Synthesis and Computational and X-ray Structure of 2, 3, 5-Triphenyl Tetrazolium, 5-Ethyl-5-phenylbarbituric Acid Salt
Abstract
1. Introduction
2. Experimental
2.1. Chemistry
2.1.1. General
2.1.2. 5-Ethyl-5-phenylbarbituric acid Triphenyl Tetrazolium Salt (4)
2.2. X-ray Crystallography
General
2.3. Computational Study
3. Results and Discussion
3.1. Chemistry
3.2. X-ray Crystallography
3.3. Computational Study
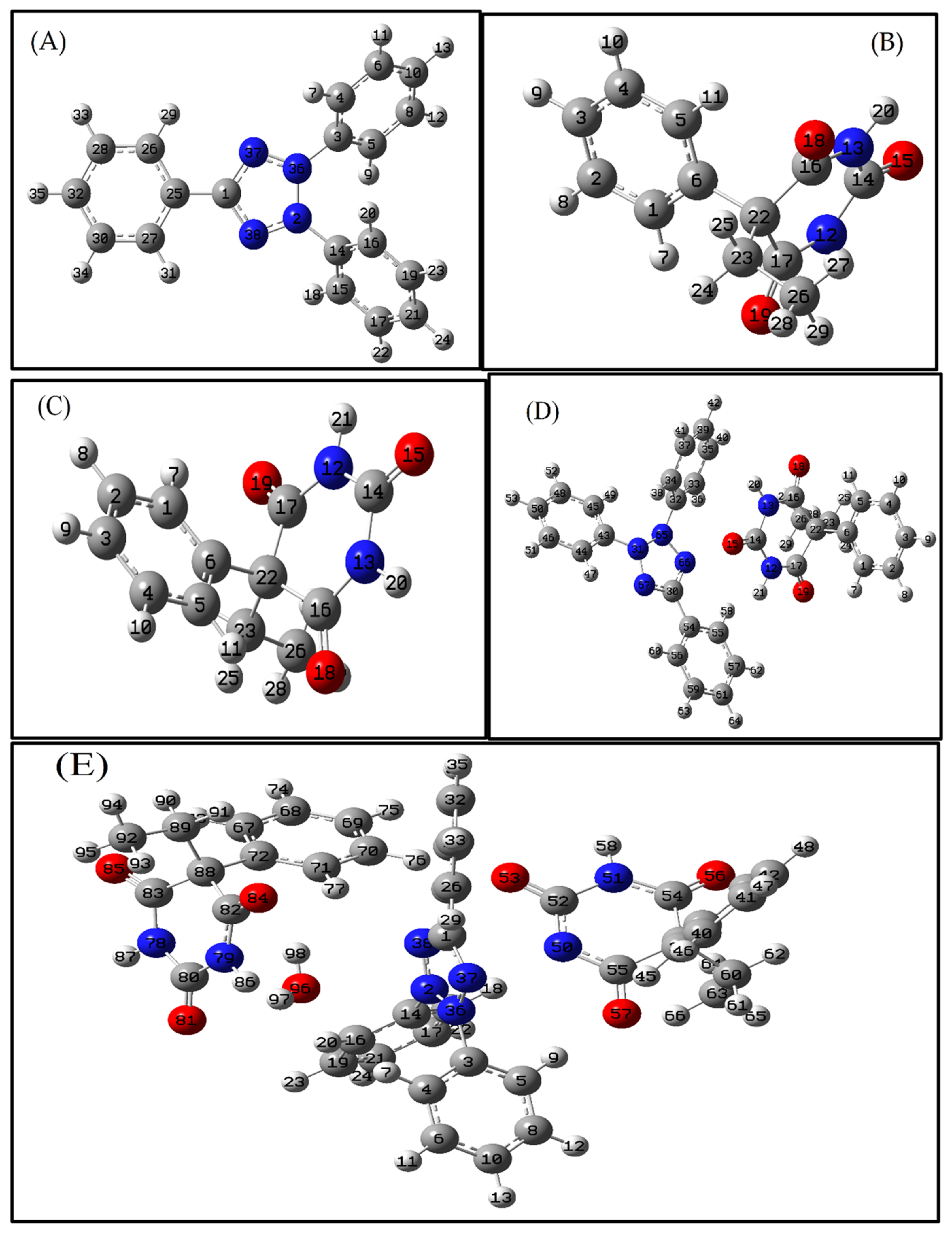
3.3.1. Molecular Geometry
3.3.2. Interaction Energies (IE)
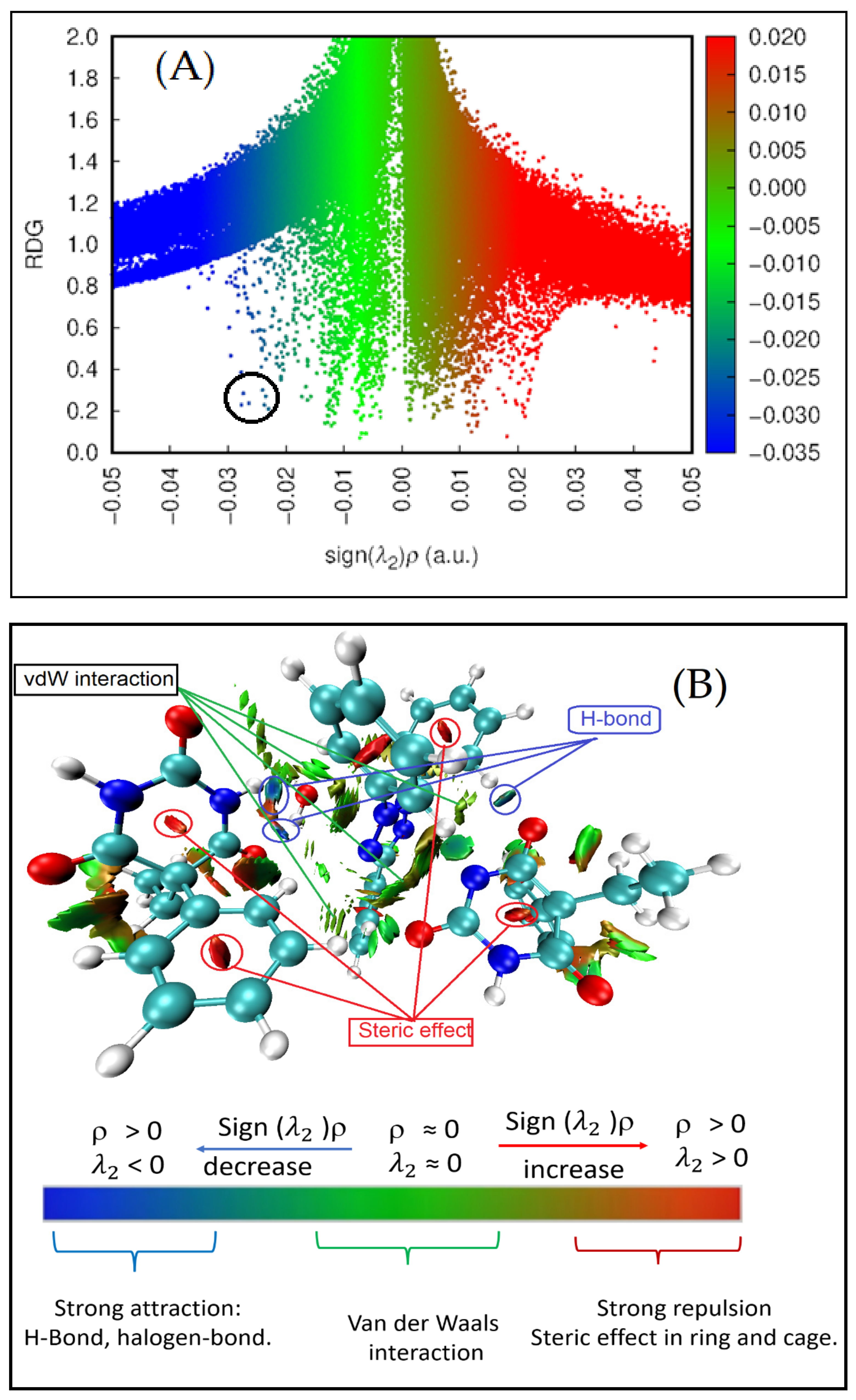
3.3.3. Non-Covalent Interaction (NCI) Index
3.3.4. Quantum Theory of Atoms in Molecules (QTAIM)
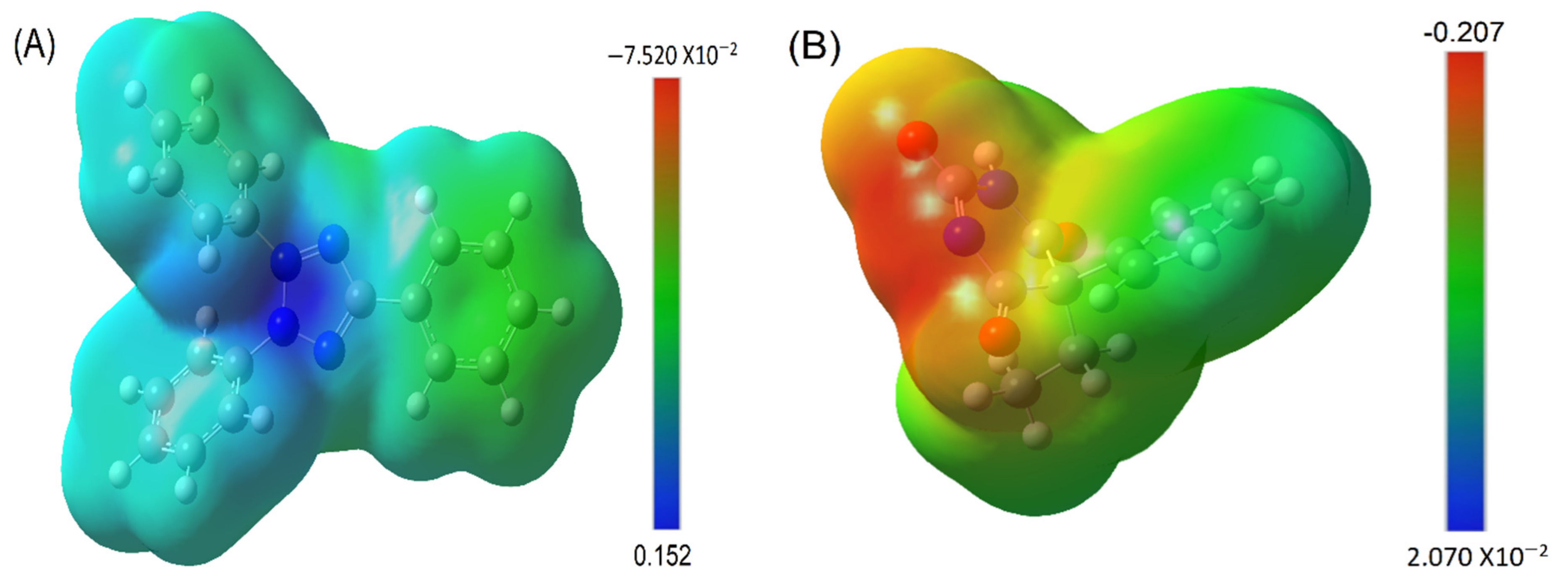
3.4. MESP Analysis
3.5. Reactivity Descriptors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gennaro, A.R. Remington: The Science and Practice of Pharmacy, 19th ed.; Easton, P.A., Ed.; Mack Publishing Co.: London, UK, 1995; Volume II, pp. 1164–1165. [Google Scholar]
- Schmidt, D.; Ilse, E.; Ferdinand, H. The influence of seizure type on the efficacy of plasma concentrations of phenytoin, phenobarbital, and carbamazepine. Arch. Neurol. 1986, 43, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Ilangaratne, N.B.; Nilanka, N.M.; Gail, S.B.; Josemir, W.S. Phenobarbital: Missing in action. Bull. World Health Organ. 2012, 90, 871−871a. [Google Scholar] [CrossRef] [PubMed]
- Gavazov, K.B.; Atanas, N.; Vanya, D.L. The use of tetrazolium salts in inorganic analysis. Russ. Chem. Rev. 2007, 76, 169–179. [Google Scholar] [CrossRef]
- Abbas, M.N.; Mostafa, G.A.E. New triiodomercurate-modified carbon paste electrode for the potentiometric determination of mercury. Anal. Chim. Acta 2003, 478, 329–335. [Google Scholar] [CrossRef]
- Hassanien, M.M.; Abou-El-Sherbini, K.S.; Mostafa, G.A.E. A novel tetrachlorothallate(III)-PVC membrane sensor for the potentiometric determination of thallium (III). Talanta 2003, 59, 383–392. [Google Scholar] [CrossRef]
- Mostafa, G.A.E. PVC matrix membrane sensor for potentiometric determination of triphenyl tetrazolium chloride and ascorbic acid. Ann. Di Chim. 2007, 97, 1247–1256. [Google Scholar] [CrossRef]
- Fenniri, H.; Mathivanan, P.; Vidale, K.L.; Sherman, D.M.; Hallenga, K.; Wood, K.V.; Stowell, J.G. Helical rosette nanotubes: Design, self-assembly, and characterization. J. Am. Chem. Soc. 2001, 123, 3854–3855. [Google Scholar] [CrossRef]
- Kruse, P.; Johnson, E.R.; DiLabio, G.A.; Wolkow, R.A. Patterning of vinylferrocene on H−Si (100) via self-directed growth of molecular lines and STM-induced decomposition. Nano Lett. 2002, 2, 807–810. [Google Scholar] [CrossRef]
- Sheiko, S.S.; Sun, F.C.; Randall, A.; Shirvanyants, D.; Rubinstein, M.; Lee, H.-i.; Matyjaszewski, K. Adsorption-induced scission of carbon–carbon bonds. Nature 2006, 440, 191–194. [Google Scholar] [CrossRef]
- Johnson, E.R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A.J.; Yang, W. Revealing noncovalent interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef]
- DiLabio, G.A.; Piva, P.G.; Kruse, P.; Wolkow, R.A. Dispersion interactions enable the self-directed growth of linear alkane nanostructures covalently bound to silicon. J. Am. Chem. Soc. 2004, 126, 16048–16050. [Google Scholar] [CrossRef] [PubMed]
- Kollman, P.A. Noncovalent interactions. Acc. Chem. Res. 1977, 10, 365–371. [Google Scholar] [CrossRef]
- Keinan, S.; Ratner, M.A.; Marks, T.J. Molecular zippers–designing a supramolecular system. Chem. Phys. Lett. 2004, 392, 291–296. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2007, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Becke, A. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785. [Google Scholar] [CrossRef]
- Ghabbour, H.A.; Bakheit, A.H.; Ezzeldin, E.; Mostafa, G.A.E. Synthesis Characterization and X-ray Structure of 2-(2,6-Dichlorophenylamino)-2-imidazoline Tetraphenylborate: Computational Study. Appl. Sci. 2022, 12, 3568. [Google Scholar] [CrossRef]
- Ditchfield, R.; Hehre, W.J.; Pople, J.A. Self-consistent molecular-orbital methods. IX. An extended Gaussian-type basis for molecular-orbital studies of organic molecules. J. Chem. Phys. 1971, 54, 724–728. [Google Scholar] [CrossRef]
- Mostafa, G.A.E.; Bakheit, A.; AlMasoud, N.; AlRabiah, H. Charge Transfer Complexes of Ketotifen with 2,3-Dichloro-5,6-dicyano-p-benzoquinone and 7,7,8,8-Tetracyanoquodimethane: Spectroscopic Characterization Studies. Molecules 2021, 26, 2039. [Google Scholar] [CrossRef]
- Solovyov, S.A. Categorical foundations of variety-based topology and topological systems. Fuzzy Sets Syst. 2012, 192, 176–200. [Google Scholar] [CrossRef]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
- Grabowski, S.J. Theoretical studies of strong hydrogen bonds. Annu. Rep. Sect. C Phys. Chem. 2006, 102, 131–165. [Google Scholar] [CrossRef]
- Espinosa, E.; Molins, E.; Lecomte, C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998, 285, 170–173. [Google Scholar] [CrossRef]
- Espinosa, E.; Alkorta, I.; Rozas, I.; Elguero, J.; Molins, E. About the evaluation of the local kinetic, potential and total energy densities in closed-shell interactions. Chem. Phys. Lett. 2001, 336, 457–461. [Google Scholar] [CrossRef]
- Bader, R.F.; Essén, H. The characterization of atomic interactions. J. Chem. Phys. 1984, 80, 1943–1960. [Google Scholar] [CrossRef]
- Espinosa, E.; Alkorta, I.; Elguero, J.; Molins, E. From weak to strong interactions: A comprehensive analysis of the topological and energetic properties of the electron density distribution involving X–H⋯ F–Y systems. J. Chem. Phys. 2002, 117, 5529–5542. [Google Scholar] [CrossRef]
- Weinhold, F. Nature of H-bonding in clusters, liquids, and enzymes: An ab initio, natural bond orbital perspective. J. Mol. Struct. THEOCHEM 1997, 398, 181–197. [Google Scholar] [CrossRef]
- Bader, R.F. Atoms in molecules. Acc. Chem. Res. 1985, 18, 9–15. [Google Scholar] [CrossRef]
- Bader, R.F. A quantum theory of molecular structure and its applications. Chem. Rev. 1991, 91, 893–928. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; Taie, H.A.A.; Bakheit, A.H.; Marzouk, M.; Abdellatif, M.M.; Al-Salahi, R. Biological Evaluation of 4-(1H-triazol-1-yl)benzoic Acid Hybrids as Antioxidant Agents: In Vitro Screening and DFT Study. Appl. Sci. 2021, 11, 11642. [Google Scholar] [CrossRef]
- Pakiari, A.; Fakhraee, S. Electron density analysis of weak van der Waals complexes. J. Theor. Comput. Chem. 2006, 5, 621–631. [Google Scholar] [CrossRef]
- Koch, U.; Popelier, P.L. Characterization of CHO hydrogen bonds on the basis of the charge density. J. Phys. Chem. 1995, 99, 9747–9754. [Google Scholar] [CrossRef]
- Popelier, P. Characterization of a dihydrogen bond on the basis of the electron density. J. Phys. Chem. A 1998, 102, 1873–1878. [Google Scholar] [CrossRef]
- Keith, T.A. AIMAll (Version 10.05. 04, Professional); TK Gristmill Software: Overland Park, KS, USA, 1997. [Google Scholar]
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView, Version 6.1; Semichem Inc.: Shawnee Mission, KS, USA, 2016. [Google Scholar]
- Contreras-García, J.; Johnson, E.R.; Keinan, S.; Chaudret, R.; Piquemal, J.-P.; Beratan, D.N.; Yang, W. NCIPLOT: A program for plotting noncovalent interaction regions. J. Chem. Theory Comput. 2011, 7, 625–632. [Google Scholar] [CrossRef]
- Moffat, A.C.; Osselton, M.D.; Brian, W.; Clarke, E.G.C. Clarke’s analysis of drugs and poisons. In Pharmaceuticals, Body Fluids and Postmortem Materials; Pharmaceutical Press: London, UK, 2004; Volume 1, p. 1431. [Google Scholar]
- Gjikaj, M.; Xie, T.; Brockner, W. Uncommon compounds in antimony pentachloride-ionic liquid systems: Synthesis, crystal structure and vibrational Spectra of the Complexes [TPT][SbCl6] and [Cl-EMIm][SbCl6]. Z. Für Anorg. Und Allg. Chem. 2009, 635, 1036–1040. [Google Scholar] [CrossRef]

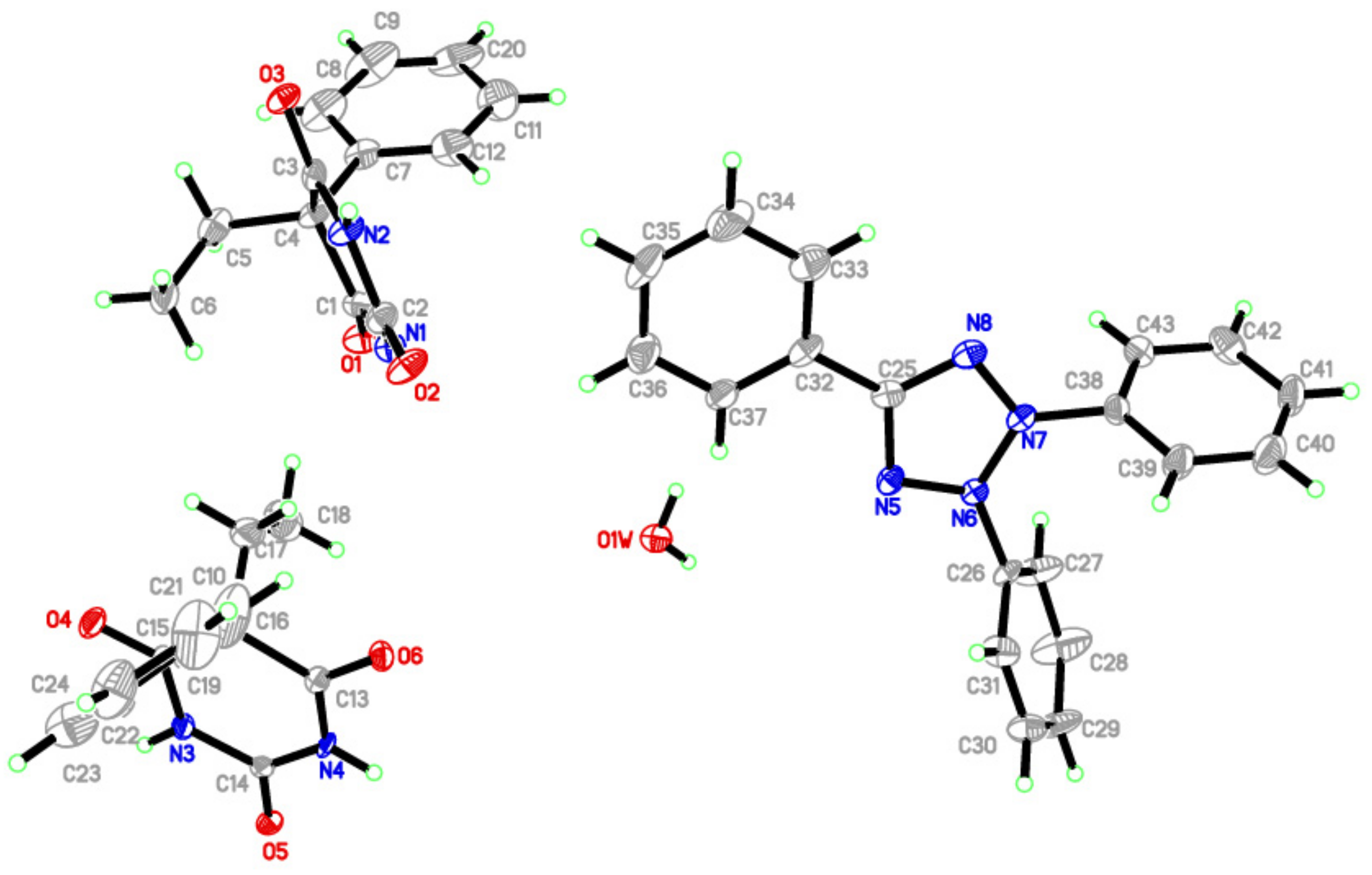
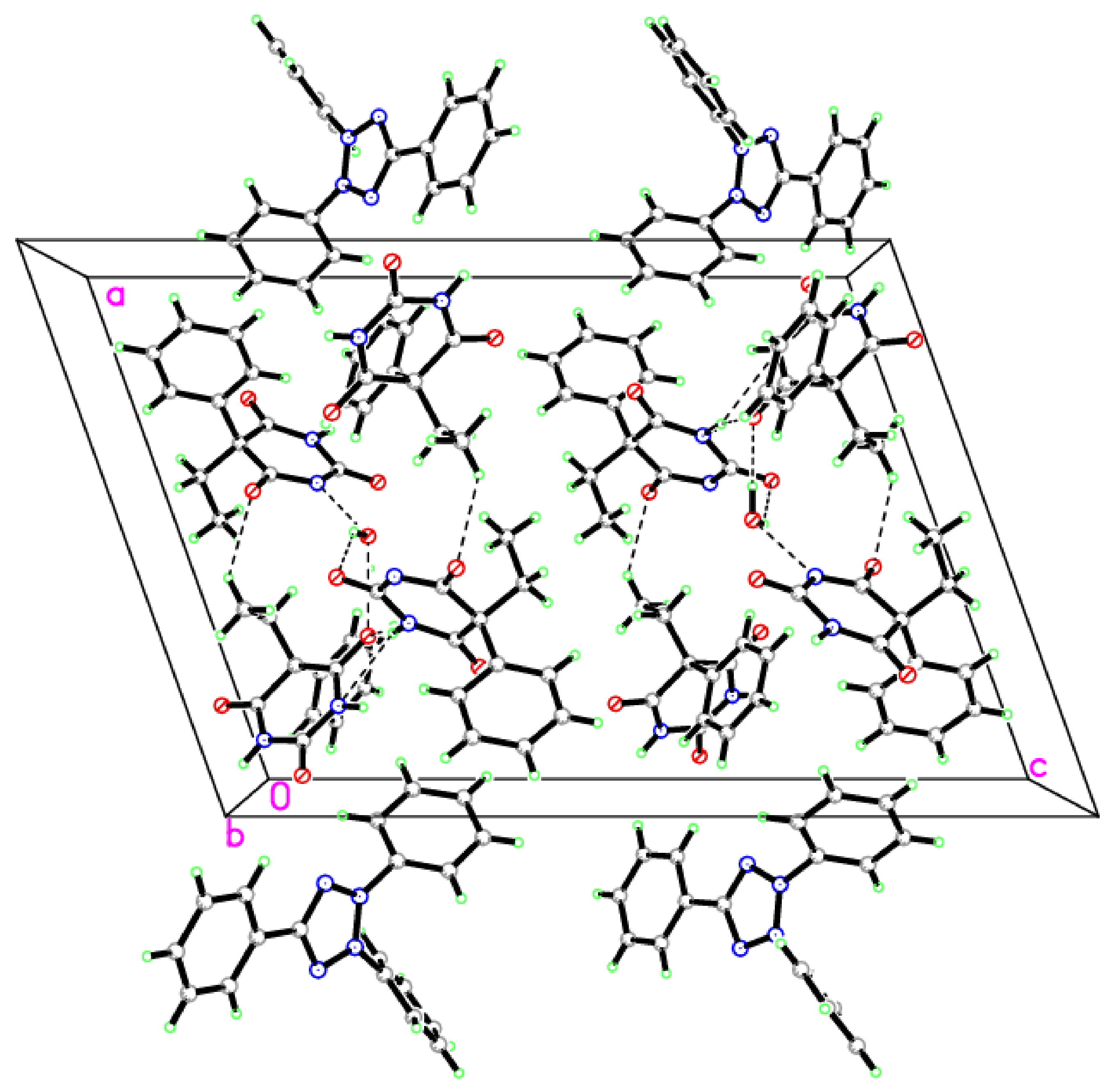
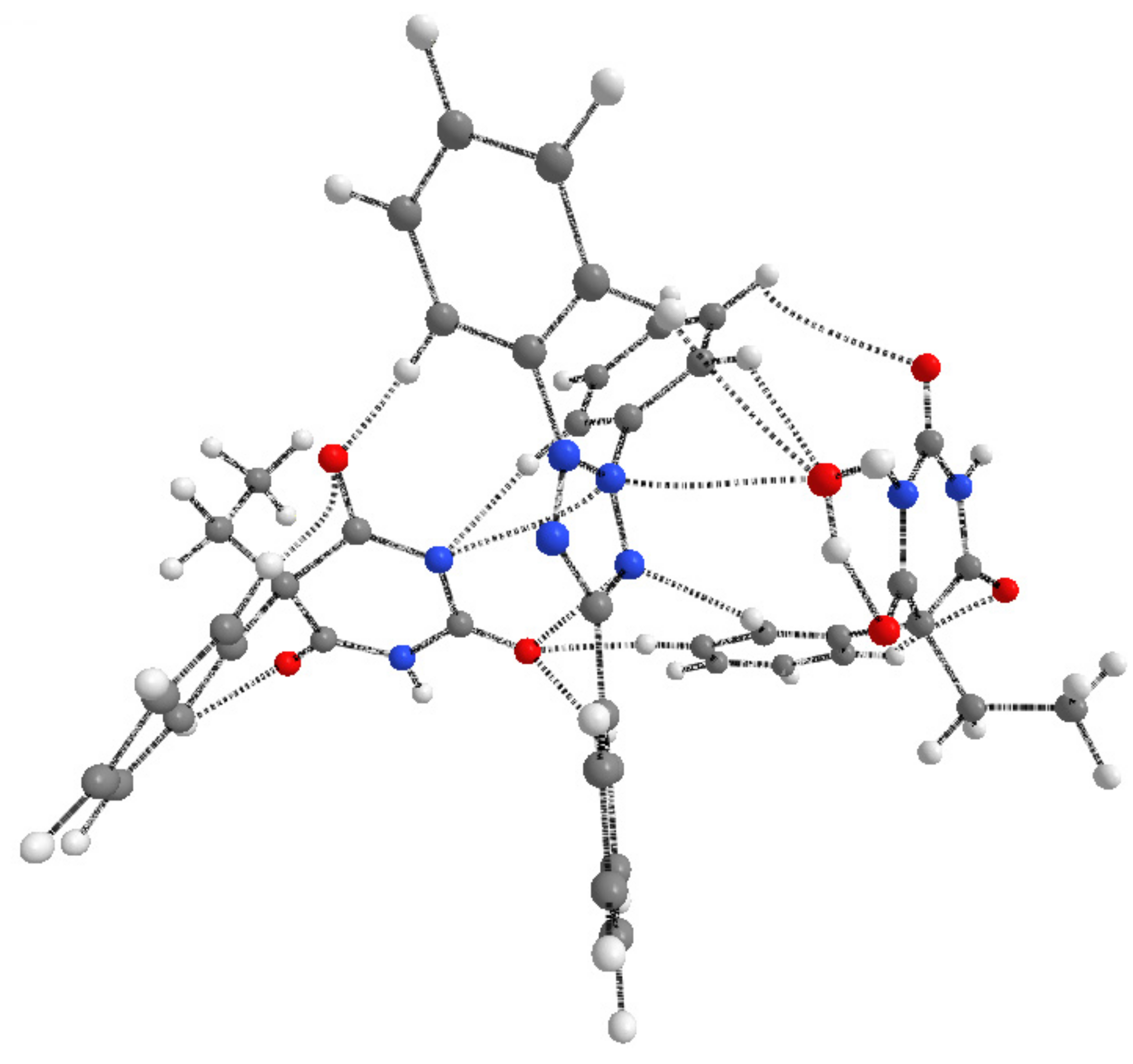

| Crystal Data | |
|---|---|
| Chemical formula | C43H40N8O7 |
| Molecular weight | 780.83 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, and c (Å) | 15.3678 (9), 12.2710 (7), and 21.8514 (13) |
| α, β, and γ (°) | 90.00, 109.867 (2), and 90.00 |
| V (Å3) | 3875.5 (4) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.09 |
| Crystal size (mm) | 0.36 × 0.14 × 0.05 |
| Data collection | |
| Diffractometer | Bruker APEX-II D8 venture diffractometer |
| Absorption correction | Multi-scan SADABS Bruker 2014 |
| Tmin and Tmax | 0.873 and 0.891 |
| No. of measured, independent, and observed [I > 2σ(I)] reflections | 68448, 6820, and 4452 |
| Rint | 0.156 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), and S | 0.086, 0.216, and 1.07 |
| No. of reflections | 6820 |
| No. of parameters | 536 |
| No. of restraints | 0 |
| H atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax and Δρmin (e Å−3) | 1.00 and −0.59 |
| O4—C15 | 1.221 (6) | N7—C38 | 1.460 (6) |
| O5—C14 | 1.242 (5) | N8—C25 | 1.342 (6) |
| O6—C13 | 1.256 (5) | N3—C15 | 1.367 (6) |
| O1—C1 | 1.212 (5) | N3—C14 | 1.400 (5) |
| O2—C2 | 1.217 (6) | N4—C14 | 1.334 (5) |
| O3—C3 | 1.218 (5) | N4—C13 | 1.328 (6) |
| N5—N6 | 1.314 (5) | N1—C1 | 1.374 (6) |
| N5—C25 | 1.346 (6) | N1—C2 | 1.371 (6) |
| N6—C26 | 1.445 (6) | N2—C3 | 1.366 (6) |
| N6—N7 | 1.328 (6) | N2—C2 | 1.383 (7) |
| N7—N8 | 1.317 (6) | ||
| N6—N5—C25 | 103.5 (4) | O6—C13—N4 | 120.2 (4) |
| N5—N6—C26 | 123.8 (4) | O6—C13—C16 | 116.7 (4) |
| N7—N6—C26 | 126.0 (4) | N4—C13—C16 | 123.1 (4) |
| N5—N6—N7 | 110.2 (4) | N3—C14—N4 | 121.3 (4) |
| N6—N7—C38 | 124.7 (4) | O5—C14—N4 | 122.4 (4) |
| N8—N7—C38 | 125.2 (4) | O5—C14—N3 | 116.3 (4) |
| N6—N7—N8 | 110.2 (4) | O4—C15—C16 | 122.9 (4) |
| N7—N8—C25 | 103.5 (4) | O4—C15—N3 | 120.6 (4) |
| C14—N3—C15 | 125.2 (4) | N3—C15—C16 | 116.6 (4) |
| C13—N4—C14 | 120.4 (4) | O1—C1—N1 | 120.4 (4) |
| C1—N1—C2 | 125.6 (4) | O1—C1—C4 | 121.2 (4) |
| C2—N2—C3 | 126.7 (4) | N1—C1—C4 | 118.4 (3) |
| N5—C25—N8 | 112.6 (4) | N1—C2—N2 | 116.6 (4) |
| N8—C25—C32 | 124.5 (4) | O2—C2—N2 | 122.1 (4) |
| N5—C25—C32 | 122.9 (4) | O2—C2—N1 | 121.3 (4) |
| N6—C26—C31 | 118.0 (4) | N2—C3—C4 | 117.8 (4) |
| N6—C26—C27 | 118.8 (4) | O3—C3—N2 | 121.0 (4) |
| N7—C38—C39 | 118.2 (4) | N7—C38—C43 | 117.9 (4) |
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| O1W—H1OW···O2i | 0.85 (8) | 2.33 (9) | 3.017 (5) | 139 (7) |
| O1W—H2OW···O6ii | 0.90 (8) | 1.85 (8) | 2.733 (6) | 167 (6) |
| N2—H1N2···O6iii | 0.91 (7) | 1.94 (7) | 2.827 (5) | 163 (5) |
| N2—H1N2···N4iii | 0.91 (7) | 2.60 (6) | 3.361 (5) | 141 (6) |
| N3—H3A···O5iv | 0.8600 | 1.9300 | 2.793 (5) | 179.00 |
| C18—H18A···O1 | 0.9600 | 2.5700 | 3.319 (7) | 135.00 |
| C20—H20A···O5v | 0.9300 | 2.5100 | 3.348 (8) | 150.00 |
| C27—H27A···O4ii | 0.9300 | 2.5400 | 3.253 (7) | 133.00 |
| C28—H28A···O1vi | 0.9300 | 2.4800 | 3.109 (7) | 125.00 |
| C29—H29A···O1vi | 0.9300 | 2.5700 | 3.151 (6) | 121.00 |
| C34—H34A···O5iii | 0.9300 | 2.5900 | 3.307 (7) | 134.00 |
| Symmetry codes: (i) x, y − 1, z; (ii) −x + 1, y−1/2, −z−1/2; (iii) −x + 1, y + 1/2, −z − 1/2; (iv) −x + 2, −y + 1, −z; (v) x − 1, y, z; and (vi) x, −y + 1/2, z − 1/2. | ||||
| Bond Distance | Exp. | DFT-B3LYP | Bond Distance | Exp. | DFT-B3LYP |
|---|---|---|---|---|---|
| O4—C15 | 1.221 (6) | 1.2185 | N7—C38 | 1.460 (6) | 1.4461 |
| O5—C14 | 1.242 (5) | 1.2297 | N8—C25 | 1.342 (6) | 1.3431 |
| O6—C13 | 1.256 (5) | 1.2402 | N3—C15 | 1.367 (6) | 1.3686 |
| O1—C1 | 1.212 (5) | 1.2083 | N3—C14 | 1.400 (5) | 1.4181 |
| O2—C2 | 1.217 (6) | 1.2078 | N4—C14 | 1.334 (5) | 1.3506 |
| O3—C3 | 1.218 (5) | 1.2194 | N4—C13 | 1.328 (6) | 1.3493 |
| N5—N6 | 1.314 (5) | 1.3029 | N1—C1 | 1.374 (6) | 1.3909 |
| N5—C25 | 1.346 (6) | 1.3457 | N1—C2 | 1.371 (6) | 1.3859 |
| N6—C26 | 1.445 (6) | 1.4402 | N2—C3 | 1.366 (6) | 1.3761 |
| N6—N7 | 1.328 (6) | 1.3436 | N2—C2 | 1.383 (7) | 1.3899 |
| N7—N8 | 1.317 (6) | 1.3068 | |||
| Bond Angle | Exp. | DFT-B3LYP | Bond Angle | Exp. | DFT-B3LYP |
| N6—N5—C25 | 103.5 (4) | 104.5042 | O6—C13—N4 | 120.2 (4) | 121.1666 |
| N5—N6—C26 | 123.8 (4) | 124.0503 | O6—C13—C16 | 116.7 (4) | 117.3011 |
| N7—N6—C26 | 126.0 (4) | 126.336 | N4—C13—C16 | 123.1 (4) | 121.4691 |
| N5—N6—N7 | 110.2 (4) | 109.5968 | N3—C14—N4 | 121.3 (4) | 118.9716 |
| N6—N7—C38 | 124.7 (4) | 124.0266 | O5—C14—N4 | 122.4 (4) | 124.4207 |
| N8—N7—C38 | 125.2 (4) | 126.336 | O5—C14—N3 | 116.3 (4) | 116.5377 |
| N6—N7—N8 | 110.2 (4) | 109.8148 | O4—C15—C16 | 122.9 (4) | 123.6163 |
| N7—N8—C25 | 103.5 (4) | 104.491 | O4—C15—N3 | 120.6 (4) | 121.6167 |
| C14—N3—C15 | 125.2 (4) | 126.8903 | N3—C15—C16 | 116.6 (4) | 114.7192 |
| C13—N4—C14 | 120.4 (4) | 121.1432 | O1—C1—N1 | 120.4 (4) | 120.4475 |
| C1—N1—C2 | 125.6 (4) | 127.2581 | O1—C1—C4 | 121.2 (4) | 122.1526 |
| C2—N2—C3 | 126.7 (4) | 127.2992 | N1—C1—C4 | 118.4 (3) | 117.3323 |
| N5—C25—N8 | 112.6 (4) | 111.5839 | N1—C2—N2 | 116.6 (4) | 114.1451 |
| N8—C25—C32 | 124.5 (4) | 124.1659 | O2—C2—N2 | 122.1 (4) | 123.1933 |
| N5—C25—C32 | 122.9 (4) | 124.1751 | O2—C2—N1 | 121.3 (4) | 122.6287 |
| N6—C26—C31 | 118.0 (4) | 117.1484 | N2—C3—C4 | 117.8 (4) | 116.1018 |
| N6—C26—C27 | 118.8 (4) | 119.8471 | O3—C3—N2 | 121.0 (4) | 120.4432 |
| N7—C38—C39 | 118.2 (4) | 117.7853 | N7—C38—C43 | 117.9 (4) | 118.9108 |
| Complexes | ∆ECorrected (kcal/mol) | ∆EBSSE |
|---|---|---|
| TPT–PBT | −1.02 | 0.0044 |
| TPT–PBT–(PBT−1)–H2O | −89.88 | 0.0234 |
| TPT–PBT–PBT−1 | −75.85 | 0.0104 |
| BCP | Bond | Bond Length | ρ(r) (a.u.) | K(r) (a.u.) | V(r) (a.u.) | H(r) (a.u.) | ∇2ρ(r) (a.u.) | G(r) (a.u.) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 121 | 31(H) -- 53(O) | 2.31059 | 0.0124 | −0.0016 | −0.008 | 0.0016 | 0.0446 | 0.0096 | 0.8373 | 0.129 |
| 138 | 53(O) -- 76(H) | 2.30255 | 0.0116 | −0.0016 | −0.0076 | 0.0016 | 0.0434 | 0.0092 | 0.8239 | 0.138 |
| 156 | 53(O) -- 38(N) | 3.13278 | 0.0071 | −0.0007 | −0.0043 | 0.0007 | 0.023 | 0.005 | 0.8536 | 0.099 |
| 163 | 77(H) -- 38(N) | 2.72730 | 0.0062 | −0.0008 | −0.0032 | 0.0008 | 0.0193 | 0.004 | 0.801 | 0.129 |
| 174 | 84(O) -- 98(H) | 1.91750 | 0.0285 | −0.0011 | −0.0235 | 0.0011 | 0.1025 | 0.0246 | 0.957 | 0.039 |
| 177 | 50(N) -- 2(N) | 3.24489 | 0.0073 | −0.0007 | −0.0044 | 0.0007 | 0.023 | 0.0051 | 0.8702 | 0.096 |
| 187 | 50(N) -- 18(H) | 2.25773 | 0.0172 | −0.0017 | −0.0102 | 0.0017 | 0.0541 | 0.0119 | 0.8584 | 0.099 |
| 191 | 2(N) -- 96(O) | 3.23042 | 0.0056 | −0.0007 | −0.0041 | 0.0007 | 0.0214 | 0.0047 | 0.8617 | 0.125 |
| 197 | 96(O) -- 86(H) | 2.01802 | 0.0239 | −0.00138 | −0.019 | 0.00138 | 0.0869 | 0.0203 | 0.93596 | 0.058 |
| 204 | 57(O) -- 9(H) | 2.00690 | 0.024 | −0.002 | −0.0179 | 0.002 | 0.0876 | 0.0199 | 0.8983 | 0.083 |
| 207 | 96(O) -- 7(H) | 3.03756 | 0.0029 | −0.0006 | −0.0016 | 0.0006 | 0.0112 | 0.0022 | 0.7356 | 0.207 |
| 209 | 96(O) -- 20(H) | 2.58144 | 0.0079 | −0.001 | −0.0049 | 0.001 | 0.0274 | 0.0059 | 0.8368 | 0.127 |
| 225 | 81(O) -- 23(H) | 2.94830 | 0.0039 | −0.0007 | −0.0022 | 0.0007 | 0.0146 | 0.003 | 0.7622 | 0.179 |
| Compounds | E(N) | HOMO (N) | HOMO (N + 1) | HOMO (N − 1) | VerticalEA | VerticalIP | χ | μ | η | S (eV−1) | ω |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Triphenyltetrazolium | −25910.0 | −3.27 | 1.28 | −9.93 | −0.05 | 4.82 | 2.39 | −2.39 | 4.87 | 0.21 | 0.58 |
| Phenobarbitone | −21767.7 | −7.23 | 1.75 | −11.51 | −0.06 | 9.08 | 4.51 | −4.51 | 9.15 | 0.11 | 1.11 |
| Phenobarbitoneanion | −21748.4 | −7.35 | −1.86 | −12.02 | 3.88 | 9.86 | 6.87 | −6.87 | 5.99 | 0.17 | 3.94 |
| Water | −2080.2 | −8.10 | 5.58 | −22.71 | −3.16 | 12.46 | 4.65 | −4.65 | 15.63 | 0.06 | 0.69 |
| Complex | −71508.1 | −5.85 | −0.02 | −9.034 | 1.34 | 7.37 | 4.36 | −4.36 | 6.02 | 0.17 | 1.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakheit, A.H.; Ghabbour, H.A.; Hussain, H.; Al-Salahi, R.; Ali, E.A.; Mostafa, G.A.E. Synthesis and Computational and X-ray Structure of 2, 3, 5-Triphenyl Tetrazolium, 5-Ethyl-5-phenylbarbituric Acid Salt. Crystals 2022, 12, 1706. https://doi.org/10.3390/cryst12121706
Bakheit AH, Ghabbour HA, Hussain H, Al-Salahi R, Ali EA, Mostafa GAE. Synthesis and Computational and X-ray Structure of 2, 3, 5-Triphenyl Tetrazolium, 5-Ethyl-5-phenylbarbituric Acid Salt. Crystals. 2022; 12(12):1706. https://doi.org/10.3390/cryst12121706
Chicago/Turabian StyleBakheit, Ahmed H., Hazem A. Ghabbour, Hadayt Hussain, Rashad Al-Salahi, Essam A. Ali, and Gamal A. E. Mostafa. 2022. "Synthesis and Computational and X-ray Structure of 2, 3, 5-Triphenyl Tetrazolium, 5-Ethyl-5-phenylbarbituric Acid Salt" Crystals 12, no. 12: 1706. https://doi.org/10.3390/cryst12121706
APA StyleBakheit, A. H., Ghabbour, H. A., Hussain, H., Al-Salahi, R., Ali, E. A., & Mostafa, G. A. E. (2022). Synthesis and Computational and X-ray Structure of 2, 3, 5-Triphenyl Tetrazolium, 5-Ethyl-5-phenylbarbituric Acid Salt. Crystals, 12(12), 1706. https://doi.org/10.3390/cryst12121706









