Glycosylation: A “Last Word” in the Protein-Mediated Biomineralization Process
Abstract
1. Introduction
2. Glycoproteins in Biomineralization: An Overview
2.1. Enamelin
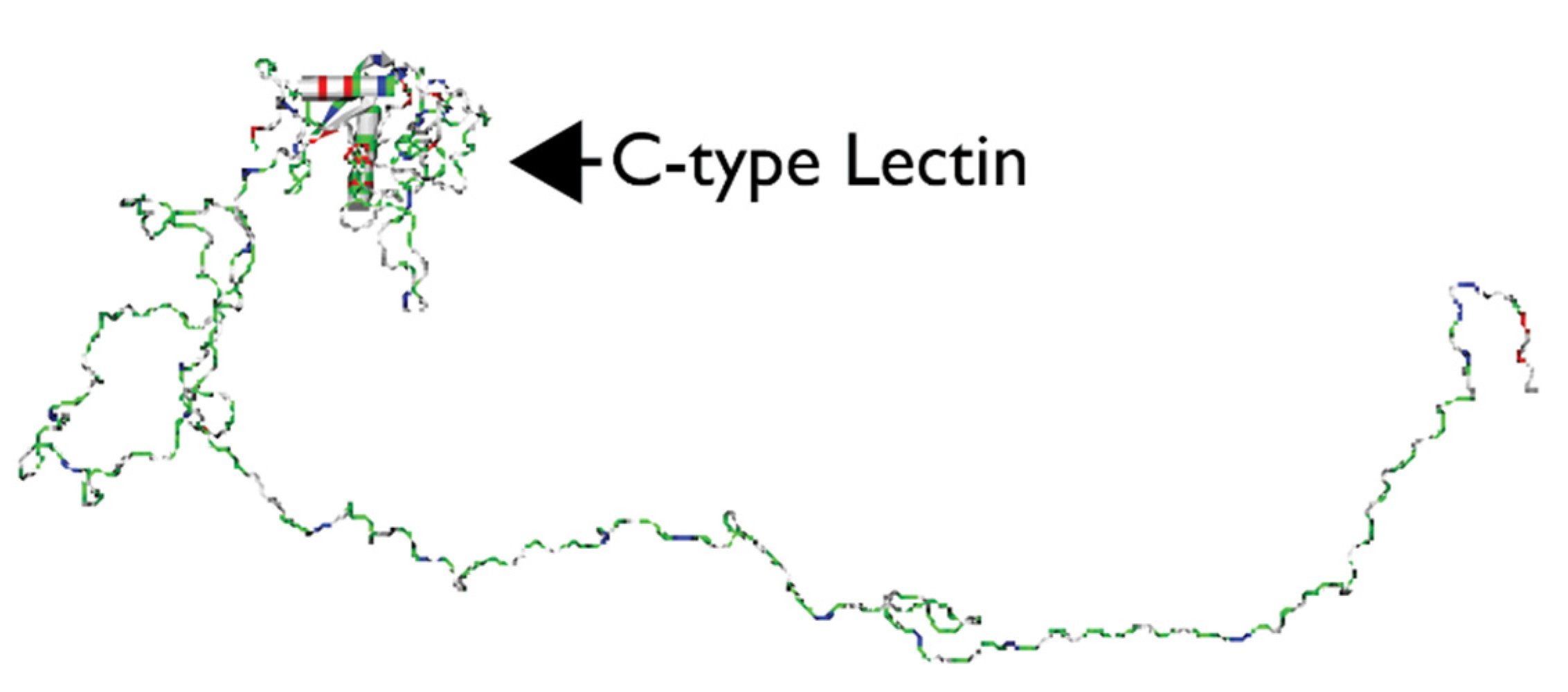
2.2. EDIL3, MFGE8
2.3. Proteoglycans
2.4. SIBLING Family
2.5. SpSM30A-F
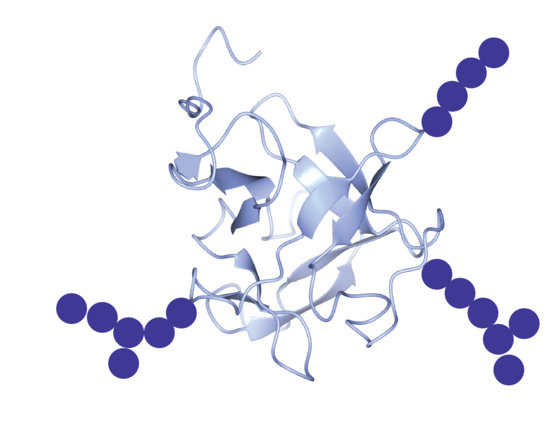
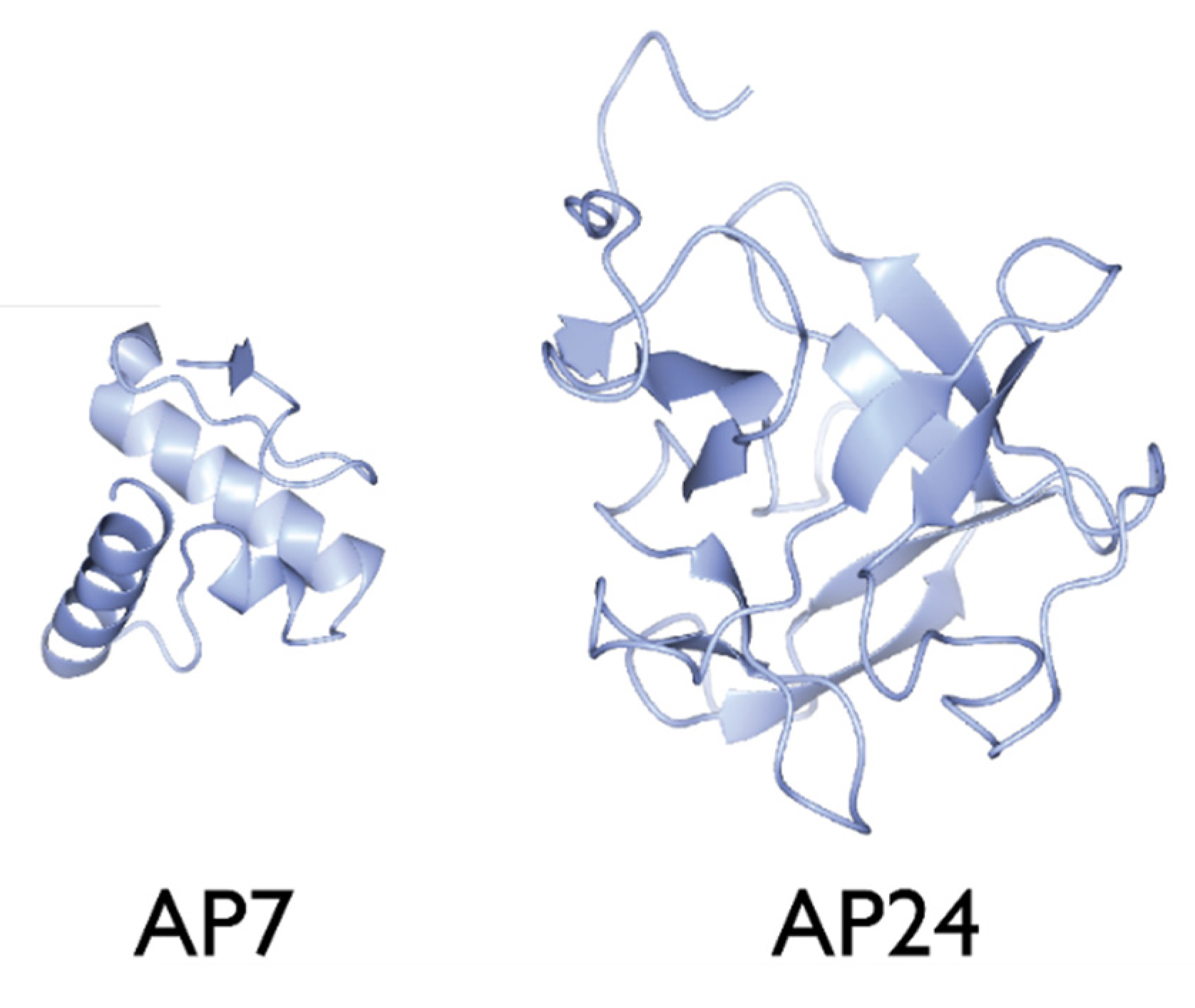
2.6. AP24
3. The Impact of Glycosylation on Protein Function
3.1. The Nacre Glycoprotein AP24
3.2. The Spicule Matrix Glycoprotein SpSM30B/C
3.3. How Does Glycosylation Impact Function?
4. The Impact of Glycosylation on Protein–Protein Interaction (Matrix Formation)
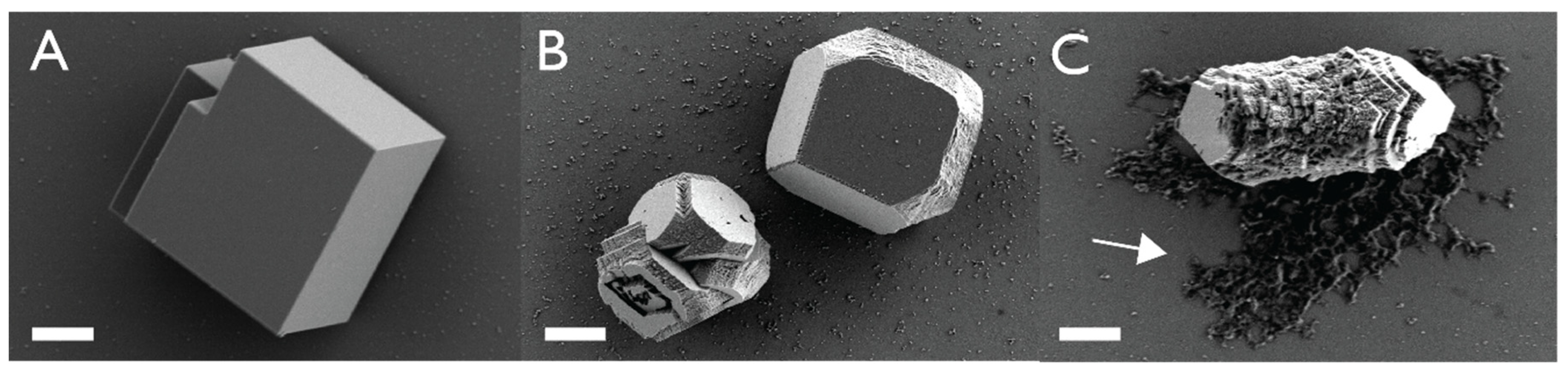
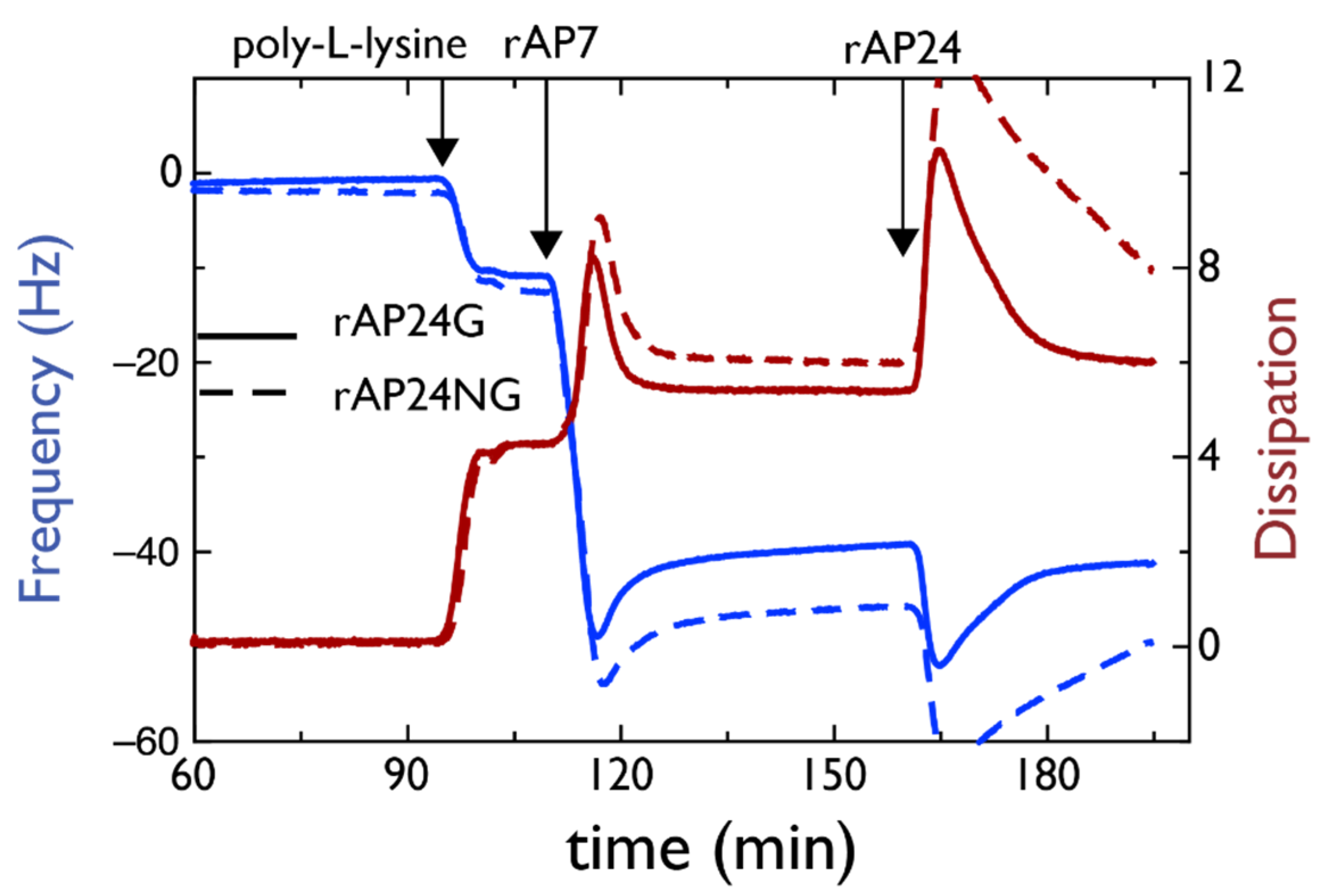
4.1. AP7–AP24 Complex
4.2. SpSM50–SpSM30B/C Complex
5. Summary and Future Directions
5.1. A More Aggressive Approach to Glycoprotein Isolation and Identification
5.2. Improvements in Glycoprotein Purification and Structure Determination
5.3. Improvements in Glycoprotein Localization
5.4. Improvements in Understanding the Role of Variations in Glycosylation
Funding
Acknowledgments
Conflicts of Interest
References
- Studart, A.R. Towards high-performance bioinspired composites. Adv. Mater. 2012, 24, 5024–5044. [Google Scholar] [CrossRef] [PubMed]
- Lowenstam, H.A.; Weiner, S. On Biomineralization; Oxford University Press: New York, NY, USA, 1989; pp. 1–134. ISBN 0-19-504977-2. [Google Scholar]
- Mann, S. Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry; Oxford University Press: New York, NY, USA, 2001; pp. 6–9, 24–108. [Google Scholar]
- Wallace, A.F.; Hedges, L.O.; Fernandez-Martinez, A.; Raiteri, P.; Gale, J.D.; Waychunas, G.A.; Whitelam, S.; Banfield, J.F.; De Yoreo, J.J. Microscopic evidence for liquid-liquid separation in supersaturated calcium carbonate solutions. Science 2013, 341, 885–889. [Google Scholar] [CrossRef]
- De Yoreo, J.J.; Gilbert, P.U.P.A.; Sommerdijk, N.A.J.M.; Penn, R.L.; Whitelam, S.; Joester, D.; Zhang, H.; Rimer, J.D.; Navrotsky, A.; Banfield, J.F.; et al. Crystallization by particle attachment in synthetic, biogenic, and geologic environments. Science 2015, 349, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Perovic, I.; Chang, E.P.; Verch, A.; Rao, A.; Cölfen, H.; Kroeger, R.; Evans, J.S. An oligomeric C-RING nacre protein influences pre-nucleation events and organizes mineral nanoparticles. Biochemistry 2014, 53, 7259–7268. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Xiang, L.; Sun, J.; Zheng, G.; Zhang, G.; Wang, H.; Xie, L.; Zhang, R. The role of matrix proteins in the control of nacreous layer deposition during pearl formation. Proc. R. Soc. B 2012, 279, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Sea Urchin Genome Sequencing Consortium; Sodergren, E.; Weinstock, G.M.; Davidson, E.H.; Cameron, R.A.; Gibbs, R.A.; Angerer, R.C.; Angerer, L.; Arnone, M.I.; Burgess, D.; et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006, 314, 941–952. [Google Scholar] [CrossRef]
- Tirichine, L.; Rastogi, A.; Bowler, C. Recent progress in diatom genomics and epigenomics. Curr. Op. Plant Biol. 2017, 36, 46–55. [Google Scholar] [CrossRef]
- Uversky, V.N. Post-Translational Modification. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Stanley, M., Kelly, H., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 425–430. [Google Scholar]
- Seo, J.; Lee, K.J. Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004, 37, 35–44. [Google Scholar] [CrossRef]
- Wang, Y.C.; Peterson, S.E.; Loring, J.F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014, 24, 143–160. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Krasnova, L.; Wong, C.H. Understanding the chemistry and biology of glycosylation with glycan synthesis. Ann. Rev. Biochem. 2016, 85, 599–630. [Google Scholar] [CrossRef] [PubMed]
- Dwek, R.A. Chapter 7: Glycobiology, a quantum leap in carbohydrate chemistry. Found. Mod. Biochem. 1996, 2, 153–202. [Google Scholar]
- Iijima, M.; Fan, D.; Bromley, K.; Sun, Z.; Moradian-Oldak, J. Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacalcium phosphate crystals. Cryst. Growth Des. 2011, 19, 4815–4822. [Google Scholar] [CrossRef] [PubMed]
- Yamakoshi, Y. Carbohydrate moieties of porcine 32 kDa enamelin. Calcif. Tissue Int. 1996, 56, 323–330. [Google Scholar] [CrossRef]
- Stapane, L.; Roy, N.; Hincke, M.T.; Gautron, J. The glycoproteins EDIL3 and MFGE8 regulate vesicle-mediated eggshell calcification in a new model for avain biomineralization. J. Biol. Chem. 2019, 294, 14256–14545. [Google Scholar] [CrossRef]
- Couchman, J.R.; Pataki, C.A. An introduction to proteoglycans and their localization. J. Histochem. Cytochem. 2012, 60, 885–897. [Google Scholar] [CrossRef]
- Ozawa, H.; Hoshi, K.; Amizuka, N. Current concepts in bone biomineralization. J. Oral Biosci. 2008, 50, 1–14. [Google Scholar] [CrossRef]
- Staines, K.A.; MacRae, V.E.; Farquharson, C. The importance of the SIBLING family of proteins on skeletal mineralization and bone remodeling. J. Endocrinol. 2012, 214, 241–255. [Google Scholar] [CrossRef]
- Bellahcene, A.; Castronovo, V.; Ogbureke, K.U.E.; Fisher, L.W.; Fedarko, N.S. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): Multifunctional proteins in cancer. Natl. Rev. Cancer 2008, 8, 212–226. [Google Scholar] [CrossRef]
- Killian, C.; Croker, L.; Wilt, F.H. SpSM30 gene family expression in embryonic and adult biomineralized tissues of the sea urchin, strongylocentrotus purpuratus. Gene Expr. Patterns 2010, 10, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Michenfelder, M.; Fu, G.; Lawrence, C.; Weaver, J.C.; Wustman, B.A.; Taranto, L.; Evans, J.S.; Morse, D.E. Characterization of two molluscan crystal-modulating biomineralization proteins and identification of putative mineral binding domains. Biopolymers 2003, 70, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.P.; Perovic, I.; Rao, A.; Cölfen, H.; Evans, J.S. Insect cell glycosylation and its impact on the functionality of a recombinant intracrystalline nacre protein, AP24. Biochemistry 2016, 55, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Jain, G.; Pendola, M.; Rao, A.; Cölfen, H.; Evans, J.S. A model sea urchin spicule matrix protein self-associates to form mineral-modifying protein hydrogels. Biochemistry 2016, 55, 4410–4421. [Google Scholar] [CrossRef]
- Juan-Colas, J.; Jung, Y.S.; Johnson, S.; Evans, J.S. A complicated relationship: Glycosylation, Ca(II), and primary sequence affect the interactions and kinetics between two model mollusk shell intracrystalline nacre proteins. Biochemistry 2020, 59, 346–350. [Google Scholar] [CrossRef]
- Jain, G.; Pendola, M.; Koutsoumpeli, E.; Johnson, S.; Evans, J.S. Glycosylation fosters interactions between model sea urchin spicule matrix proteins. Implications for embryonic spiculogenesis and biomineralization. Biochemistry 2018, 57, 3032–3035. [Google Scholar] [CrossRef]
- Pendola, M.; Jain, G.; Huang, Y.-C.; Gebauer, D.; Evans, J.S. Secrets of the sea urchin spicule revealed: Protein cooperativity is responsible for ACC transformation, intracrystalline incorporation, and guided mineral particle assembly in biocomposite material formation. ACS Omega 2018, 3, 11823–11830. [Google Scholar] [CrossRef]
- Le Pabic, C.; Marie, A.; Marie, B.; Percot, A.; Bonnaud-Ponticelli, L.; Lopez, P.J.; Luquet, G. First proteomic analysis of the dorsal and ventral parts of the Sepia officinalis cuttlebone. J. Proteom. 2017, 150, 63–73. [Google Scholar] [CrossRef]
- Albeck, S.; Weiner, S.; Addadi, L. Polysaccharides of intracrystalline proteins modulate calcite growth in vitro. Chem. A Eur. J. 1996, 2, 278–284. [Google Scholar] [CrossRef]
- Arivalagan, J.; Yarra, T.; Marie, B.; Sleight, V.A.; Duvernois-Berthet, E.; Clark, M.S.; Marie, A.; Berland, S. Insights from the shell proteome: Biomineralization to adaptation. Mol. Biol. Evol. 2017, 34, 66–77. [Google Scholar] [CrossRef]
- Tweedie, E.P.; Coblentz, F.E.; Shafer, T.H. Purification of a soluble glycoprotein from the uncalcified ecdysial cuticle of the blue crab Callinectes sapidus and its possible role in initial mineralization. J. Expt. Zool. 2004, 207, 2589–2598. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kamiya, H.; Jimbo, M.; Yako, H.; Muramoto, K. Participation of the C-type hemolymph lectin in mineralization of the acorn barnacle Megabalanus rosa. Mar. Biol. 2002, 140, 1235–1240. [Google Scholar]
- Kroger, N.; Bergsdorf, C.; Sumper, M. A new calcium binding glycoprotein family constitutes a major diatom cell wall component. EMBO J. 1994, 13, 4676–4683. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, M.; Ren, D.; Herrera, S.; Pan, S.; Tamura, T.; Inagaki, K.; Kisailus, D. Integrative transcriptomic and proteomic analyses of a molecular mechanism of radular teeth biomineralization in Cryptochiton Stelleri. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Gerard, C. Purification of glycoproteins. Methods Enzymol. 1990, 182, 529–539. [Google Scholar]
- Dang, H.V.; Chan, Y.P.; Park, Y.J.; Snijder, J.; Da Silva, S.C.; Vu, B.; Yan, L.; Feng, Y.R.; Rockx, B.; Geisbert, T.W.; et al. An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections. Nat. Struct. Mol. Biol. 2019, 26, 980–987. [Google Scholar] [CrossRef]
- Malaker, S.A.; Pedram, K.; Ferracane, M.J.; Bensing, B.A.; Krishnan, V.; Pett, C.; Yu, J.; Woods, E.C.; Kramer, J.R.; Westerlind, U.; et al. The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. USA 2019, 116, 7278–7287. [Google Scholar] [CrossRef]
- Mallajosyula, S.S.; Jo, S.; Im, W.; MacKerell, A.D., Jr. Molecular dynamics simulations of glycoproteins using CHARMM. Methods Mole Biol. 2015, 1273, 407–429. [Google Scholar]
| Post-Translational Modification | Type of Modification | Site(s) of Modification |
|---|---|---|
| Disulfide bond formation | Thiol oxidation to form -S-S- bond | Cys |
| Hydroxylation | Addition of -OH group | Pro, Glu |
| Ubiquitination | Addition of ubiquitin protein(s) | Lysine |
| Lipidation | Esterification to lipid group | Cys, Lys, N-terminal Gly |
| SUMOylation | Small Ubiquitin-like Modifier protein | Lys |
| Acetylation | Addition of acetyl group | N-terminus, Lys, Ser |
| Methylation | Addition of methyl group | Lys, Arg, termini |
| Phosphorylation | Addition of phosphate group | Ser, Thr, Tyr |
| Glycosylation | Addition of carbohydrate group(s) | Asn, Thr, Ser |
| Nitration | Addition of nitrogen | Tyr |
| Acylation | Addition of acyl chain | Cys, Gly, Ser, Thr, Lys |
| Protein | Organism | Tissue | Associated Mineral Phase |
|---|---|---|---|
| SpSM30 A-F | S. purpuratus (sea urchin) | Embryonic spicule | Magnesium Calcite (CaMgCO3) |
| AP24 | H. rufescens (abalone) | Shell nacre | Aragonite (CaCO3) |
| Enamelin | Vertebrates | Tooth enamel | Hydroxyapatite (CaPO4) |
| SIBLING Family | Vertebrates | Bone, tooth dentine | Hydroxyapatite (CaPO4) |
| EDIL3 | Avian | Eggshell | Calcite (CaCO3) |
| MFGE8 | Avian | Eggshell | Calcite (CaCO3) |
| Proteoglycans | Vertebrates | Bone, tooth dentine | Hydroxyapatite (CaPO4) |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evans, J.S. Glycosylation: A “Last Word” in the Protein-Mediated Biomineralization Process. Crystals 2020, 10, 818. https://doi.org/10.3390/cryst10090818
Evans JS. Glycosylation: A “Last Word” in the Protein-Mediated Biomineralization Process. Crystals. 2020; 10(9):818. https://doi.org/10.3390/cryst10090818
Chicago/Turabian StyleEvans, John Spencer. 2020. "Glycosylation: A “Last Word” in the Protein-Mediated Biomineralization Process" Crystals 10, no. 9: 818. https://doi.org/10.3390/cryst10090818
APA StyleEvans, J. S. (2020). Glycosylation: A “Last Word” in the Protein-Mediated Biomineralization Process. Crystals, 10(9), 818. https://doi.org/10.3390/cryst10090818