Molecular Dynamics Investigation of Phenolic Oxidative Coupling Protein Hyp-1 Derived from Hypericum perforatum
Abstract
1. Introduction
2. Materials and Methods
2.1. Hyp-1 Structure Model
2.2. Molecular Dynamics Simulation
2.3. Comparison between Experimental and Simulated Structure Model
2.4. Application of MD Nanoscale Simulation to Calculate ADPs Distribution
2.5. Stereochemical Constraints of Main- and Side-Chain Conformations
2.6. Comparison between Experimental and Calculated Distributions of Side-Chain Dihedral Angles
3. Results and Discussion
3.1. Impact of MD Simulation Parameters and Possible Artifacts
3.2. Accuracy of Experimental and Simulated Hyp-1 Structure Model
3.3. Actual and Experimental ADPs Comparison
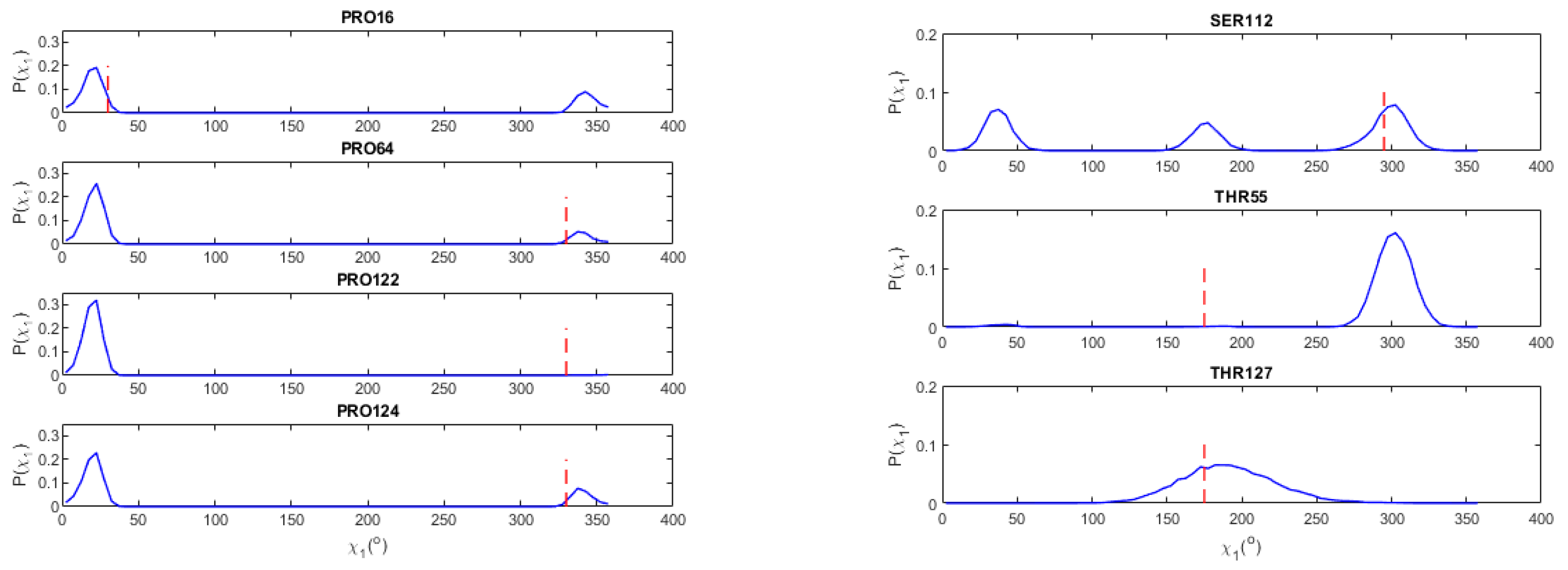
3.4. Side-Chain Angles Probability Distribution P(χ1) for Pro, Ser, Cys
3.5. Val and Thr
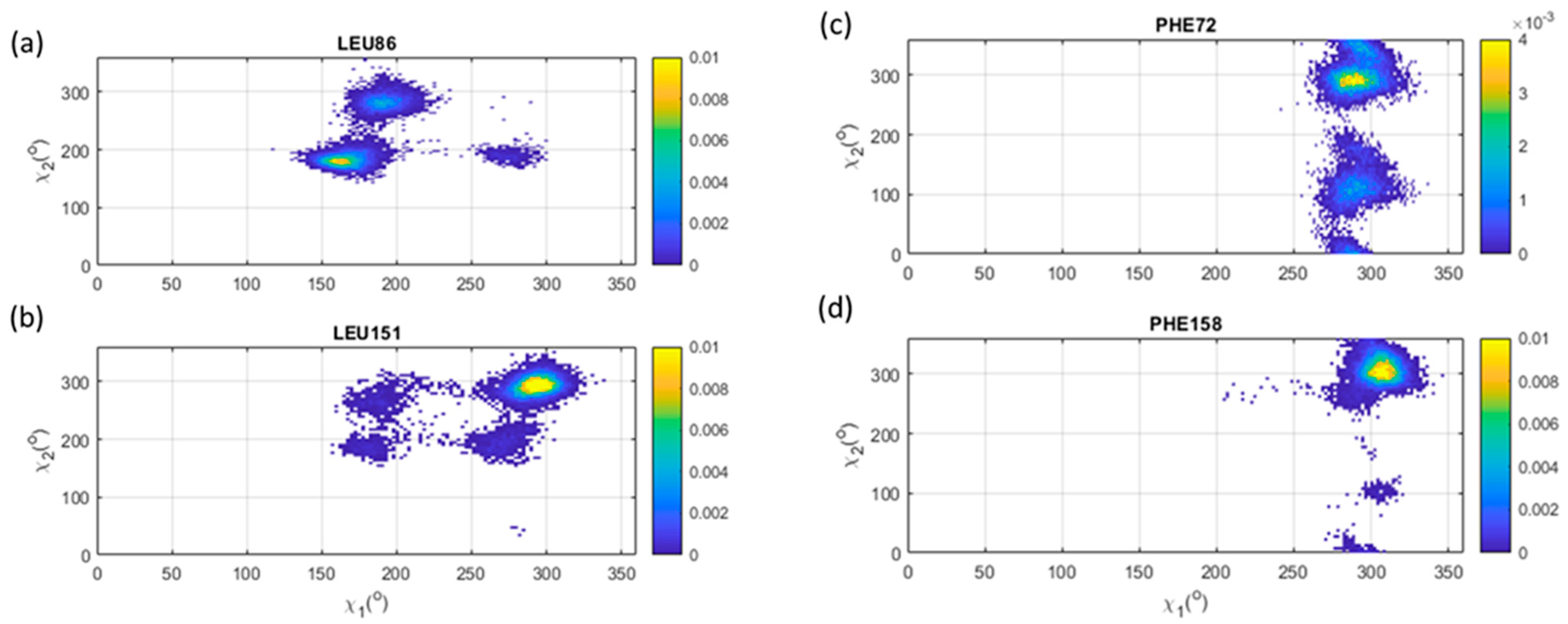
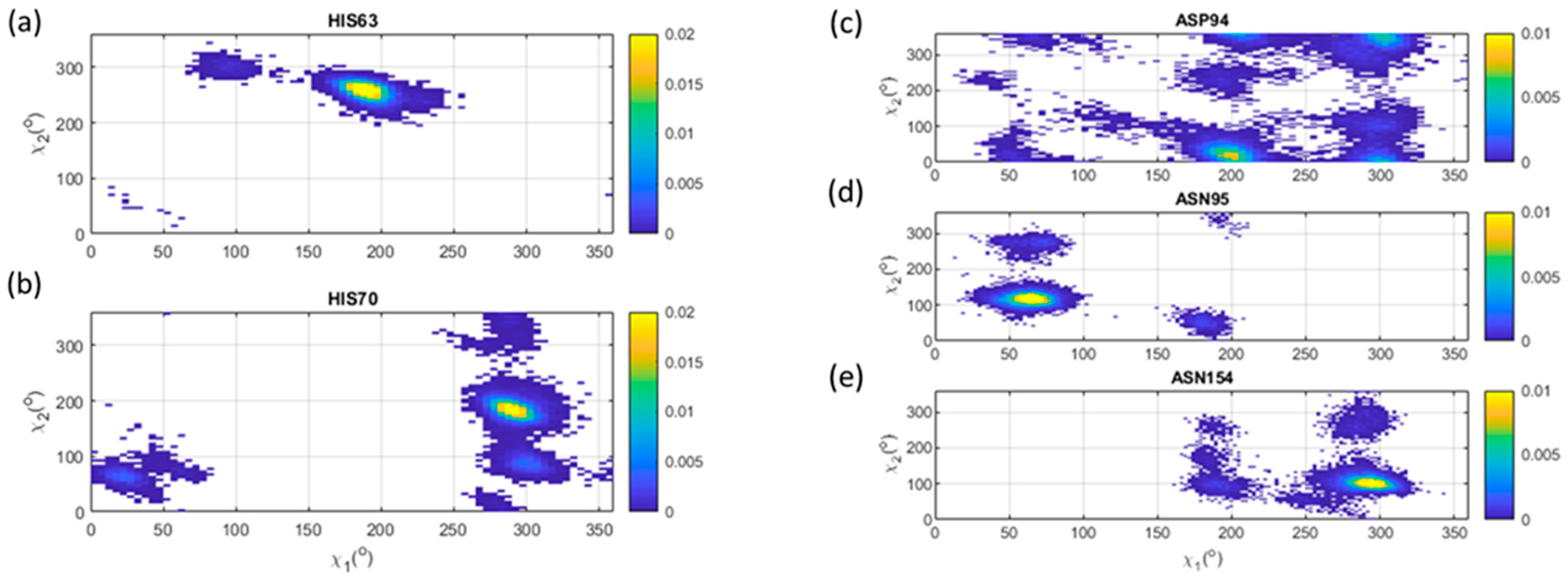
3.6. Leu, Phe, Tyr, His
3.7. Asp, Asn
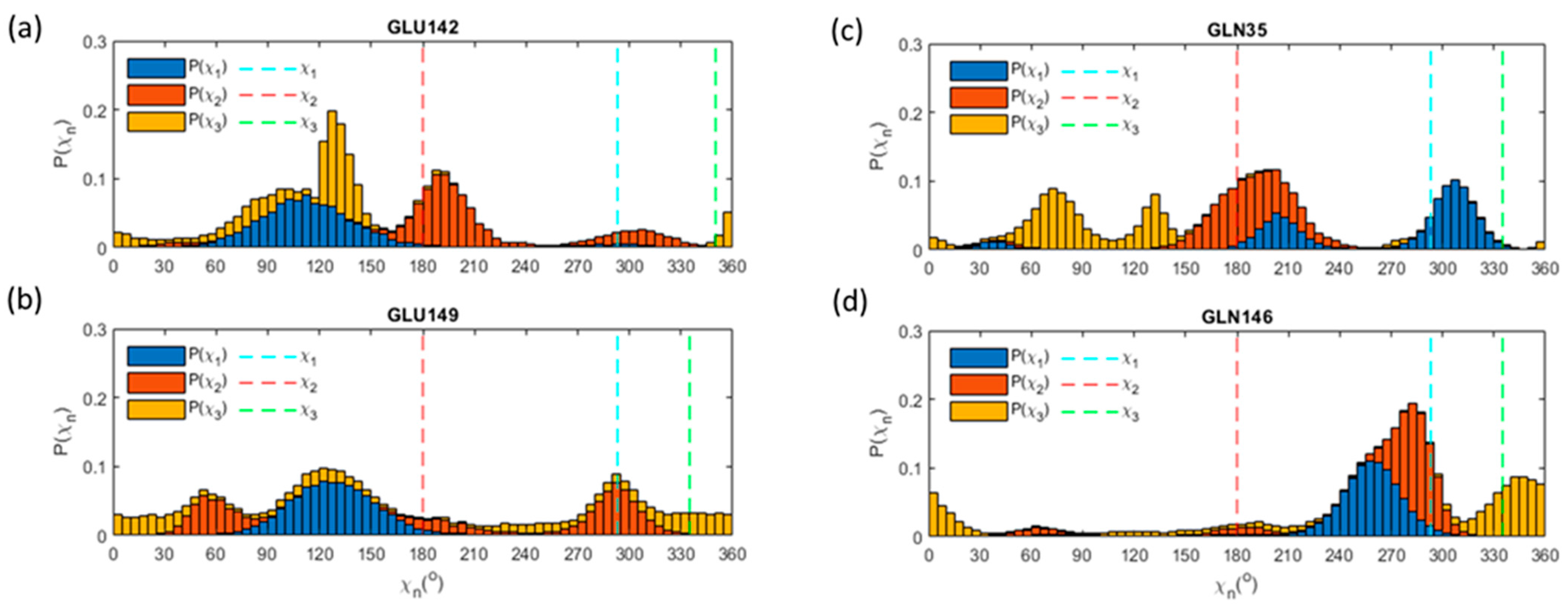
3.8. Glu, Gln
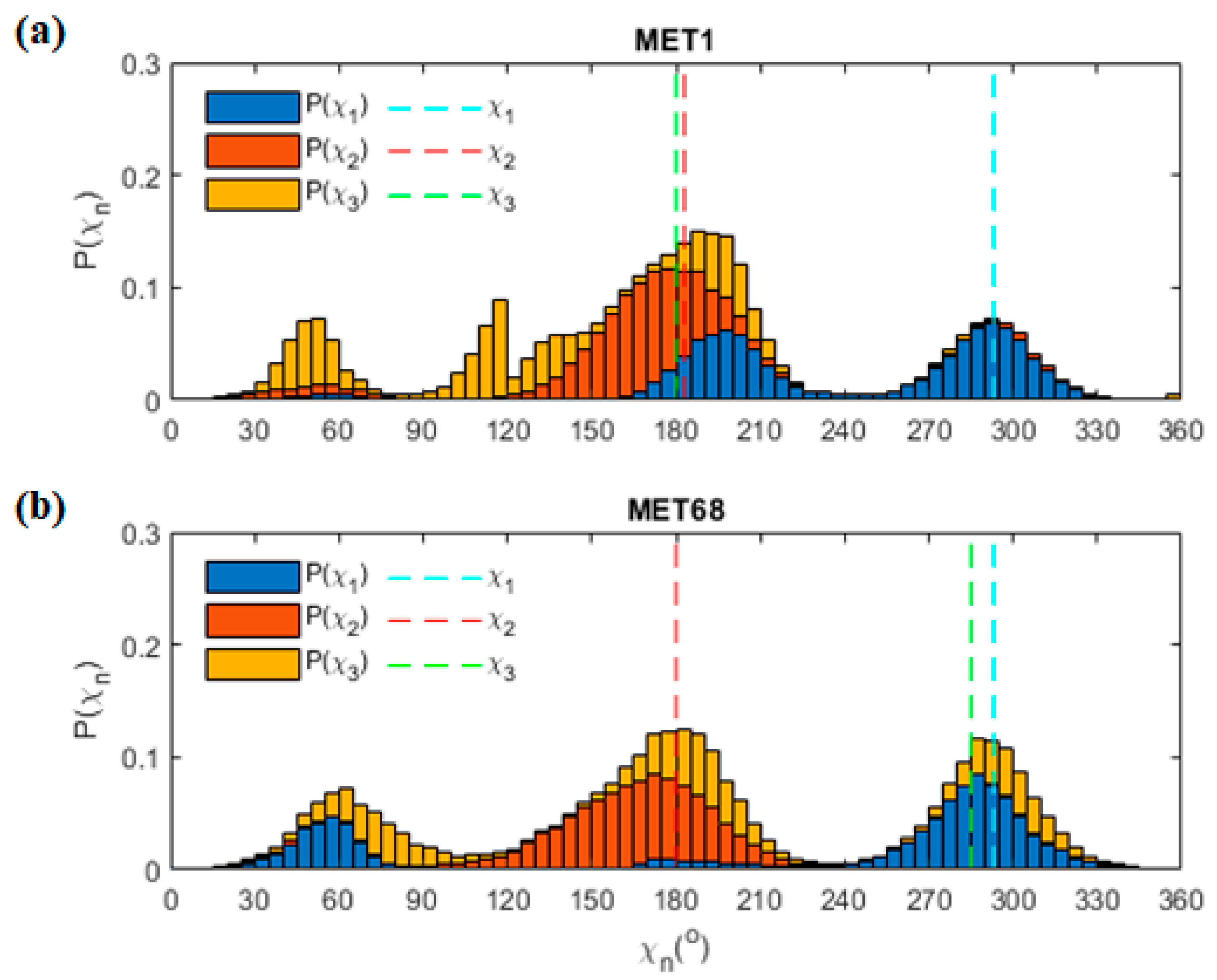
3.9. Met
3.10. Lys, Arg
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- McCammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nat. Cell Biol. 1977, 267, 585–590. [Google Scholar] [CrossRef]
- Gaalswyk, K.; Muniyat, M.I.; Maccallum, J.L. The emerging role of physical modeling in the future of structure determination. Curr. Opin. Struct. Biol. 2018, 49, 145–153. [Google Scholar] [CrossRef]
- Patodia, S.; Bagaria, A.; Chopra, D. Molecular Dynamics Simulation of Proteins: A Brief Overview. J. Chem. Phys. Biophys. 2014, 4. [Google Scholar] [CrossRef]
- Karplus, M.; Kuriyan, J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. USA 2005, 102, 6679–6685. [Google Scholar] [CrossRef]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef]
- Freddolino, P.L.; Arkhipov, A.S.; Larson, S.B.; McPherson, A.; Schulten, K. Molecular Dynamics Simulations of the Complete Satellite Tobacco Mosaic Virus. Structure 2006, 14, 437–449. [Google Scholar] [CrossRef]
- Perez, A.; Morrone, J.A.; Simmerling, C.; Dill, K.A. Advances in free-energy-based simulations of protein folding and ligand binding. Curr. Opin. Struct. Biol. 2016, 36, 25–31. [Google Scholar] [CrossRef]
- Allen, F.; Almasi, G.; Andreoni, W.; Beece, D.; Berne, B.J.; Bright, A.; Brunheroto, J.; Cascaval, C.; Castanos, J.; Coteus, P.; et al. Blue Gene: A vision for protein science using a petaflop supercomputer. IBM Syst. J. 2001, 40, 310–327. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Lee, M.R.; Tsai, J.; Baker, D.; Kollman, P.A. Molecular dynamics in the endgame of protein structure prediction. J. Mol. Biol. 2001, 313, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Dror, R.O.; Dirks, R.M.; Grossman, J.; Xu, H.; Shaw, D.E. Biomolecular Simulation: A Computational Microscope for Molecular Biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef]
- Chen, J.; Brooks, C.L. Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins Struct. Funct. Bioinform. 2007, 67, 922–930. [Google Scholar] [CrossRef]
- Kuzmanic, A.; Pannu, N.S.; Zagrovic, B. X-ray refinement significantly underestimates the level of microscopic heterogeneity in biomolecular crystals. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Piana, S. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011, 101, 1015. [Google Scholar] [CrossRef][Green Version]
- Izmailov, S.A.; Podkorytov, I.S.; Skrynnikov, N.R. Simple MD-based model for oxidative folding of peptides and proteins. Sci. Rep. 2017, 7, 9293. [Google Scholar] [CrossRef]
- Ensign, D.L.; Kasson, P.M.; Pande, V.S. Heterogeneity Even at the Speed Limit of Folding: Large-scale Molecular Dynamics Study of a Fast-folding Variant of the Villin Headpiece. J. Mol. Biol. 2007, 374, 806–816. [Google Scholar] [CrossRef]
- Stumpff-Kane, A.W.; Maksimiak, K.; Lee, M.S.; Feig, M. Sampling of near-native protein conformations during protein structure refinement using a coarse-grained model, normal modes, and molecular dynamics simulations. Proteins Struct. Funct. Bioinform. 2007, 70, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Wereszczynski, J.; McCammon, J.A. Statistical mechanics and molecular dynamics in evaluating thermodynamic properties of biomolecular recognition. Q. Rev. Biophys. 2011, 45, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Adiyaman, R.; McGuffin, L.J. Methods for the Refinement of Protein Structure 3D Models. Int. J. Mol. Sci. 2019, 20, 2301. [Google Scholar] [CrossRef] [PubMed]
- Wong-Ekkabut, J.; Karttunen, M. The good, the bad and the user in soft matter simulations. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2529–2538. [Google Scholar] [CrossRef]
- Bernardi, R.C.; Melo, M.C.R.; Schulten, K. Enhanced sampling techniques in molecular dynamics simulation of biological systems. Biochim. Biophys. Acta 2015, 1850, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Feig, M.; Mirjalili, V. Protein structure refinement via molecular-dynamics simulations: What works and what does not? Proteins Struct. Funct. Bioinform. 2015, 84, 282–292. [Google Scholar] [CrossRef]
- Pang, Y.-P. Use of multiple picosecond high-mass molecular dynamics simulations to predict crystallographic B-factors of folded globular proteins. Heliyon 2016, 2. [Google Scholar] [CrossRef]
- Masmaliyeva, R.C.; Murshudov, G.N. Analysis and validation of macromolecular B values. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 505–518. [Google Scholar] [CrossRef]
- Caldararu, O.; Kumar, R.; Oksanen, E.; Logan, D.T.; Ryde, U. Are crystallographic B-factors suitable for calculating protein conformational entropy? Phys. Chem. Chem. Phys. 2019, 21, 18149–18160. [Google Scholar] [CrossRef]
- Parthasarathy, S.; Murthy, M. Analysis of temperature factor distribution in high-resolution protein structures. Protein Sci. 2008, 6, 2561–2567. [Google Scholar] [CrossRef]
- Carugo, O.; Argos, P. Correlation between side chain mobility and conformation in protein structures. Protein Eng. Des. Sel. 1997, 10, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Meinhold, L.; Smith, J.C. Fluctuations and Correlations in Crystalline Protein Dynamics: A Simulation Analysis of Staphylococcal Nuclease. Biophys. J. 2005, 88, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Kuriyan, J.; Weis, W.I. Rigid protein motion as a model for crystallographic temperature factors. Proc. Natl. Acad. Sci. USA 1991, 88, 2773–2777. [Google Scholar] [CrossRef]
- Soheilifard, R.; Makarov, D.E.; Rodin, G.J. Critical evaluation of simple network models of protein dynamics and their comparison with crystallographic B-factors. Phys. Biol. 2008, 5, 026008. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Isupovb, M.N.; Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. 2001, 57, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O. How large B-factors can be in protein crystal structures. BMC Bioinform. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nakayoshi, T.; Fukuyoshi, S.; Kurimoto, E.; Oda, A. Validation of Molecular Dynamics Simulations for Prediction of Three-Dimensional Structures of Small Proteins. Molecules 2017, 22, 1716. [Google Scholar] [CrossRef]
- Hintze, B.J.; Lewis, S.M.; Richardson, J.S.; Richardson, D.C. Molprobity’s Ultimate Rotamer-Library Distributions for Model Validation. Proteins 2016, 84, 1177–1189. [Google Scholar] [CrossRef]
- Towse, C.-L.; Rysavy, S.J.; Vulovic, I.M.; Daggett, V. New Dynamic Rotamer Libraries: Data-Driven Analysis of Side-Chain Conformational Propensities. Structure 2016, 24, 187–199. [Google Scholar] [CrossRef]
- Harder, T.; Boomsma, W.; Paluszewski, M.; Frellsen, J.; Johansson, K.E.; Hamelryck, T. Beyond rotamers: A generative, probabilistic model of side chains in proteins. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef]
- Haddad, J.; Vojtech, A.; Heger, Z. Rotamer Dynamics: Analysis of Rotamers in Molecular Dynamics Simulations of Proteins. Biophys. J. 2019, 116, 2062–2072. [Google Scholar] [CrossRef] [PubMed]
- Butterweck, V. Mechanism of action of St. John’s wort in depression: What is known? CNS Drugs 2003, 17, 539–562. [Google Scholar] [CrossRef] [PubMed]
- Michalska, K.; Fernandes, H.; Sikorski, M.M.; Jaskolski, M. Crystal structure of Hyp-1, a St. John’s wort protein implicated in the biosynthesis of hypericin. J. Struct. Biol. 2010, 169, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Sliwiak, J.; Dauter, Z.; Jaskolski, M. Hyp-1 protein from St John’s wort as a PR-10 protein. Biotechnology 2013, 1, 47–50. [Google Scholar] [CrossRef]
- Sliwiak, J.; Dauter, Z.; Jaskolski, M. Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin. Front. Plant Sci. 2016, 7, 668. [Google Scholar] [CrossRef]
- Sliwiak, J.; Dauter, Z.; Kowiel, M.; McCoy, A.; Read, R.J.; Jaskolski, M. ANS complex of St. John’s wort PR-10 protein with 28 copies in the asymmetric unit: A fiendish combination of pseudosymmetry with tetartohedral twinning. Acta Cryst. 2015, D71, 829–843. [Google Scholar] [CrossRef]
- Berendsen, H.; Van Der Spoel, D.; Van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Lindahl, E.; Hess, B.; Van Der Spoel, D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001, 7, 306–317. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; Van Der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Van Der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Pall, S.; Abraham, M.J.; Kutzner, C.; Hess, B.; Lindahl, E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. In Solving Software Challenges for Exascale; Markidis, S., Laure, E., Eds.; Springer: Cham, Switzerland, 2015; pp. 3–27. [Google Scholar]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B.; GROMACS Development Team. GROMACS User Manual Version 2018. Available online: www.gromacs.org (accessed on 2 November 2020).
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; McDonald, N.A. Development of an all-atom force field for heterocycles. Properties of liquid pyridine and diazenes. J. Mol. Struct. Theochem 1998, 424, 145–155. [Google Scholar] [CrossRef]
- McDonald, N.A.; Jorgensen, W.L. Development of an All-Atom Force Field for Heterocycles. Properties of Liquid Pyrrole, Furan, Diazoles, and Oxazoles. J. Phys. Chem. B 1998, 102, 8049–8059. [Google Scholar] [CrossRef]
- Rizzo, R.C.; Jorgensen, W.L. OPLS All-Atom Model for Amines: Resolution of the Amine Hydration Problem. J. Am. Chem. Soc. 1999, 121, 4827–4836. [Google Scholar] [CrossRef]
- Price, M.L.P.; Ostrovsky, D.; Jorgensen, W.L. Gas-phase and liquid-state properties of esters, nitriles, and nitro compounds with the OPLS-AA force field. J. Comput. Chem. 2001, 22, 1340–1352. [Google Scholar] [CrossRef]
- Watkins, E.K.; Jorgensen, W.L. Perfluoroalkanes: Conformational Analysis and Liquid-State Properties from ab Initio and Monte Carlo Calculations. J. Phys. Chem. A 2001, 105, 4118–4125. [Google Scholar] [CrossRef]
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Horn, H.W.; Swope, W.C.; Pitera, J.W.; Madura, J.D.; Dick, T.J.; Hura, G.L.; Head-Gordon, T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J. Chem. Phys. 2004, 120, 9665–9678. [Google Scholar] [CrossRef]
- Hohenberg, P.; Kohn, W. Inhomogeneous electron gas. Phys. Rev. 1964, 136, B864–B871. [Google Scholar] [CrossRef]
- Kohn, W.; Sham, L.J. Self-Consistent Equations Including Exchange and Correlation Effects. Phys. Rev. 2002, 140, A1133–A1138. [Google Scholar] [CrossRef]
- EMSL Basis Set Exchange. Available online: https://bse.pnl.gov/bse/portal (accessed on 5 November 2020).
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Valiev, M.; Bylaska, E.J.; Govind, N.; Kowalski, K.; Straatsma, T.P.; van Dam, H.J.J.; Wang, D.; Nieplocha, J.; Apra, E.; Windus, T.L.; et al. NWChem: A comprehensive and scalable open-source solution for large scale molecular simulations. Comp. Phys. Commun. 2010, 181, 1477–1489. [Google Scholar] [CrossRef]
- Darden, T.A.; York, D.M.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Krissinel, E. Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 2012, 1, 76–85. [Google Scholar]
- Pang, Y.-P. At least 10% shorter C–H bonds in cryogenic protein crystal structures than in current AMBER forcefields. Biochem. Biophys. Res. Commun. 2015, 458, 352–355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Engh, R.A.; Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. Sect. A Found. Crystallogr. 1991, 47, 392–400. [Google Scholar] [CrossRef]
- Lovell, S.C.; Word, J.M.; Richardson, J.S.; Richardson, D.C. The penultimate rotamer library. Proteins 2000, 40, 389–408. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Zhou, A.Q.; O’Hern, C.S.; Regan, L.J. Predicting the side-chain dihedral angle distributions of nonpolar, aromatic, and polar amino acids using hard sphere models. Proteins Struct. Funct. Bioinform. 2014, 82, 2574–2584. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Vriend, G.; Sander, C.; Abola, E.E. Errors in protein structures. Nature 1996, 381, 272. [Google Scholar] [CrossRef]
- Steinbach, P.J.; Brooks, B.R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994, 15, 667–683. [Google Scholar] [CrossRef]
- Da Silveira, C.H.; Pires, D.E.H.; Minardi, R.C.; Ribeiro, C.; Veloso, C.J.M.; Lopes, J.C.D.; Meira, W.; Neshich, G.; Ramos, C.H.I.; Habesch, R.; et al. Protein cutoff scanning: A comparative analysis of cutoff dependent and cutoff free methods for prospecting contacts in proteins. Proteins 2009, 74, 727–743. [Google Scholar] [CrossRef]
- Reif, M.M.; Krautler, V.; Kastenholz, M.A.; Daura, X.; Hunenberger, P.H. Molecular dynamics simulations of a reversibly folding beta-heptapeptide in methanol: Influence of the treatment of long-range electrostatic interactions. J. Phys. Chem. B 2009, 113, 3112–3128. [Google Scholar] [CrossRef]
- Piana, S.; Lindorff-Larsen, K.; Dirks, R.M.; Salmon, J.K.; Dror, R.O.; Shaw, D.E. Evaluating the Effects of Cutoffs and Treatment of Long-range Electrostatics in Protein Folding Simulations. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Strzalka, R.; Buganski, I.; Smietanska, J.; Wolny, J. Structural Disorder in Quasicrystals. Arch. Metall. Mater. 2020, 65, 291–294. [Google Scholar]
- Wolny, J.A.; Buganski, I.; Kuczera, P.; Strzałka, R. Pushing the limits of crystallography. J. Appl. Crystallogr. 2016, 49, 2106–2115. [Google Scholar] [CrossRef]
- Buganski, I.; Strzalka, R.; Wolny, J. Phason-flips refinement of and multiple-scattering correction for the d-AlCuRh quasicrystal. Acta Cryst. 2019, A75, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Mantsyzov, A.B.; Savelyev, O.Y.; Ivantcova, P.M.; Bräse, S.; Kudryavtsev, K.V.; Polshakov, V.I. Theoretical and NMR Conformational Studies of β-Proline Oligopeptides with Alternating Chirality of Pyrrolidine Units. Front. Chem. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Caballero, D.; Smith, W.W.; O’Hern, C.S.; Regan, L.J. Equilibrium transitions between side-chain conformations in leucine and isoleucine. Proteins Struct. Funct. Bioinform. 2015, 83, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Shapovalov, M.V.; Dunbrack, R.L. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 2011, 19, 844–858. [Google Scholar] [CrossRef]
- Virrueta, A.; O’Hern, C.S.; Regan, L.J. Understanding the physical basis for the side-chain conformational preferences of methionine. Proteins Struct. Funct. Bioinform. 2016, 84, 900–911. [Google Scholar] [CrossRef]




| Simulation Step | ID | Number of Steps/Time Step (fs) |
|---|---|---|
| Energy minimization | EM | 100,000/1 |
| NVT equilibration | NVT | 100,000/1 |
| NPT equilibration | NPT | 1,000,000/1 |
| Simulation of rotamer 1 | ROT1 | 100/200 |
| Simulation of rotamer 2 | ROT2 | 10/20 |
| Simulation of rotamer 3 | ROT3 | 1/2 |
| Cooling down | CD | 1,200,000/1 |
| RMSF gathering | RMSF | 1,000,000/1 |
| Initial Torsion Angle | Intermediate Torsion Angles | Terminal Torsion Angle | |
|---|---|---|---|
| Arg | N-CA-CB-CG | CA-CB-CG-CD CB-CG-CD-NE | CG-CD-NE-CZ |
| Asn | N-CA-CB-CG | CA-CB-CG-OD1 | |
| Asp | N-CA-CB-CG | CA-CB-CG-OD1 | |
| Cys | N-CA-CB-SG | ||
| Gln | N-CA-CB-CG | CA-CB-CG-CD | CB-CG-CD-OE1 |
| Glu | N-CA-CB-CG | CA-CB-CG-CD | CB-CG-CD-OE1 |
| His | N-CA-CB-CG | CA-CB-CG-ND1 | |
| Leu | N-CA-CB-CG | CA-CB-CG-CD1 | |
| Lys | N-CA-CB-CG | CA-CB-CG-CD CB-CG-CD-CE | CG-CD-CE-NZ |
| Met | N-CA-CB-CG | CA-CB-CG-SD | CB-CG-SD-CE |
| Phe | N-CA-CB-CG | CA-CB-CG-CD1 | |
| Pro | N-CA-CB-CG | ||
| Ser | N-CA-CB-OG | ||
| Thr | N-CA-CB-OG1 | ||
| Tyr | N-CA-CB-CG | CA-CB-CG-CD1 | |
| Val | N-CA-CB-CG1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smietanska, J.; Kozik, T.; Strzalka, R.; Buganski, I.; Wolny, J. Molecular Dynamics Investigation of Phenolic Oxidative Coupling Protein Hyp-1 Derived from Hypericum perforatum. Crystals 2021, 11, 43. https://doi.org/10.3390/cryst11010043
Smietanska J, Kozik T, Strzalka R, Buganski I, Wolny J. Molecular Dynamics Investigation of Phenolic Oxidative Coupling Protein Hyp-1 Derived from Hypericum perforatum. Crystals. 2021; 11(1):43. https://doi.org/10.3390/cryst11010043
Chicago/Turabian StyleSmietanska, Joanna, Tomasz Kozik, Radoslaw Strzalka, Ireneusz Buganski, and Janusz Wolny. 2021. "Molecular Dynamics Investigation of Phenolic Oxidative Coupling Protein Hyp-1 Derived from Hypericum perforatum" Crystals 11, no. 1: 43. https://doi.org/10.3390/cryst11010043
APA StyleSmietanska, J., Kozik, T., Strzalka, R., Buganski, I., & Wolny, J. (2021). Molecular Dynamics Investigation of Phenolic Oxidative Coupling Protein Hyp-1 Derived from Hypericum perforatum. Crystals, 11(1), 43. https://doi.org/10.3390/cryst11010043





