Abstract
Herein, we described the synthesis and X-ray crystal structure of the new [Pd(3)2Cl2] complex with 1,2,3-triazole-based ligand (3). In the unit cell, there are two [Pd(3)2Cl2] molecules, and the asymmetric unit comprised half of this formula due to the presence of an inversion symmetry element at the Pd(II) center. The monoclinic unit cell volume is 1327.85(6) Å3, with crystal parameters of a = 10.7712(2) Å, b = 6.8500(2) Å, and c = 18.2136(6) Å, while β = 98.851(2)°. The structure comprised two trans triazole ligand units coordinated to the Pd(II) ion via one of the N-atoms of the triazole moiety. In addition, the Pd(II) is further coordinated with two trans chloride groups, where each of the trans bonds is equidistant. The crystal structure of the [Pd(3)2Cl2] complex was compared with that for free triazole ligand 3. It was found that the coordinated ligand showed less twist around the C–N bond compared to free triazole ligand 3. The molecular packing of the latter is found controlled by short O…H, N…H, C…N, and C…C interactions in addition to the short Cl…F interhalogen and π–π interactions. H…H (23.5%), Cl…H (14.4%), N…H (14.3%), and O…H (11.2%) are the most dominant contacts. In the [Pd(3)2Cl2] complex, no significant interhalogen or π–π interactions were detected. In this case, Cl…H (31.1%), H…H (16.7%), O…H (11.6%), and F…H (9.7%) are the most dominant contacts.
1. Introduction
The development of a nicely described functional ligand system is a hot topic in the field of inorganic and organometallic chemistry. Indeed, 1,2,3-triazoles are among the important heterocyclic functional ligands that are demonstrated to be interesting building blocks in several organic molecules and inorganic compounds. Moreover, these heterocycles are considered as subunit linkers for many target compounds having important pharmacological effects, including xanthine oxidase (XO) and IDO inhibitors, anti-inflammatory, anti-HIV, anticancer, antibacterial, antifungal, antitubercular, and antiallergic properties [1,2,3,4,5]. Due to its high aromatic stability, this ring has good stability towards the hydrolysis and oxidation/reduction reactions in basic or acidic media [4]. Exploring the chemistry of this scaffold has attracted much attention from researchers. Notably, the 1,2,3-triazole unit was a key motif in several compounds that have been reported for many applications in drug discovery, material science, and agrochemical and polymer chemistry [6,7].
The chemistry of palladium metal complexes has emerged for many years as a hot topic of research for many organic/inorganic and medicinal chemists due to the wide importance and applications in many fields, such as catalysis, material sciences, agrochemical, and drug discovery [8,9,10,11,12,13,14,15,16,17,18]. Palladium catalysis has engaged in many asymmetric/symmetric synthetic transformations, such as carbon–carbon and carbon–nitrogen bond formation, which is the most common technique for bond formation in process chemistry and medicinal chemistry. Many reactions are well known to feature palladium metal as a catalyst, such as Suzuki, Heck, Buchwald–Hartwig, and Stille cross-coupling reactions; Tsuji–Trost allylation and Wacker transformation; and many processes, such as carbonylation, hydrogenation, pericyclic reactions, and cycloisomerization [19,20,21,22,23,24,25].
In drug discovery research, recently, several palladium complexes exhibited high efficacy against tumor cell lines and showed high correlation between the reactivity and lipophilicity of the Pd-complexes. In such cases, the Pd-complexes are considered to be lead organometallic compounds for cancer research due to significant anti-cancer activity with fewer side effects compared to platinum complexes, such as cisplatin [26,27,28,29,30]. To design organometallic-complex-derived Pd(II) and 1,2,3-triazole scaffold as a new material science for different application disciplines is still a challenge. Therefore, the aim of this work was to introduce a new Pd(II) complex based on the 1,2,3-tirazole ligand.
In light of this information, the synthesis, characterization, single-crystal diffraction analysis, and Hirshfeld analyses for both the free triazole ligand and its Pd(II) complex were investigated (Figure 1).
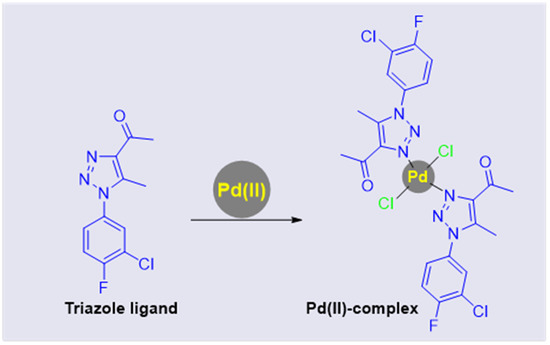
Figure 1.
Triazole framework and its Pd(II) complex.
2. Materials and Methods
All experimental details are described in the Supplementary Materials. The synthesis of the triazole ligand and the Pd(II)-complex followed the reported literature method [31,32,33,34,35,36].
2.1. Synthesis of Triazole Ligand 3
To a stirring solution of 3-chloro-4-fluoroaniline 1 (4.466 g, 0.0308 mol) in dil. HCl (40 mL, 3 M), a solution of sodium nitrite (2.34 g, 0.0339 mol, 25 mL, 1 M) treated at temperature < 10 °C was added. The progress of the reaction is confirmed by liberation of brown gases (1 h). When the brown gases crases, a solution of sodium azide (4.006 g, 0.0616 mol) is added to the stirred cold reaction mass. The formation of product is confirmed by TLC (1 h). The reaction mixture was extracted using diethyl ether. A brown viscous liquid confirmed the product 1.
A solution of the appropriate azide 2 (10.0 mmol, 1.71 gm) and acetylacetone (2.0 g, 20.0 mmol) in CHCl3 (20 mL) was treated by adding Et3N (2.02 g, 20.0 mmol) and DBU (1,8-Diazabicyclo[5.4.0]undec-7-ene) (0.76 g, 5.00 mmol). The reaction mixture was stirred at room temperature for 1–2 h. The solvent was removed and water added, then extracted with Et2O, dried with Na2SO4, evaporated, and the solid product was washed with n-hexane to provide pure compound of triazole ligand 3 (Scheme 1). A suitable single crystal for X-ray diffraction analysis was grown in Et2O/hexane.
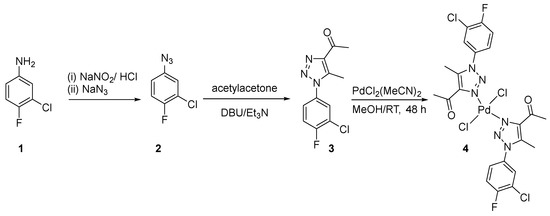
Scheme 1.
Synthetic route for Pd(II)-complex 4.
2.2. Synthesis of [Pd(3)2Cl2] Complexcomplex, 4
To a CH3OH solution of (126.5 mg, 0.5 mmol), 3 64.74 mg (0.25 mmol) of [PdCl2(MeCN)2] was added and the resulting solution was stirred at room temperature for 48 h. Subsequently, the reaction mixture was kept for slow evaporation to provide the complex as yellow solid in a crystalline form, which is suitable for single-crystal X-ray diffraction analysis. Yield = 0.16 g (83%). 1H NMR (400 MHz, CDCl3) δ 7.55 (d, J = 6.0 Hz, 1H, Ph), 7.34 (d, J = 6.6 Hz, 2H, Ph), 2.71 (s, 3H, CH3), 2.57 (s, 3H, CH3); 13C NMR (101 MHz, CDCl3) δ 194.34, 194.28, 160.22, 157.70, 143.78, 137.63, 131.87, 128.84, 127.09, 124.58, 122.84, 117.10, 28.59, 10.73.
2.3. X-ray Structure Determinations
The technical procedures and the software [37,38,39,40,41,42] employed for solving the X-ray structures of triazole ligand 3 and Pd(II)-complex 4 are provided in the Supplementary Materials.
2.4. Hirshfeld Surface Analysis
Analysis of molecular packing was performed using Crystal Explorer 17.5 program [43].
3. Results and Discussion
The triazole ligand required for this study was synthesized according to the reported method [31,32,33,34,35,36], and the spectral data were found matched with those reported in the literature. The triazole was mixed with the [PdCl2(MeCN)2] in MeOH at room temperature for 2 days to provide the desired Pd(II)-complex 4, as depicted in Scheme 1. Slow evaporation of the metal complex solution afforded a suitable crystalline material, which assigned unambiguously by single-crystal X-ray-diffraction analysis to confirm the chemical structure of the target Pd(II)-complex 4.
3.1. Crystal Structure Description of Triazole Ligand 3
The structure of free ligand 3 is shown in Figure 2. Free ligand 3 crystallized in an orthorhombic crystal system (Table 1). There is one molecule of 3 as an asymmetric unit, and z = 4. The unit cell volume is 1149.53(3) Å3, with crystal parameters of a = 12.99270(10) Å, b = 13.1421(2) Å, and c = 6.73220(10) Å. The structure comprised two planar rings, which are the phenyl and triazole moieties connected by a C6–N1 bond. The two rings are not coplanar with each other, where the twist angle between the two moieties is 64.15°.
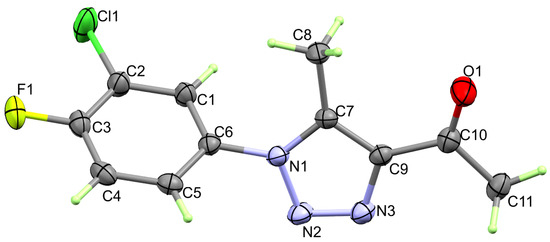
Figure 2.
X-ray structure of triazole ligand 3. List of some bond important distances and angles is given in Table 2.

Table 1.
Crystal data for triazole ligand 3 and Pd(II)-complex 4.
The molecules of triazole ligand 3 are connected with each other by weak C1-H1...N3 and C4-H4...O1 interactions (Figure 3A). The hydrogen bond parameters are listed in Table 3, while a view of molecular packing for triazole ligand 3 is shown in Figure 3B. In addition, the packing is controlled by the interhalogen and π–π stacking interactions shown in Figure 2C. Cl1...F1, C3...C9, and C7...N2 are the shortest interactions. The respective atom...atoms distances are 3.128, 3.379, and 3.197 Å.
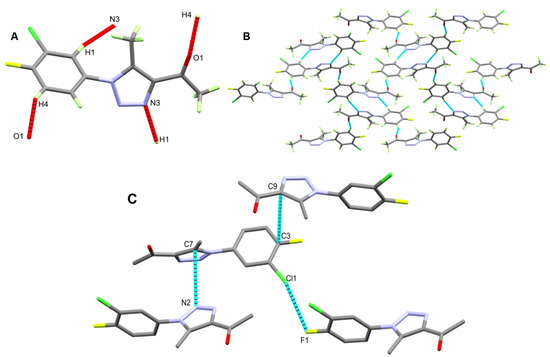
Figure 3.
Possible contacts (A) and molecular packing view via C–H…N and C–H…O (B) and interhalogen/π–π interactions (C) of triazole ligand 3.

Table 3.
Hydrogen bonds for triazole ligand 3 and its Pd(II) complex 4 [Å and °].
3.2. Crystal Structure Description of [Pd(3)2Cl2] Complex 4
The X-ray structure of the corresponding Pd(II) complex 4 of triazole ligand 3 is shown in Figure 4. It crystallized in a lower symmetry monoclinic crystal system, and P21/c is the space group. There are two [Pd(3)2Cl2] molecules per unit cell, and the asymmetric unit comprised half of this formula. The unit cell volume is 1327.85(6) Å3, with crystal parameters of a = 10.7712(2) Å, b = 6.8500(2) Å, and c = 18.2136(6) Å, while β = 98.851(2)°. A list of some bond distances and angles is provided in Table 4. The structure comprised two trans triazole ligands coordinated to the Pd(II) ion via one of the nitrogen atoms of the triazole moiety. In addition, the Pd(II) is further coordinated with two trans chloride groups. Because of the presence of an inversion center located at the Pd(II), each of the trans bonds is equidistant (Table 4). The structure of the coordinated ligand showed less twist around the C–N bond compared to free triazole ligand 3. In the former, the twist angle between the two moieties is 44.96°, indicating that the two rings are also not coplanar but to a lesser extent compared to free triazole ligand 3.
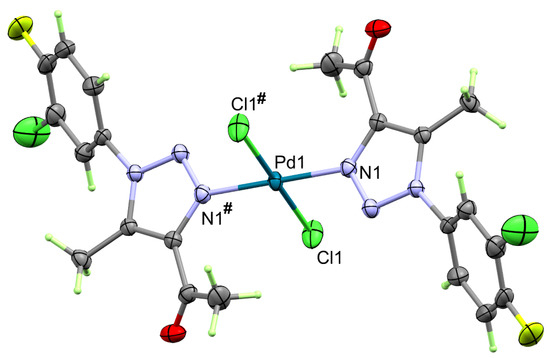
Figure 4.
Structure of [Pd(3)2Cl2] complex 4. Symmetry code: #1 − x + 1, −y, −z + 1.

Table 4.
Selected bond lengths [Å] and angles [°] for [Pd(3)2Cl2] complex 4.
The supramolecular structure of [Pd(3)2Cl2] is controlled by weak C5-H5A…Cl1, C7-H7...Cl1, and C10-H10...O1 interactions. The hydrogen–acceptor distances are 2.79, 2.77, and 2.37 Å, respectively, while the donor–acceptor distances are 3.7337(19), 3.6827(19), and 3.294(2) Å, respectively (Table 3). The packing view of the crystal structure of the [Pd(3)2Cl2] is shown in Figure 5.
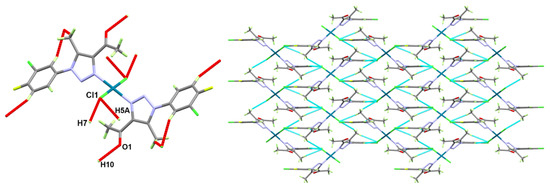
Figure 5.
Packing view of [Pd(3)2Cl2] via C–H...O and C–H...Cl interactions.
3.3. Hirshfeld Surface Analysis
Analysis of molecular packing is further investigated using Hirshfeld topology calculations in order to detect all possible intermolecular contacts that control the molecular packing in free triazole ligand 3 and its Pd(II) complex 4. For the free ligand, the different Hirshfeld surfaces are presented in Figure 6.
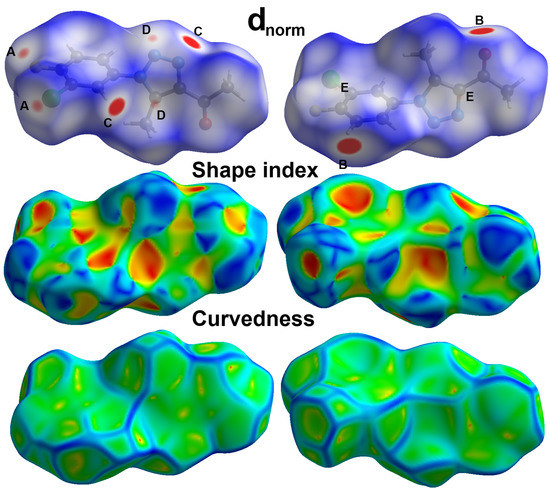
Figure 6.
Hirshfeld surfaces for free triazole ligand 3; A (Cl…F), B (O…H), C (N…H), D (C…N), and E (C…C).
As can be clearly seen from the dnorm map, there are many red spots. These red regions are indicative of the presence of short contacts having interaction distances shorter than the vdWs radii sum of interacting atoms, which are Cl…F, O…H, N…H, C…N, and C…C contacts. Cl1…F1 (3.128 Å), O1…H4 (2.167 Å), N3…H1 (2.336 Å), C7…N2 (3.197 Å), and C3…C9 (3.379 Å) are the shortest contacts, respectively. In addition to the conventional O…H and N…H hydrogen bonding interactions, the short Cl…F contacts confirmed the presence of interhalogen interactions among the neighboring molecular units. Moreover, the short C…C and C…N contacts are good indicators of the π–π interactions between the stacked triazole moieties. The sum of the C…C and C…N contacts is 6.7%, which are related to the π–π stacking interactions.
The percentages of all possible contacts occurring in the crystal structure of the free ligand are presented in Figure 6. The dominant contacts are H…H (23.5%), Cl…H (14.4%), N…H (14.3%), and O…H (11.2%) interactions. The interhalogen interactions contributed 4.5% from the whole contacts, where there are two types of this interaction. The significantly short Cl…F contacts contribute 3.5% from the whole interactions and the relatively long Cl…Cl contacts (1.0%). The latter appeared in the dnorm as a blue color, indicating long-distance interaction (Figure 7).
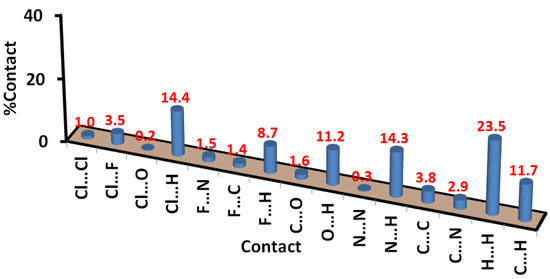
Figure 7.
Percentages of intermolecular interactions in 3.
In addition, the decomposed fingerprint plots for all short contacts are presented in Figure 8. The area of the fingerprint plot represents the frequency of each contact, which provides the percentages of each contact (Figure 7). In addition, the pattern of the fingerprint plot provided an idea about the strength of the corresponding interaction. Sharp spikes reveal short distance and strong interaction. The Cl…F, O…H, N…H, C…N, and C…C interactions clearly showed these features.
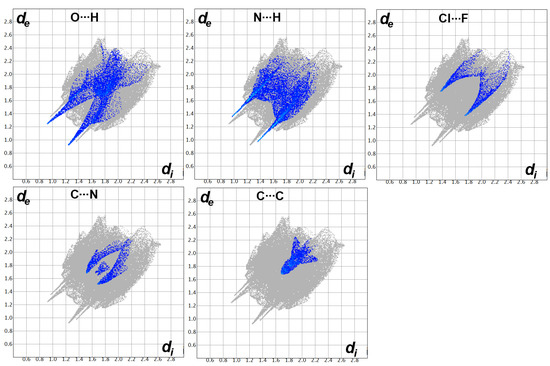
Figure 8.
Fingerprint plots for the important contacts in free triazole ligand 3.
For Pd(II) complex 4, the coordination between the Pd(II) and the organic ligand significantly affects the intermolecular interactions. The first observation is the disappearance of the interhalogen interactions where no significant Cl...F or Cl...Cl interactions were detected. Only a small amount of long distance Cl...Cl (2.7%) interactions were detected with contact distances of 3.947 Å (Cl1...Cl2). This interaction is significantly longer than twice the vdWs radii of chlorine atom. Additionally, there are no short C…C or C…N contacts detected in the Pd(II) complex. Their percentages are 6.0 and 0.0, respectively, and no red regions are related to these interactions in the dnorm map. The shortest C...C contact distances are 3.481 Å (C11…C9) and 3.433 Å (C10…C9). A summary of all the observed contacts and their percentages is shown in Figure 9.
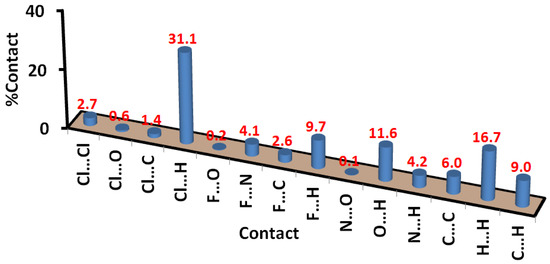
Figure 9.
Intermolecular contacts and their percentages in Pd(II) complex 4.
The most dominant contacts are Cl...H (31.1%), H...H (16.7%), O...H (11.6%), and F...H (9.7%) interactions. Other less contributing contacts were detected, and their percentages are depicted in Figure 8. Among these interactions, Cl...H, O...H, and Cl...C interactions are the most significant; only these contacts appeared as red regions in the dnorm (Figure 10). For the Cl...H interactions, Cl1…H7 (2.645 Å), Cl1…H5A (2.688 Å), and Cl1…H5B (2.799 Å) are the shortest. Unlike the free ligand, all these interactions are related to the coordinated chloride ion, while no significant Cl...H interactions related to the chlorine atom attached to the phenyl ring were detected. Moreover, O1…H10 (2.246 Å) and Cl1…C2 (3.29 Å) have significantly short contact distances. All these interactions are clearly observed from the corresponding fingerprint plot as sharp spikes, which confirm their significance (Figure 11).
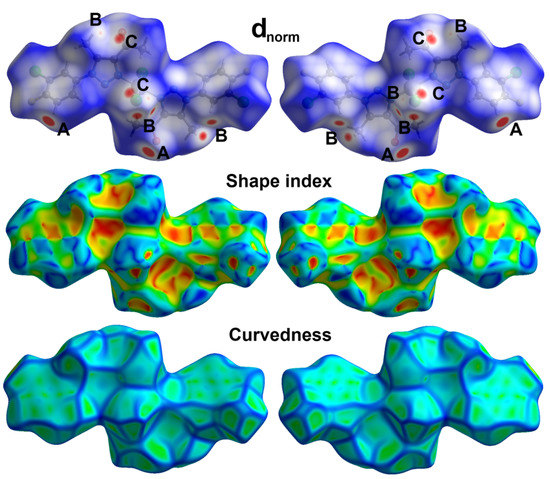
Figure 10.
Hirshfeld surfaces of Pd(II) complex 4; A (O…H), B (Cl…H), and C (Cl…C).
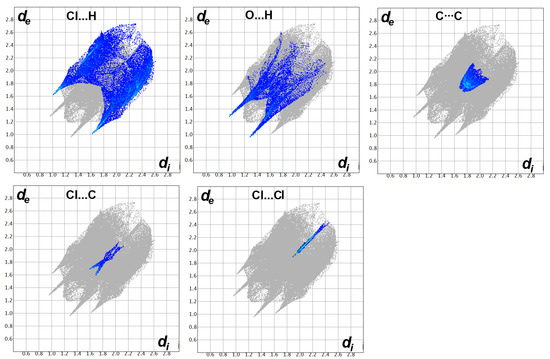
Figure 11.
Fingerprint plots of the short contacts in Pd(II) complex 4.
4. Conclusions
Two single crystals of the free triazole ligand named 1-(1-(3-chloro-4-fluorophenyl)-5-methyl-1H-1,2,3-triazol-4-yl)ethan-1-one (3) and its Pd(II) complex [Pd(3)2Cl2] were prepared and their single-crystal structures were presented. The [Pd(3)2Cl2] complex comprised a tetracoordinated Pd(II) ion with 3 and Cl¯, with both acting as monodentate ligands. There is a center of symmetry located at the Pd(II), so the trans bonds are equidistant. The molecular packing of free triazole ligand 3 is controlled by short Cl…F, O…H, N…H, C…N, and C…C interactions. The presence of short Cl…F contacts indicates the presence of interhalogen interactions, while the short C…C and C…N contacts are good indicators of π–π interactions. In this case, the dominant contacts are H…H (23.5%), Cl…H (14.4%), N…H (14.3%), and O…H (11.2%) interactions. In contrast, the packing of [Pd(3)2Cl2] showed no significant interhalogen or π–π interactions. In this case, the most dominant contacts are Cl...H (31.1%), H...H (16.7%), O...H (11.6%), and F...H (9.7%) interactions, while Cl...H, O...H, and Cl…C interactions are the most significant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst12101335/s1. General remarks about the instruments used in this study and also the X-ray structure determinations technical protocol.
Author Contributions
Conceptualization, A.B.; methodology, H.H.A.-R., A.M.A.-M., and M.A.; software, M.H. and S.M.S.; validation, S.M.S. and A.E.-F.; formal analysis, H.H.A.-R. and M.A.; investigation, H.H.A.-R., A.M.A.-M., and M.A.; resources, A.B.; data curation, M.H. and S.M.S.; writing—original draft preparation, H.H.A.-R., M.H., S.M.S., A.M.A.-M., M.A., A.E.-F., and A.B.; writing—review and editing, A.E.-F., S.M.S., and A.B.; supervision, A.B.; funding acquisition, A.B. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to extend their sincere appreciation to the Researchers Supporting Project (RSP-2021/64), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are not available on request from the corresponding author.
Acknowledgments
The authors would like to extend their sincere appreciation to the Researchers Supporting Project (RSP-2021/64), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kumar, S.; Sharma, B.; Mehra, V.; Kumar, V. Recent accomplishments on the synthetic/biological facets of pharmacologically active 1H-1,2,3-triazoles. Eur. J. Med. Chem. 2021, 212, 113069. [Google Scholar] [CrossRef] [PubMed]
- Aouad, M.R.; Almehmadi, M.A.; Rezki, N.; Al-blewi, F.F.; Messali, M.; Ali, I. Design, click synthesis, anticancer screening and docking studies of novel benzothiazole-1,2,3-triazoles appended with some bioactive benzofused heterocycles. J. Mol. Struct. 2019, 1188, 153–164. [Google Scholar] [CrossRef]
- Tan, A. Novel 1,2,3-triazole compounds: Synthesis, In vitro xanthine oxidase inhibitory activity, and molecular docking studies. J. Mol. Struct. 2020, 1211, 128060. [Google Scholar] [CrossRef]
- Haider, S.; Alam, M.S.; Hamid, H. 1,2,3-Triazoles: Scaffold with medicinal significance. Inflamm. Cell Signal 2014, 1, e95. [Google Scholar]
- Totobenazara, J.; Burke, A.J. New click-chemistry methods for 1,2,3-triazoles synthesis: Recent advances and applications. Tetrahedron Lett. 2015, 56, 2853–2859. [Google Scholar] [CrossRef]
- Sajja, Y.; Vanguru, S.; Vulupala, H.R.; Bantu, R.; Yogeswari, P.; Sriram, D.; Nagarapu, L. Design, synthesis and in vitro anti-tuberculosis activity of benzo [6,7] cyclohepta [1,2-b] pyridine 1,2,3-triazole derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 5119–5121. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Hu, X.; Duan, C.; Liu, P.; Liu, S.; Lan, L.; Chen, D.; Ying, L.; Su, S.; Gong, X. A series of new medium-bandgap conjugated polymers based on naphtho [1,2-c:5,6-c] bis (2-octyl-[1,2,3] triazole) for high-performance polymer solar cells. Adv. Mater. 2013, 25, 3683–3688. [Google Scholar] [CrossRef] [PubMed]
- Beccalli, E.M.; Broggini, G.; Martinelli, M.; Sottocornola, S. C−C, C−O, C−N bond formation on sp 2 carbon by Pd (II)-catalyzed reactions involving oxidant agents. Chem. Rev. 2007, 107, 5318–5365. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Bulger, P.G.; Sarlah, D. Palladium-Catalyzed Cross-Coupling Reactions in Total Synthesis. Angew. Chem. Int. Ed. 2005, 44, 4442–4489. [Google Scholar] [CrossRef]
- Ruiz-Castillo, P.; Buchwald, S.L. Applications of Palladium-Catalyzed C–N Cross-Coupling Reactions. Chem. Rev. 2016, 1163, 12564–12649. [Google Scholar] [CrossRef]
- Leone, A.K.; Mueller, E.A.; McNeil, A.J. The History of Palladaium-Catalyzed CrossCouplings Should Inspire the Future of Catalyst-Transfer Polymerization. J. Am. Chem. Soc. 2018, 140, 15126–15139. [Google Scholar] [CrossRef] [PubMed]
- Leone, A.K.; Goldberg, P.K.; McNeil, A.J. Ring-Walking in Catalyst-Transfer Polymerization. J. Am. Chem. Soc. 2018, 140, 7846–7850. [Google Scholar] [CrossRef] [PubMed]
- Devendar, P.; Qu, R.-Y.; Kang, W.-M.; He, B.; Yang, G.-F. Palladium-Catalyzed CrossCoupling Reactions: A Powerful Tool for the Synthesis of Agrochemicals. J. Agric. Food Chem. 2018, 66, 8914–8934. [Google Scholar] [CrossRef]
- Torborg, C.; Beller, M. Recent Applications of PalladiumCatalyzed Coupling Reactions in the Pharmaceutical, Agrochemical, and Fine Chemical Industries. Adv. Synth. Catal. 2009, 351, 3027–3043. [Google Scholar] [CrossRef]
- Brown, D.G.; Boström, J. Analysis of Past and Present Synthetic Methodologies on Medicinal Chemistry: Where Have All the New Reactions Gone? J. Med. Chem. 2016, 59, 4443–4458. [Google Scholar] [CrossRef]
- Gesmundo, N.J.; Sauvagnat, B.; Curran, P.J.; Richards, M.P.; Andrews, C.L.; Dandliker, P.J.; Cernak, T. Nanoscale synthesis and affinity ranking. Nature 2018, 557, 228–232. [Google Scholar] [CrossRef]
- Goodyear, A.; Linghu, X.; Bishop, B.; Chen, C.; Cleator, E.; McLaughlin, M.; Sheen, F.J.; Stewart, G.W.; Xu, Y.; Yin, J. Process Development and Large-Scale Synthesis of MK06186, a Non-Nucleoside Reverse Transcriptase Inhibitor for the Treatment of HIV. Org. Process Res. Dev. 2012, 16, 605–611. [Google Scholar] [CrossRef]
- Rayadurgam, J.; Sana, S.; Sasikumar, M.; Gu, A. Palladium catalyzed C–C and C–N bond forming reactions: An update on the synthesis of pharmaceuticals from 2015–2020. Org. Chem. Front. 2021, 8, 384–414. [Google Scholar] [CrossRef]
- Negishi, E. (Ed.) Handbook of Organopalladium Chemistry for Organic Synthesis; Wiley-Interscience: New York, NY, USA, 2002. [Google Scholar]
- Tsuji, J. Palladium Reagents and Catalysts: Innovations in Organic Synthesis; Wiley and Sons: New York, NY, USA, 1995. [Google Scholar]
- Tsuji, J. Palladium Reagents and Catalysts: New Perspectives for the 21st Century; Wiley and Sons: New York, NY, USA, 2003. [Google Scholar]
- Tsuji, J. Palladium in Organic Synthesis; Springer: Berlin, Germany, 2005. [Google Scholar]
- Heck, R.F. Palladium Reagents in Organic Synthesis; Academic Press: New York, NY, USA, 1985. [Google Scholar]
- Li, J.J.; Gribble, G.W. Palladium in Heterocyclic Chemistry; Pergamon: New York, NY, USA, 2000. [Google Scholar]
- Heumann, A.; Jens, K.-J.; Reglier, M. Progress in Inorganic Chemistry. Karlin, K.D., Ed.; Wiley and Sons: New York, NY, USA, 1994; Volume 42, pp. 483–576. [Google Scholar]
- Galanski, M. Recent developments in the field of anticancer platinum complexes. Recent Pat. Anti-Cancer Drug Discov. 2006, 1, 285–295. [Google Scholar] [CrossRef]
- Gao, E.; Liu, C.; Zhu, M.; Lin, H.; Wu, Q.; Liu, L. Current development of Pd(II) complexes as potential antitumor agents. Anti-Cancer Agents Med. Chem. 2009, 9, 356–368. [Google Scholar] [CrossRef]
- Abu-Surrah, A.S.; Safieh, K.A.A.; Ahmad, I.M.; Abdalla, M.Y.; Ayoub, M.T.; Qaroush, A.K.; Abu-Mahtheieh, A.M. New palladium(II) complexes bearing pyrazole-based Schiff base ligands: Synthesis, characterization and cytotoxicity. Eur. J. Med. Chem. 2010, 45, 471–475. [Google Scholar] [CrossRef]
- Gill, D.S. Structure activity relationship of antitumor palladium complexes. In Platinum Coordination Complexes in Cancer Chemotherapy; Hacker, M.P., Douple, E.B., Krakoff, I.H., Eds.; Developments in Oncology; Springer: New York, NY, USA, 1984; Volume 17, pp. 267–278. [Google Scholar]
- Al-Masoudi, N.A.; Abdullah, B.H.; Essa, A.H.; Loddo, R.; LaColla, P. Platinum and palladium-triazole complexes as highly potential antitumor agents. Arch. Pharm. 2010, 343, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Santosh, R.; Selvam, M.K.; Kanekar, S.U.; Nagaraja, G.K. Synthesis, characterization, antibacterial and antioxidant studies of some heterocyclic compounds from triazole-linked chalcone derivatives. ChemistrySelect 2018, 3, 6338–6343. [Google Scholar] [CrossRef]
- Shafran, Y.M.; Beryozkina, T.V.; Efimov, I.V.; Bakulev, V.A. Synthesis of β-azolyl-and β-azolylcarbonylenamines and their reactions with aromatic azides. Chem. Heterocycl. Compd. 2019, 55, 704–715. [Google Scholar] [CrossRef]
- Elgogary, S.R.; Khidre, R.E.; El-Telbani, E.M. Regioselective synthesis and evaluation of novel sulfonamide 1,2,3-triazole derivatives as antitumor agents. J. Iran. Chem. Soc. 2020, 17, 765–776. [Google Scholar] [CrossRef]
- Singh, H.; Sindhu, J.; Khurana, J.M. Synthesis of biologically as well as industrially important 1,4,5-trisubstituted-1,2,3-triazoles using a highly efficient, green and recyclable DBU–H2O catalytic system. RSC Adv. 2013, 3, 22360–22366. [Google Scholar] [CrossRef]
- Amoah, C.; Obuah, C.; Ainooson, M.K.; Muller, A. Synthesis, characterization and fluorescent properties of ferrocenyl pyrazole and triazole ligands and their palladium complexes. J. Organomet. Chem. 2021, 935, 121664. [Google Scholar] [CrossRef]
- Altowyan, M.S.; Soliman, S.M.; Haukka, M.; Al-Shaalan, N.H.; Alkharboush, A.A.; Barakat, A. Synthesis and Structure Elucidation of Novel Spirooxindole Linked to Ferrocene and Triazole Systems via [3+2] Cycloaddition Reaction. Molecules 2022, 27, 4095. [Google Scholar] [CrossRef]
- Rikagu Oxford Diffraction. CrysAlisPro. Rikagu Oxford Diffraction, Inc.: Oxfordshire, UK, 2022. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In Methods in Enzymology, Volume 276, Macromolecular Crystallography, Part A; Carter, C.W., Sweet, J., Eds.; Academic Press: New York, NY, USA, 1997; pp. 307–326. [Google Scholar]
- Sheldrick, G.M. SADABS—Bruker Nonius Scaling and Absorption Correction; Bruker AXS, Inc.: Madison, WI, USA, 2012. [Google Scholar]
- Sheldrick, G.M. SHELXT-Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Crystallogr. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer17. University of Western Australia: Crawley, WA, Australia, 2017; Available online: http://hirshfeldsurface.net (accessed on 20 May 2017).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).