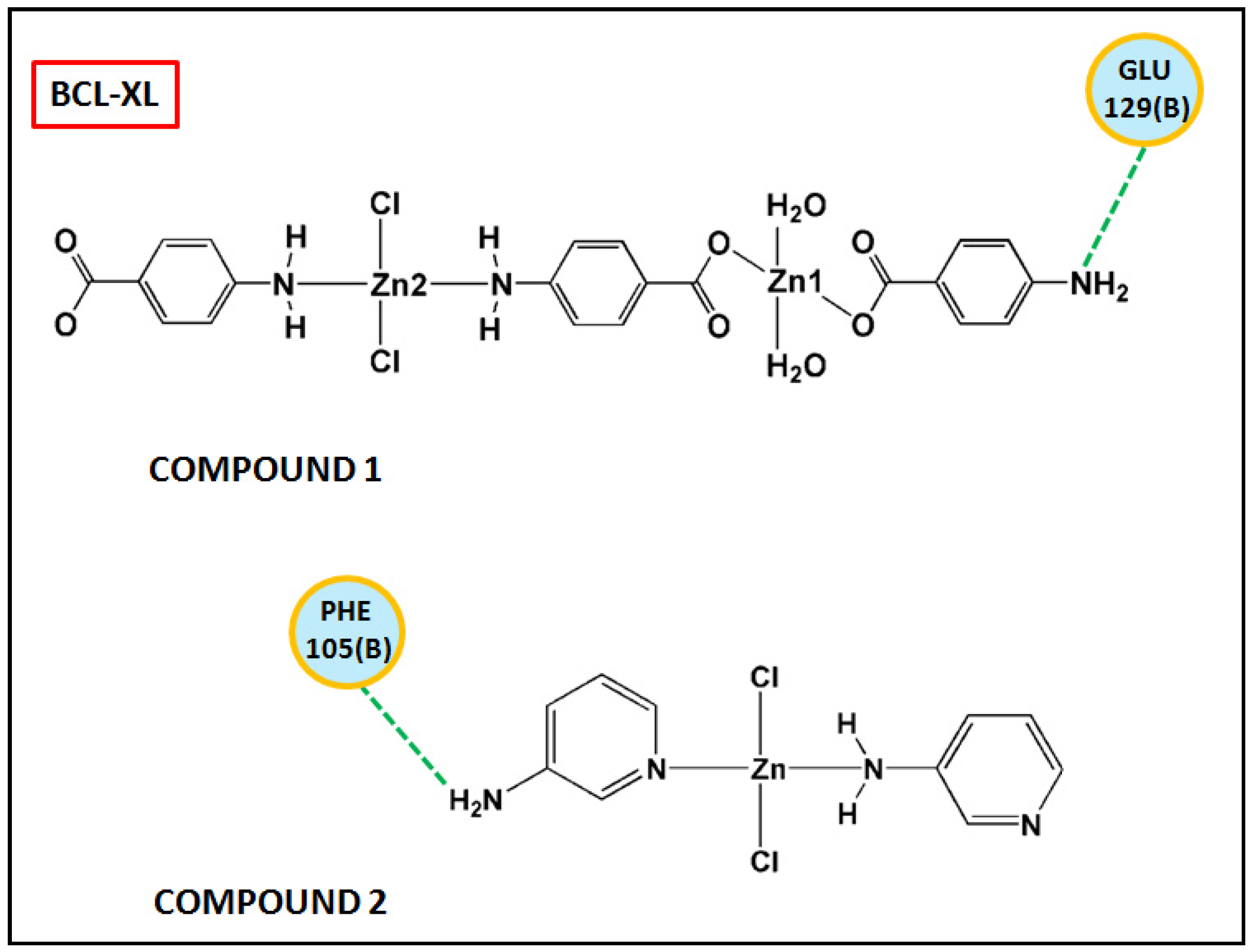
Figure 17 and Figure 18 represent the chemical interactions between the compounds and the amino acid residues with the active sites of antiapoptotic proteins. The docking score of compound 1 for BCL-2 and BCL-XL proteins were −99.92 and −86.29, respectively, whereas, for compound 2, the scores are −59.78 and −62.59, respectively. Compound 1 interacted strongly with the active sites of BCL-2 protein having amino acid residues Tyr19, Arg95, Arg106, Gln96, Asp99, and Asp100, whereas, single interaction was observed in the active sites of BCL-XL protein having Glu129(B) residue (Figure 17 and Figure 18). On the other hand, compound 2 interacted with four amino acid residues, viz., Arg95, Asp99, Ser102, and Arg103 in BCL-2 protein, while single interaction was observed in BCL-XL protein with Phe105(B) (Figure 17 and Figure 18). Thus, it can be stated that the compounds’ efficient ability to induce cytotoxicity and apoptosis may be due to their effective interactions with the antiapoptotic target proteins. Konakanchi et al. have supported the experimental anticancer activities of five coordination compounds of Co(II), Ni(II), Cu(II), Zn(II), and Pd(II) involving substituted aminobenzoic acid using molecular docking studies [107].
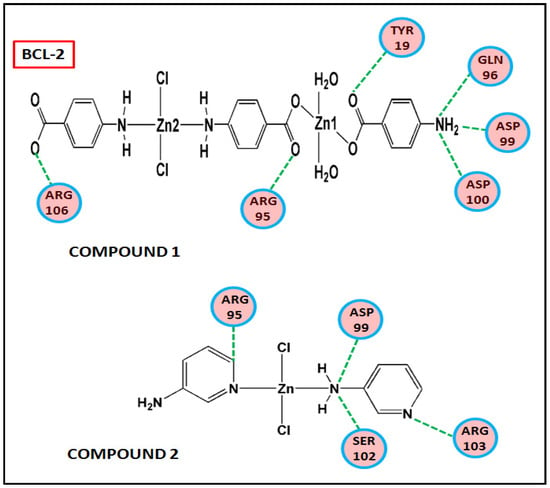
Figure 17.
Chemical interactions of modelled fragments of the compounds with BCL-2 have been represented with compounds’ atoms and interacting amino acid residues.
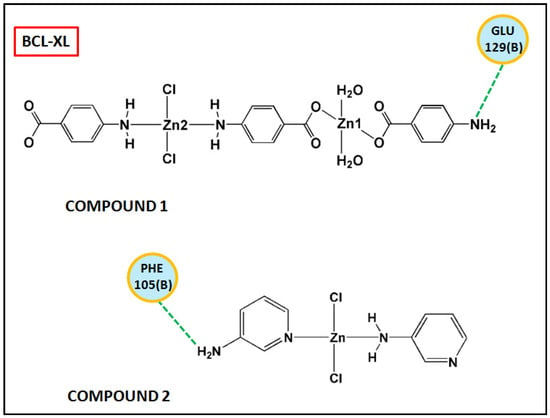
Figure 18.
Chemical interactions of modelled fragments of the compounds with BCL-XL have been represented with compounds’ atoms and interacting amino acid residues.
4. Conclusions
Two Zn(II) coordination polymers have been prepared at room temperature and characterized using elemental analysis, FT-IR, electronic spectroscopy, TGA, and single crystal XRD. Compounds 1 and 2 crystallize as 4-aminobenzoato and 3-aminopyridine bridged coordination polymers of Zn(II), respectively. Non-covalent anion-π, aromatic π-stacking, and unusual NH2(amino)⋯π contacts are observed in the crystal structures which provide rigidity to the layered assemblies. Unorthodox Type I Cl⋯Cl interactions also play a pivotal role in the stabilization of the layered assembly of compound 1. The non-covalent interactions observed in the solid-state architectures of the compounds have been further analyzed theoretically using DFT calculations. Interestingly, the MEP surface analysis evidenced interesting duality in donor-acceptor topologies of the aromatic rings of coordinated 4-AmBz and 3-AMpy moieties in the compounds revealing the concurrent formation of unusual NH2···π and anion–π interactions. DFT calculations, NCI plot index, and QTAIM analysis reveal that among various non-covalent interactions involved in the crystal packing of the compounds, H-bonds in compound 1 and π-interactions (NH2···π, π-π, anion–π) in compound 2 are energetically significant. We have carried out in vitro anticancer activities of the compounds in Dalton’s lymphoma (DL) malignant cancer cell line using trypan blue exclusion and apoptosis assays. The study reveals concentration-dependent cytotoxicity and apoptosis-inducing abilities of the compounds in cancerous cells having low effects in normal PBMC cells. The compounds efficiently bind with the active sites of antiapoptotic proteins which is evident from the molecular docking simulation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst13030382/s1, Figure S1: 1D polymeric chain of compound 1 assisted by intra-molecular O–H⋯O hydrogen bonding interactions; Figure S2: 1D polymeric chain of compound 2 stabilised by C–H⋯Cl hydrogen bonding interactions; Figure S3: FT-IR spectra of the compounds 1 and 2.; Figure S4: (a) UV-Vis-NIR spectrum of 1; (b) UV-Vis spectrum of 1 in water (10−3 M); Figure S5: (a) UV-Vis-NIR spectrum of 2; (b) UV-Vis spectrum of 2 in water (10−3 M); Figure S6: 1H-NMR spectrum of compound 1 in DMSO-d6; Figure S7: 1H-NMR spectrum of compound 2 in DMSO-d6; Figure S8: Thermogravimetric curves of the compounds 1 and 2; Table S1: Comparison of crystal parameters of compound 2 with the already reported compound; Table S2: IC50 values (in μM) of compounds 1, 2, ligands and metal salt calculated using dose response curves.
Author Contributions
Conceptualization, A.F., A.K.V. and M.K.B.; methodology, A.F., A.K.V. and M.K.B.; software, A.F., R.M.G. and A.K.V.; formal analysis, A.F.; investigation, P.S., R.M.G. and D.D.; data curation, M.B.-O.; writing—original draft preparation, P.S. and M.K.B.; writing—review and editing, M.K.B.; visualization, A.F.; supervision, M.K.B.; project administration, A.F. and M.K.B.; funding acquisition, A.F. and M.K.B. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support from ASTEC, DST, Govt. of Assam (Grant number: ASTEC/S&T/192(177)/2020-2021/43) and the Gobierno de España, MICIU/AEI (project No. PID2020-115637GB-I00 FEDER funds) is gratefully acknowledged. The authors thank IIT, Guwahati for TGA data.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Lustig, W.P.; Mukherjee, S.; Rudd, N.D.; Desai, A.V.; Li, J.; Ghosh, S.K. Metal-organic frameworks: Functional luminescent and photonic materials for sensing applications. Chem. Soc. Rev. 2017, 46, 3242–3285. [Google Scholar] [CrossRef] [PubMed]
- Dhakshinamoorthy, A.; Li, Z.; Garcia, H. Catalysis and photocatalysis by metal organic frameworks. Chem. Soc. Rev. 2018, 47, 8134–8172. [Google Scholar] [CrossRef] [PubMed]
- Marabello, D.; Antoniotti, P.; Benzi, P.; Canepa, C.; Mortati, L.; Sassi, M.P. Synthesis, structure and non-linear optical properties of new isostructural β-D-fructopyranose alkaline halide metal–organic frameworks: A theoretical and an experimental study. Acta Cryst. 2017, 73, 737–743. [Google Scholar] [CrossRef]
- Sun, Q.; Zhu, X.; Zhang, N.; Zhang, B.; Lu, J.; Liu, H. Auxiliary ligand-assisted structural variation of two Co(II) metal-organic frameworks: Syntheses, crystal structure and magnetic properties. Inorg. Chem. Commun. 2019, 99, 172–175. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, D.; Humphrey, M.G.; Meng, S.; Zaworotko, M.J.; Cifuentes, M.P.; Zhang, C. Ammonium–crown ether supramolecular cation-templated assembly of an unprecedented hetero bicluster–metal coordination polymer with enhanced NLO properties. Chem. Commun. 2016, 52, 3797–3800. [Google Scholar] [CrossRef]
- Chiniforoshan, H.; Pourrahim, N.; Tabrizi, L.; Tavakol, H.; Sabzalian, M.R.; Notash, B. Syntheses, studies and crystal structure of new coordination polymers of mercury (II) with phenylcyanamide derivative ligands. Inorg. Chim. Acta 2014, 416, 85–92. [Google Scholar] [CrossRef]
- Janiak, C. Engineering coordination polymers towards applications. Dalton Trans. 2003, 12, 2781–2804. [Google Scholar] [CrossRef]
- Croitor, L.; Coropceanu, E.B.; Duca, G.; Siminel, A.V.; Fonari, M.S. Nine Mn(II), Zn(II) and Cd(II) mixed-ligand coordination networks with rigid dicarboxylate and pyridine-n-aldoxime ligands: Impact of the second ligand in the structures’ dimensionality and solvent capacity. Polyhedron 2017, 129, 9–21. [Google Scholar] [CrossRef]
- Yunxiang, L.; Yingtao, L.; Haiying, L.; Xiang, Z.; Honglai, L.; Weiliang, Z. Energetic effects between halogen bonds and anion-π or lone pair-π interactions: A theoretical study. J. Phys. Chem. 2012, 116, 2591–2597. [Google Scholar]
- Lehn, J.M.; Atwood, J.L.; Davies, J.E.D.; MacNicol, D.D.; Vogtle, F. Comprehensive Supramolecular Chemistry; Elsevier, Pergamon: Oxford, UK, 1996. [Google Scholar]
- Gogoi, A.; Saha, U.; Dutta, D.; Bhattacharyya, M.K. A Supramolecular Dimer of Two crystallographically independent Co(II) phthalate bipyridine complexes in one single crystal: Synthesis, crystal structure, and thermal studies. J. Struct. Chem. 2019, 60, 324–332. [Google Scholar] [CrossRef]
- Cook, S.A.; Borovik, A.S. Molecular designs for controlling the local environments around metal ions. Acc. Chem. Res. 2015, 48, 2407–2414. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, J. Coordination chemistry beyond Werner: Interplay between hydrogen bonding and coordination. Chem. Soc. Rev. 2013, 42, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Thakuria, R.; Nath, N.K.; Saha, B.K. The nature and applications of π–π interactions: A perspective. Cryst. Growth Des. 2019, 19, 523–528. [Google Scholar] [CrossRef]
- Bauzá, A.; Frontera, A. Theoretical Study on the Dual Behavior of XeO3 and XeF4 toward Aromatic Rings: Lone Pair–π versus Aerogen–π Interactions. ChemPhysChem 2015, 16, 3625–3630. [Google Scholar] [CrossRef] [PubMed]
- Bibi, S.; Zhang, J. The ratio and topology effects of benzodithiophene donor–benzooxadiazole acceptor fragments on the optoelectronic properties of donor molecules toward solar cell materials. Phys. Chem. Chem. Phys. 2015, 17, 7986–7999. [Google Scholar] [CrossRef]
- Ramasubbu, N.; Parthasarathy, R.; Murray-Rust, P. Angular preferences of intermolecular forces around halogen centers: Preferred directions of approach of electrophiles and nucleophiles around carbon-halogen bond. J. Am. Chem. Soc. 1986, 108, 4308–4314. [Google Scholar] [CrossRef]
- Piracha, N.K.; Ito, F.; Nakanaga, T. Infrared depletion spectroscopy of aniline–toluene cluster: The investigation of the red shifts of the NH2 stretching vibrations of aniline–aromatic clusters. Chem. Phys. 2004, 297, 133–138. [Google Scholar] [CrossRef]
- Chernova, E.F.; Ovsyannikov, A.S.; Solovieva, S.E.; Antipin, I.S.; Kyritsakas, N.; Hosseini, M.W.; Ferlay, S. Control of dimensionality in Manganese Coordination Polymers using rigid tetrahedral-shaped [1.1.1.1]metacyclophane ligands bearing benzoate coordinating sites: From homochiral 1D to 3D diamond-like structures. Inorg. Chem. Commun. 2019, 106, 197–201. [Google Scholar] [CrossRef]
- Zhao, N.; Li, Y.; Gu, J.; Fernandes, T.A.; Kirillova, M.V.; Kirillov, A.M. New copper(II) coordination compounds assembled from multifunctional pyridine-carboxylate blocks: Synthesis, structures, and catalytic activity in cycloalkane oxidation. Molecules 2019, 24, 6. [Google Scholar] [CrossRef]
- Khan, E. Pyridine derivatives as biologically active precursors; organics and selected coordination complexes. ChemistrySelect 2021, 6, 3041–3064. [Google Scholar] [CrossRef]
- Raj, D.; Padhi, S.K. The sporadic μ-pyridine bridge in transition metal complexes: A real bond or an interaction? Coord. Chem. Rev. 2022, 450, 214238–214245. [Google Scholar] [CrossRef]
- Posa, V.; Hajdu, B.; Tóth, G.; Domotor, O.; Kowol, C.R.; Keppler, B.K.; Spengler, G.; Gyurcsik, B.; Enyedy, E.A. The coordination modes of (thio) semicarbazone copper (II) complexes strongly modulate the solution chemical properties and mechanism of anticancer activity. J. Inorg. Biochem. 2022, 231, 111786–111792. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Reddy, P.K.; Kumar, B. Trace element zinc, a nature’s gift to fight unprecedented global pandemic COVID-19. Biol. Trace Elem. Res. 2021, 199, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Ho, E. Zinc deficiency, DNA damage and cancer risk. J. Nutr. Biochem. 2004, 15, 572–578. [Google Scholar] [CrossRef]
- Prasad, A.S. Discovery of human zinc deficiency: 50 years later. J. Trace Elem. Med. Biol. 2012, 26, 66–69. [Google Scholar] [CrossRef]
- Phukan, N.; Baruah, J.B. Hydrolysis of 4-(4-oxopentan-2-ylideneamino) benzoic acid and in-situ formation of nickel (II), zinc (II) and cadmium (II) complexes of 4-aminobenzoic acid. Inorg. Chim. Acta 2013, 396, 430–435. [Google Scholar] [CrossRef]
- Dutta, D.; Islam, S.M.N.; Saha, U.; Chetry, S.; Guha, A.K.; Bhattacharyya, M.K. Structural topology of weak non-covalent interactions in a layered supramolecular coordination solid of zinc involving 3-aminopyridine and benzoate: Experimental and theoretical studies. J. Chem. Crystallogr. 2018, 48, 156–163. [Google Scholar] [CrossRef]
- Pahontu, E.; Ilies, D.; Shova, S.; Oprean, C.; Paunescu, V.; Olaru, O.T.; Radulescu, F.S.; Gulea, A.; Rosu, T.; Draganescu, D. Synthesis, characterization, antimicrobial and antiproliferative activity evaluation of Cu (II), Co (II), Zn (II), Ni (II) and Pt (II) complexes with isoniazid-derived compound. Molecules 2017, 22, 650. [Google Scholar] [CrossRef]
- Paria, D.; Kundu, S.; Singh, K.S. Synthesis, characterization and antidiabetic activity of some water soluble Zn (II) complexes with (E)-N-(Thiophen-2-ylmethylene) anilines. Asian J. Chem. 2018, 30, 1115–1119. [Google Scholar] [CrossRef]
- Psomas, G. Copper(II) and zinc(II) coordination compounds of non-steroidal anti-inflammatory drugs: Structural features and antioxidant activity. Coord. Chem. Rev. 2020, 412, 213259–213271. [Google Scholar] [CrossRef]
- Akter, J.; Hanif, M.A.; Banu, L.A. Synthesis, characterization and antimicrobial activity of zinc(II) ibuprofen complexes with nitrogen-based ligands. J. Coord. Chem. 2016, 69, 1110–1122. [Google Scholar]
- Dhaveethu, K.; Ramachandramoorthy, T.; Thirunavukkarasu, K. Spectroscopic, Thermal and Biological Studies of Zn(II), Cd(II) and Hg(II) Complexes Derived from 3-Aminopyridine and Nitrite Ion. J. Korean Chem. Soc. 2013, 57, 712–720. [Google Scholar] [CrossRef]
- Koczoń, P. Growth Inhibition Mode of Action of Selected Benzoic Acid Derivatives against the Yeast Pichia anomala. J. Food Prot. 2009, 72, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Devrim, A.K.; Arslantas, A.; Kayat, N.; Necefoglu, H. An investigation on the effects of a zinc complex on gooses genomic DNA. Asian J. Chem. 2008, 20, 5827–5833. [Google Scholar]
- Tarushi, A.; Kakoulidou, C.; Raptopoulou, C.P.; Psycharis, V.; Kessissoglou, D.P.; Zoi, I.; Papadopoulos, A.N.; Psomas, G. Zinc complexes of diflunisal: Synthesis, characterization, structure, antioxidant activity, and in vitro and in silico study of the interaction with DNA and albumins. J. Inorg. Biochem. 2017, 170, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Ferrada, E.; Arancibia, V.; Loeb, B.; Norambuena, E.; Olea-Azarand, C.; Huidobro-Toro, J.P. Stoichiometry and conditional stability constants of Cu (II) or Zn (II) clioquinol complexes; implications for Alzheimer’s and Huntington’s disease therapy. Neurotoxicology 2007, 28, 445–449. [Google Scholar] [CrossRef]
- Verma, P.K.; Singh, R.K.; Kumar, S.; Shukla, A.; Kumar, S.; Gond, M.K.; Bharty, M.K.; Acharya, A. Cobalt (III) complex exerts anti-cancer effects on T cell lymphoma through induction of cell cycle arrest and promotion of apoptosis. J. Pharm. Sci. 2022, 30, 127–138. [Google Scholar] [CrossRef]
- Priya, N.P.; Firdous, A.P.; Jeevana, R.; Aravindakshan, K.K. Cytotoxic and Antitumour Studies of Acetoacetanilide N(4)-methyl(phenyl)thiosemicarbazone and its Transition Metal Complexes. Indian J. Pharm. Sci. 2015, 77, 655–660. [Google Scholar]
- Jayendran, M.; Kurup, M.R.P. Structural, spectral, cytotoxic and biocatalytic studies of a dinuclearphenoxo bridged Zn (II) complex from NNO donor tridentate Schiff base. Chem. Data Collect. 2022, 39, 100853–100863. [Google Scholar] [CrossRef]
- APEX3, SAINT, SADABS and XP; Bruker AXS Inc.: Madison, WI, USA, 2015.
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; Streek, J.; Wood, P.A. Mercury CSD 2.0–new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Brandenburg, K. Diamond 3.1f; Crystal Impact GbR: Bonn, Germany, 2008. [Google Scholar]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104–154119. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef]
- Ahlrichs, R.; Bar, M.; Hacer, M.; Horn, H.; Kömel, C. Electronic structure calculations on workstation computers: The program system turbomole. Chem. Phys. Lett. 1989, 162, 165–169. [Google Scholar] [CrossRef]
- Contreras-Garcia, J.; Johnson, E.R.; Keinan, S.; Chaudret, R.; Piquemal, J.P.; Beratan, D.N.; Yang, W. NCIPLOT: A program for plotting non-covalent interaction regions. J. Chem. Theory Comput. 2011, 7, 625–632. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in molecules. Chem. Rev. 1991, 91, 893–928. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Humphrey, J.W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Klein, G. Comparative studies of mouse tumors with respect to their capacity for growth as “ascites tumors” and their average nucleic acid content per cell. Exp. Cell Res. 1951, 2, 518–524. [Google Scholar] [CrossRef]
- Sriram, M.I.; Kanth, S.B.M.; Kalishwaralal, K.; Gurunathan, S. Antitumor activity of silver nanoparticles in Dalton’s lymphoma ascites tumor model. Int. J. Nanomed. 2010, 5, 753–762. [Google Scholar]
- Pourahmad, J.; Salimi, A. Isolated human peripheral blood mononuclear cell (PBMC), a cost effective tool for predicting immunosuppressive effects of drugs and xenobiotics. Iran J. Pharm. Res. 2015, 14, 979–983. [Google Scholar] [PubMed]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Prot. Immunol. 2015, 111, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Avelar-Freitas, B.A.; Almeida, V.G.; Pinto, M.C.X.; Mourão, F.A.G.; Massensini, A.R.; Martins-Filho, O.A.; Rocha-Vieira, E.; Brito-Melo, G.E.A. Trypan blue exclusion assay by flow cytometry. Braz. J. Med. Biol. Res. 2014, 47, 307–315. [Google Scholar] [CrossRef]
- Verma, A.K.; Prasad, S.B. Changes in Glutathione, Oxidative Stress and Mitochondrial Membrane Potential in Apoptosis Involving the Anticancer Activity of Cantharidin Isolated from Redheaded Blister Beetles, Epicautahirticornis. Anti-Cancer Agents Med. Chem. 2013, 13, 1096–1114. [Google Scholar] [CrossRef]
- Verma, A.K.; Singh, S. Phytochemical analysis and in vitro cytostatic potential of ethnopharmacological important medicinal plants. Toxicol. Rep. 2020, 7, 443–452. [Google Scholar] [CrossRef]
- Berry, M.; Fielding, B.; Gamieldien, J. Practical considerations in virtual screening and Molecular docking. Emer. Trends Comput. Biol. 2015, 45, 487–502. [Google Scholar]
- Frenzel, A.; Grespi, F.; Chmelewskij, W.; Villunger, A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 2009, 14, 584–596. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Soderquist, R.S.; Eastman, A. BCL2 Inhibitors as Anticancer Drugs: A Plethora of Misleading BH3 Mimetics BCL2 Inhibitors as Anticancer Agents. Mol. Cancer Ther. 2016, 15, 2011–2017. [Google Scholar] [CrossRef]
- Alexandra, B. CCDC 1852819. CSD Commun. 2018. [Google Scholar] [CrossRef]
- Goher, M.A.S.; Abu-Youssef, M.A.M.; Mautner, F.A. Synthesis, spectral and structural characterization of a monomeric chloro complex of zinc (II) with picolinic acid, [Zn (C5H4NCO2H)(C5H4NCO2)Cl]. Polyhedron 1996, 15, 453–457. [Google Scholar] [CrossRef]
- Jin, S.; Wang, D.; Wang, X.; Guo, M.; Zhao, Q. Structural characterization of a Cu(I) coordination polymer constructed by weak intermolecular Cu–Cl⋯ Cl–Ar interaction. J. Inorg. Organomet. Polym. Mater. 2008, 18, 300–303. [Google Scholar] [CrossRef]
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Priimagi, A.; Resnati, G.; Terraneo, G. The halogen bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Du, Z.; Pan, Q.; Zhang, L.; Liu, D. The first 3, 5, 6-trichloropyridine-2-oxyacetate bridged manganese coordination polymer with features of π⋯π stacking and halogen⋯halogen interactions: Synthesis, crystal analysis and magnetic properties. Inorg. Chim. Acta 2020, 509, 119677–119688. [Google Scholar] [CrossRef]
- Dojer, B.; Pevec, A.; Belaj, F.; Kristl, M. Two new zinc (II) acetates with 3–and 4–aminopyridine: Syntheses and structural properties. Acta Chim. Slov. 2015, 62, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhua, H.; Wang, Y.; Liu, P.; Zhou, L.; Li, D.; Shi, Q. Novel supramolecular structures and theoretical calculation of Ag(I) and Zn(II) complexes based on a new dipyridylamine ligand. J. Mol. Struct. 2005, 779, 61–67. [Google Scholar] [CrossRef]
- Yaghoubi, A.; Khazaei, M.; Ghazvini, K.; Movaqar, A.; Avan, A.; Hasanian, S.M.; Soleimanpour, S. Peptides with Dual Antimicrobial-Anticancer Activity Derived from the N-terminal Region of H. pylori Ribosomal Protein L1 (RpL1). Int. J. Peptide Res. Ther. 2021, 27, 1057–1067. [Google Scholar] [CrossRef]
- Pedrosa, P.; Carvalho, A.; Baptista, P.V.; Fernandes, A.R. Basic Concepts Viewed from Frontier in Inorganic Coordination Chemistry; Wiley-VCH: Weinheim, Germany, 2018. [Google Scholar]
- Wang, Y.; Lin, X.M.; Bai, F.Y.; Sun, L.X. Novel vanadium complexes with rigid carboxylate ligands: Synthesis, structure and catalytic bromine dynamics of phenol red. J. Mol. Struct. 2017, 1149, 379–386. [Google Scholar] [CrossRef]
- Sharma, R.P.; Saini, A.; Kumar, J.; Kumar, S.; Venugopalan, P.; Ferretti, V. Coordination complexes of copper(II) with herbicide-trichlorophenoxyacetate: Syntheses, characterization, single crystal X-ray structure and packing analyses of monomeric [Cu(γ-pic)3(2,4,5-trichlorophenoxyacetate)]·H2O, [trans-Cu(en)2(2,4,5-trichlorophenoxyacetate)2]·2H2O and dimeric [Cu2(H2tea)2(2,4,5-trichlorophenoxyacetate)2]·2(H2O). Inorg. Chim. Acta 2017, 457, 59–68. [Google Scholar]
- Diab, M.A.; Mohamed, G.G.; Mahmoud, W.H.; El-Sonbati, A.Z.; Morgan, S.M.; Abbas, S.Y. Metal-and covalent organic frameworks as catalyst for organic transformation: Comparative overview and future perspectives. Appl. Organomet. Chem. 2019, 33, 4945–4965. [Google Scholar]
- Deacon, G.B.; Phillips, R.J. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Alves, S.R.; Santos, R.L.; Fornaciari, L.B.; Colquhoun, A.; Oliveira, D.S. A novel μ-oxo-diruthenium(III,III)-ibuprofen-(4-aminopyridine) chloride derived from the diruthenium(II,III)-ibuprofen paddlewheel metallodrug shows anticancer properties. J. Inorg. Biochem. 2021, 225, 111596. [Google Scholar] [CrossRef]
- Araujo, E.L.; Barbosa, H.F.G.; Dockal, E.R.; Cavalheiro, E.T.G. Synthesis, characterization and biological activity of Cu(II), Ni(II) and Zn(II) complexes of biopolymeric Schiff bases of salicylaldehydes and chitosan. Int. J. Biol. Macromol. 2017, 95, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.P.; Sharma, R.; Bala, R.; Venugopalan, P. Synthesis, spectroscopic and X-ray structural study of [trans-di (4-aminobenzoato) bis (ethylenediamine) cobalt (III)] 4-aminobenzoate tetrahydrate. J. Chem. Crystallogr. 2005, 35, 595–601. [Google Scholar] [CrossRef]
- Koleva, B.B.; Trendafilova, E.N.; Arnaudov, M.G. Spectral analysis and crystal structure of a new mononuclear copper (II) complex of 3-aminopyridine. Transit. Met. Chem. 2006, 31, 866–873. [Google Scholar] [CrossRef]
- Bonhomme, F.; Alam, T.M.; Celestian, A.J.; Tallant, D.R.; Boyle, T.J.; Cherry, B.R.; Tissot, R.G.; Rodriguez, M.A.; Parise, J.B.; Nyman, M. Tribasic lead maleate and lead maleate: Synthesis and structural and spectroscopic characterizations. Inorg. Chem. 2005, 44, 7394–7402. [Google Scholar] [CrossRef]
- Al-Azab, F.M.; Jamil, Y.M.S.; Maqtari, M.A.A.; Al Qadasy, M.K. Synthesis, characterization and biological activity of mixed transition metal complexes of pyrimethamine with 1, 10-phenanthroline. J. Modern Sci. Eng. 2017, 1, 9–21. [Google Scholar]
- Mchiri, C.; Edziri, H.; Hajji, H.; Bouachrine, M.; Acherar, S.; Frochot, C.; Eldine, H.O.B.; Moussa, S.B.; Nasri, H. Microwave assisted synthesis and characterizations of zn (ii) metal complex with bio-active isoniazid and benzoate ions. J. Chem. Sci. 2022, 134, 22–44. [Google Scholar] [CrossRef]
- Rosales-Vázquez, L.D.; Valdes-Garcia, J.; Bazany-Rodriguez, I.J.; Germán-Acacio, J.M.; Martínez-Otero, D.; Vilchis-Néstor, F.; Morales-Luckie, R.; Sánchez-Mendieta, V.; Dorazco-González, A. A sensitive photoluminescentchemosensor for cyanide in water based on a zinc coordination polymer bearing ditert-butyl-bipyridine. Dalton Trans. 2019, 48, 12407–12420. [Google Scholar] [CrossRef] [PubMed]
- Bora, S.J.; Das, B.K. Synthesis, structure and properties of a fumarate bridged Ni (II) coordination polymer. J. Mol. Struct. 2011, 999, 83–88. [Google Scholar] [CrossRef]
- Bhattacharyya, M.K.; Gogoi, A.; Chetry, S.; Dutta, D.; Verma, A.K.; Sarma, B.; Franconetti, A.; Frontera, A. Antiproliferative evaluation and supramolecular association in Mn (II) and Zn (II) bipyridine complexes: Combined experimental and theoretical studies. J. Inorg. Biochem. 2019, 200, 110803–110813. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Chen, S.; Gao, S. Two Mn (II) chloride complexes containing guest molecules. J. Therm. Anal. Calorim. 2007, 89, 567–571. [Google Scholar] [CrossRef]
- Goher, M.A.; Hafez, A.K.; Abu-Youssef, M.A.M.; Badr, A.M.A.; Gspan, C.; Mautner, F.A. New metal (II) complexes containing monodentate and bridging 3-aminopyridine and azido ligands. Polyhedron 2004, 23, 2349–2356. [Google Scholar] [CrossRef]
- Espinosa, E.; Molins, E.; Lecomte, C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998, 285, 170–173. [Google Scholar] [CrossRef]
- Ibrahim, M.A.A.; Saeed, R.A.A.; Shehata, M.N.I.; Ahmed, M.N.; Shawky, A.M.; Khowdiary, M.M.; Elkaeed, E.B.; Soliman, M.E.S.; Moussa, N.A.M. Autophagy-mediated regulation of different meristems in plants. Int. J. Mol. Sci. 2022, 23, 3114. [Google Scholar] [CrossRef]
- Tennant, J.R. Evaluation of the trypan blue technique for determination of cell viability. Transplantation 1964, 2, 685–694. [Google Scholar] [CrossRef]
- Altman, S.A.; Randers, L.; Rao, G. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnol. Progr. 1993, 9, 671–674. [Google Scholar] [CrossRef]
- Illán-Cabeza, N.A.; Hueso-Urena, F.; Moreno-Carretero, M.N.; Martínez-Martos, J.M.; Ramírez-Expósito, M.J. Synthesis, characterization and antiproliferative activity of metal complexes with the Schiff base derived from the condensation 1:2 of 2,6-diformyl-4-methylphenol and 5,6-diamino-1,3-dimethyluracil. J. Inorg. Biochem. 2008, 102, 647–655. [Google Scholar] [CrossRef]
- Alalawy, M.D.; Patel, U.H.; Bhatt, B.S.; Patel, N.J. Exploring qualitative and quantitative contributions of intermolecular interactions, DNA-binding and in vitro cytotoxic activity of isostructural and isomorphous Cd and Zn complexes. Polyhedron 2020, 185, 114595–114608. [Google Scholar] [CrossRef]
- Nath, H.; Dutta, D.; Sharma, P.; Frontera, A.; Verma, A.K.; Barceló-Oliver, M.; Devi, M.; Bhattacharyya, M.K. Adipato bridged novel hexanuclear Cu(ii) and polymeric Co(ii) coordination compounds involving cooperative supramolecular assemblies and encapsulated guest water clusters in a square grid host: Antiproliferative evaluation and theoretical studies. Dalton Trans. 2020, 49, 9863–9881. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Valdivia, A.A.; Cepeda, J.; Fernandez, B.; Medina-O’donnell, M.; Oyarzabal, I.; Parra, J.; Jannus, F.; Choquesillo-Lazarte, D.; García, J.A.; Lupiáñez, J.A.; et al. 5-Aminopyridine-2-carboxylic acid as appropriate ligand for constructing coordination polymers with luminescence, slow magnetic relaxation and anti-cancer properties. J. Inorg. Biochem. 2020, 207, 11105–11125. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, H.; Yuan, Y.; Wang, X.; Tang, J.; Chu, P.; Zhang, Y. Two novel nanostructured La(III) coordination polymers were synthesized by sonochemical: Crystal structures along with anticancer activity on human nasopharyngeal carcinoma cells. J. Iran. Chem. Soc. 2020, 17, 2307–2316. [Google Scholar] [CrossRef]
- Li, W.; Chen, X. Sulphated alumina tungstic acid (SATA): A highly efficient and novel heterogeneous mesostructured catalyst for the synthesis of pyrazolecarbonitrile derivatives and evaluation of green metrics. Inorg. Nano-Met. Chem. 2017, 47, 1682–1685. [Google Scholar] [CrossRef]
- Das, A.; Sharma, P.; Gomila, R.M.; Frontera, A.; Verma, A.K.; Sarma, B.; Bhattacharyya, M.K. Synthesis, structural topologies and anticancer evaluation of phenanthroline-based 2, 6-pyridinedicarboxylato Cu (II) and Ni (II) compounds. Polyhedron 2021, 45, 115632–115641. [Google Scholar] [CrossRef]
- D’Aguanno, S.; Bufalo, D.D. Inhibition of anti-apoptotic Bcl-2 proteins in preclinical and clinical studies: Current overview in cancer. Cells 2020, 9, 1287. [Google Scholar] [CrossRef]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007, 26, 1324–1337. [Google Scholar] [CrossRef]
- Harris, M.H.; Thompson, C.B. The role of the Bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell Death Differ. 2000, 7, 1182–1191. [Google Scholar] [CrossRef]
- Bruncko, M. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-Xl. J. Med. Chem. 2007, 50, 641–662. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Scoring functions for protein-ligand docking. Curr. Protein Peptide Sci. 2006, 7, 407–420. [Google Scholar]
- Konakanchi, R.; Pamidimalla, G.S.; Prashanth, J.; Naveen, T.; Kotha, L.R. Structural elucidation, theoretical investigation, biological screening and molecular docking studies of metal(II) complexes of NN donor ligand derived from 4-(2-aminopyridin-3-methylene)aminobenzoic acid. BioMetals 2021, 34, 529–556. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).