Microbial Biosynthesis of Antibacterial Chrysoeriol in Recombinant Escherichia coli and Bioactivity Assessment
Abstract
1. Introduction
2. Results
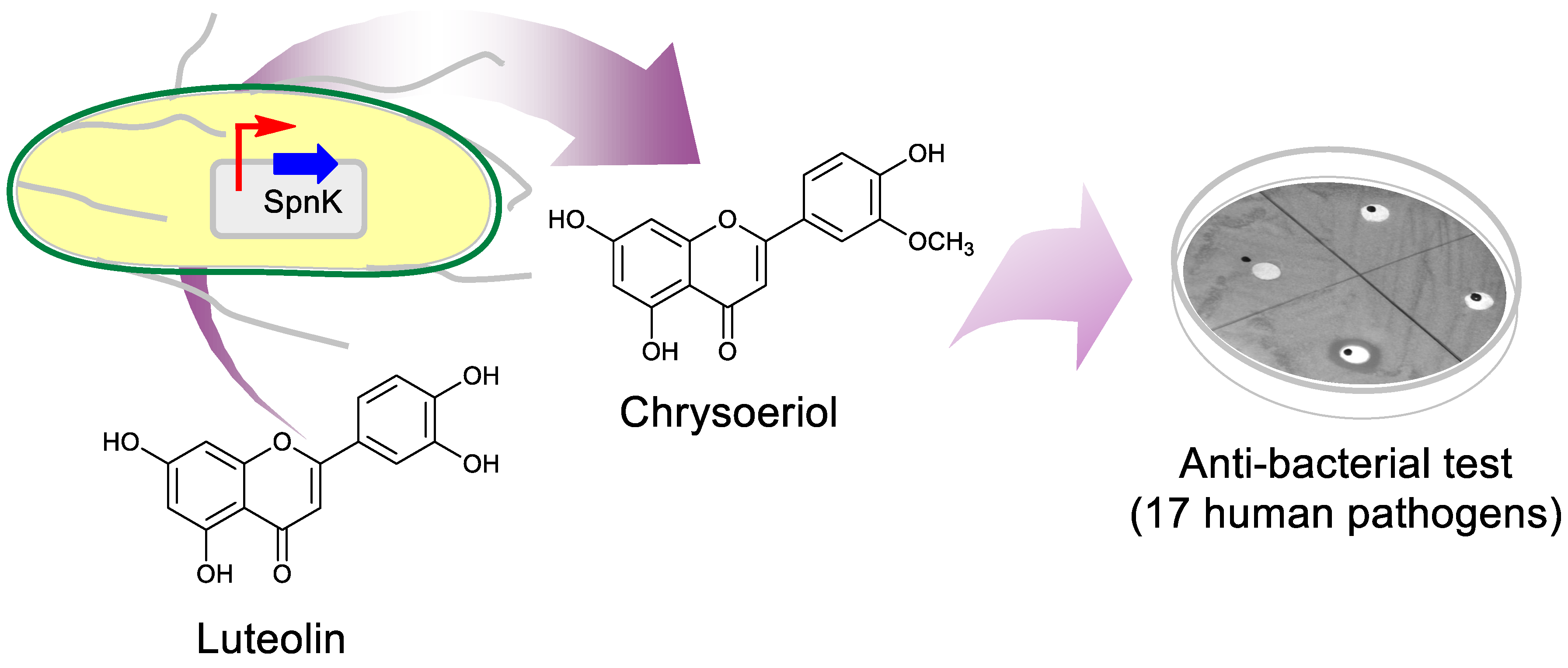
2.1. Biosynthesis of Luteolin O-methoxide
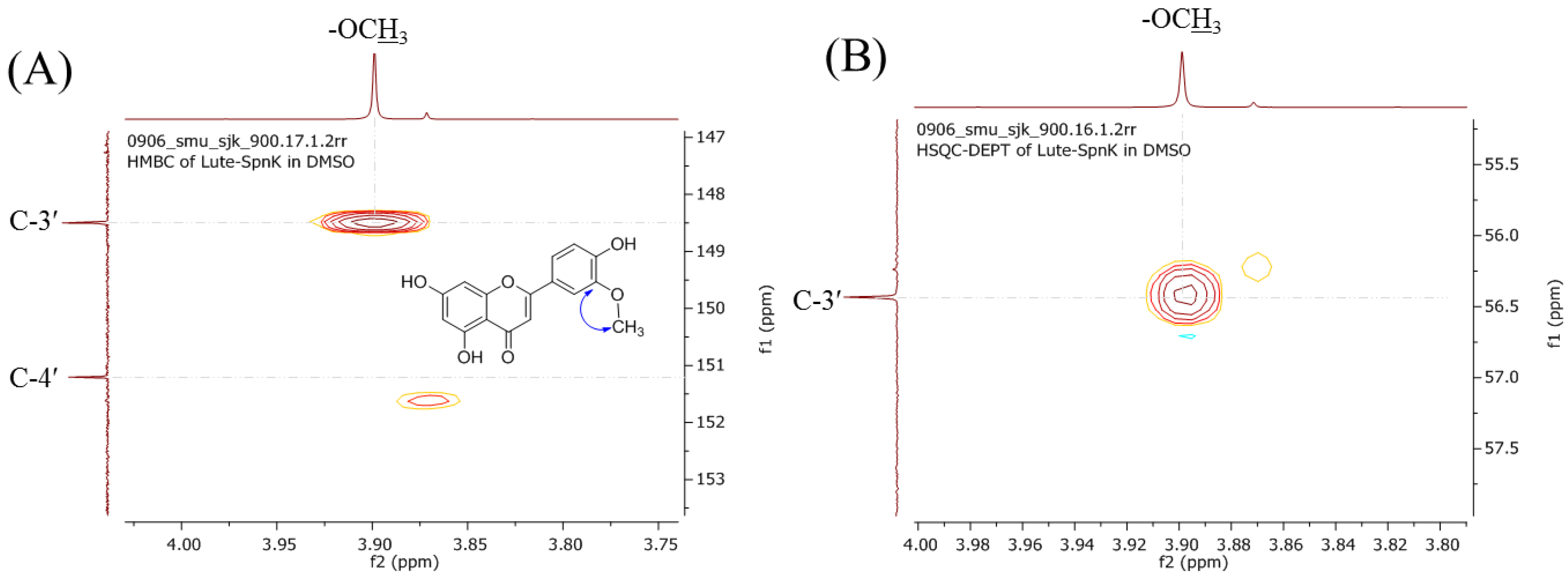
2.2. Purification and Structural Elucidation of the Metabolite
2.3. Antimicrobial Activity of Luteolin, Chrysoeriol, and Diosmetin
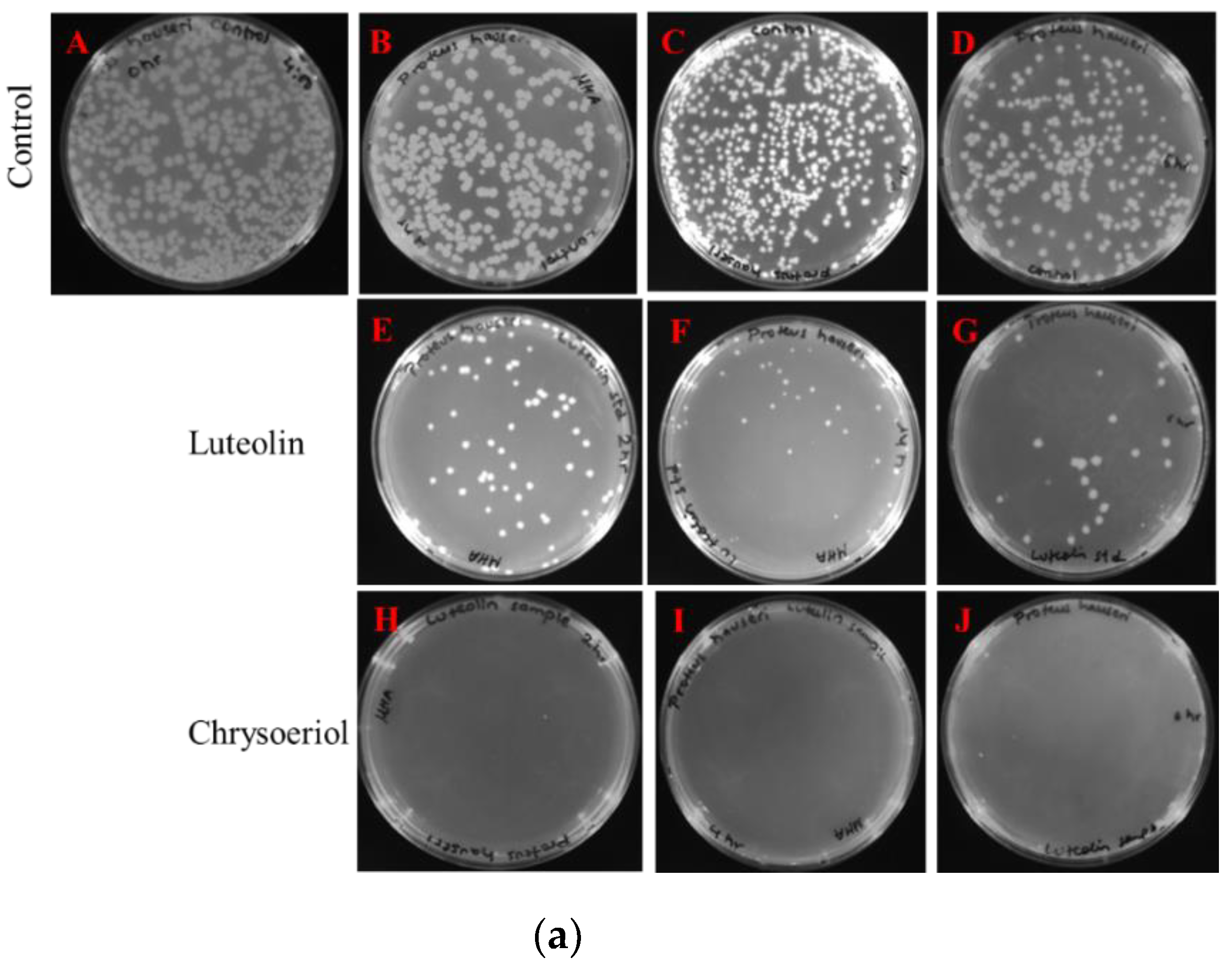
2.3.1. Disc Diffusion Assay
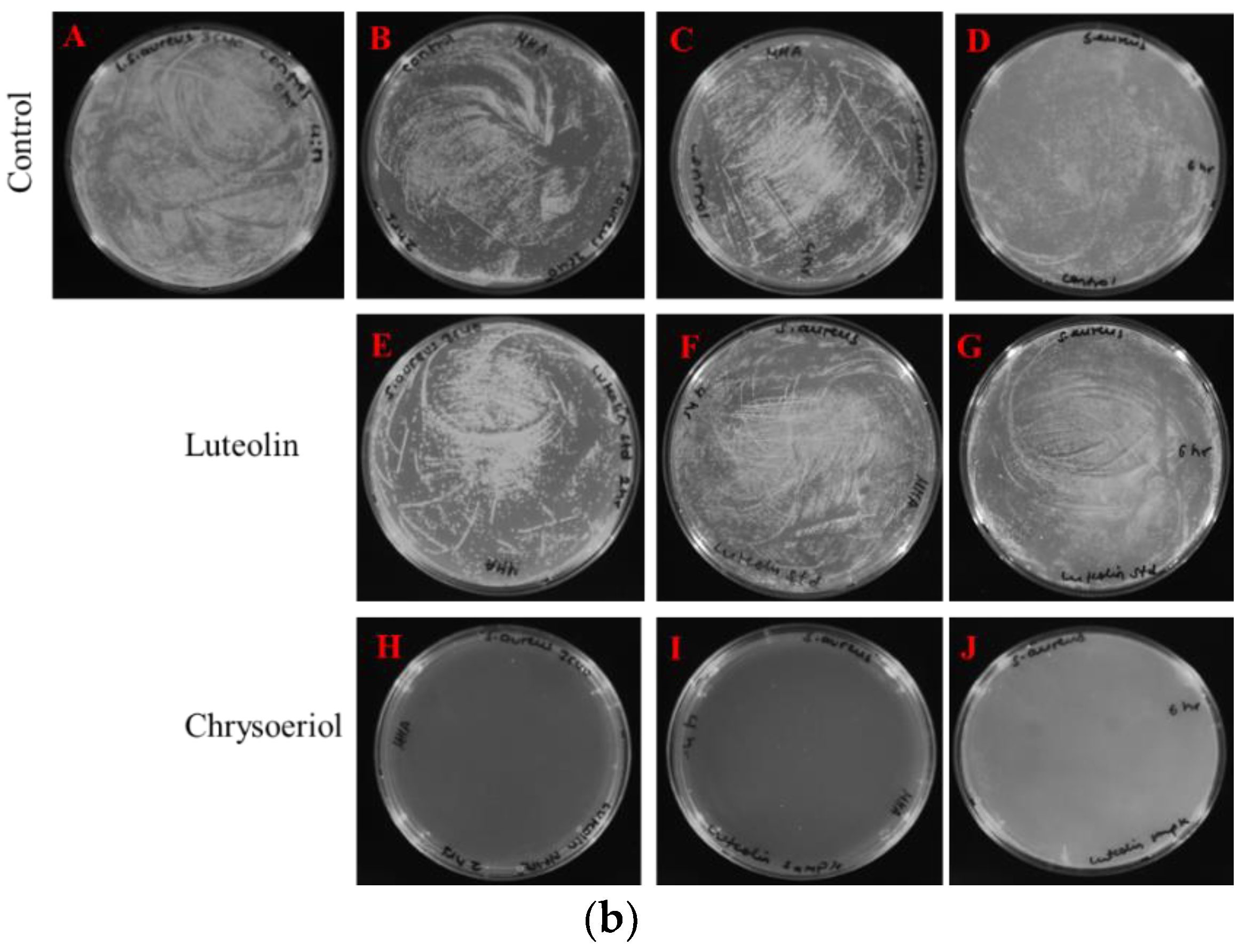
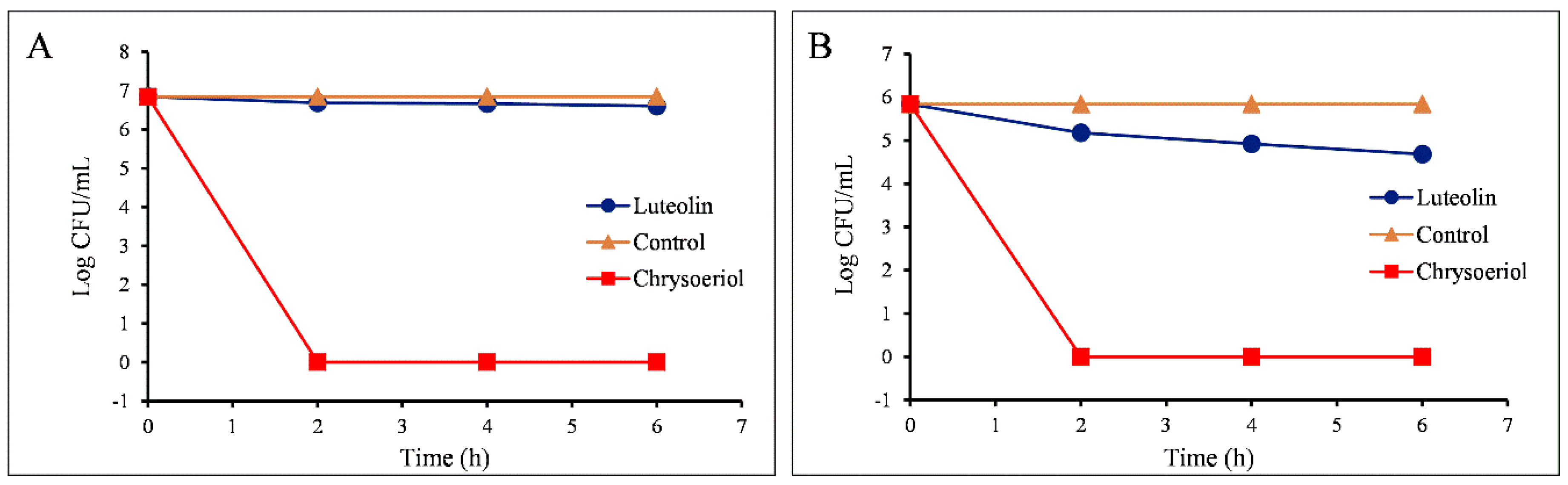
2.3.2. Measurements of Colony-Forming Units
2.3.3. Measurement of MIC Values
3. Discussion
4. Materials and Methods
4.1. General Procedures
4.2. Whole-Cell Biotransformation and Validation
4.3. Analytical Methods
4.4. Biological Activities
4.4.1. Disc Diffusion Assay
4.4.2. Measurement of the Colony-Forming Unit (CFU)
4.4.3. Measurement of Minimum Inhibitory Concentration (MIC)
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lin, Y.; Shi, R.; Wang, X.; Shen, H.M. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr. Cancer Drug Targets 2008, 8, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Braidy, N.; Gortzi, O.; Sobarzo-Sanchez, E.; Daglia, M.; Skalicka-Woźniak, K.; Nabavi, S.M. Luteolin as an anti-inflammatory and neuroprotective agent: A brief review. Brain Res. Bull. 2015, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Shang, P.; Li, D. Luteolin: A flavonoid that has multiple cardio-protective effects and its molecular mechanisms. Front. Pharmacol. 2017, 8, 692. [Google Scholar] [CrossRef] [PubMed]
- Tuorkey, M.J. Molecular targets of luteolin in cancer. Eur. J. Cancer Prev. 2016, 25, 65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Qiu, Y.; Luo, Q.; Zhao, L.; Yan, X.; Ding, Q.; Jiang, H.; Yang, H. The mechanism by which luteolin disrupts the cytoplasmic membrane of methicillin-resistant Staphylococcus aureus. J. Phys. Chem. B 2018, 122, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Baier, A.; Nazaruk, J.; Galicka, A.; Szyszka, R. Inhibitory influence of natural flavonoids on human protein kinase CK2 isoforms: Effect of the regulatory subunit. Mol. Cell Biochem. 2018, 444, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Dai, Y.Q.; Kong, S.S.; Song, F.F.; Li, L.P.; Ye, J.F.; Wang, R.W.; Zeng, S.; Zhou, H.; Jiang, H.D. Luteolin is a rare substrate of human catechol-O-methyltransferase favoring a para-methylation. Mol. Nutr. Food Res. 2013, 57, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Arroo, R.R.J.; Beresford, K.; Bhambra, A.S.; Boarder, M.; Budriesi, R.; Cheng, Z.; Micucci, M.; Ruparelia, K.C.; Surichan, S.; Androutsopoulos, V.P. Phytoestrogens as natural prodrugs in cancer prevention: Towards a mechanistic model. Phytochem Rev. 2014, 13, 853–866. [Google Scholar] [CrossRef]
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940. [Google Scholar] [CrossRef]
- Chan, B.C.; Ip, M.; Lau, C.B.; Lui, S.L.; Jolivalt, C.; Ganem-Elbaz, C.; Litaudon, M.; Reiner, N.E.; Gong, H.; See, R.H.; et al. Synergistic effects of baicalein with ciprofloxacin against NorA over-expressed methicillin-resistant Staphylococcus aureus (MRSA) and inhibition of MRSA pyruvate kinase. J. Ethnopharmacol. 2011, 137, 767–773. [Google Scholar] [CrossRef]
- Chan, B.C.; Ip, M.; Gong, H.; Lui, S.L.; See, R.H.; Jolivalt, C.; Fung, K.P.; Leung, P.C.; Reiner, N.E.; Lau, C.B. Synergistic effects of diosmetin with erythromycin against ABC transporter over-expressed methicillin-resistant Staphylococcus aureus (MRSA) RN4220/pUL5054 and inhibition of MRSA pyruvate kinase. Phytomedicine 2013, 20, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Joung, D.K.; Lee, Y.S.; Han, S.H.; Lee, S.W.; Cha, S.W.; Mun, S.H.; Kong, R.; Kang, O.H.; Song, H.J.; Shin, D.W.; Kwon, D.Y. Potentiating activity of luteolin on membrane permeabilizing agent and ATPase inhibitor against methicillin-resistant Staphylococcus aureus. Asian Pac. J. Trop Med. 2016, 9, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, M. Antibacterial activity and mechanism of luteolin on Staphyloccus aureus. Wei Sheng Wu Xue Bao 2013, 50, 1180–1184. [Google Scholar]
- Nascimento, P.L.A.; Nascimento, T.C.E.S.; Ramos, N.S.M.; Silva, G.R.; Gomes, J.E.; Falcão, R.E.A.; Moreira, K.A.; Porto, A.L.F.; Silva, T.M.S. Quantification, antioxidant and antimicrobial activity of phenolics isolated from different extracts of Capsicum frutescens (Pimenta Malagueta). Molecules 2014, 19, 5434–5447. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.Y.; Lee, J.Y.; Kim, M.R.; Woo, E.R.; Kim, Y.G.; Kang, K.W. Chrysoeriol potently inhibits the induction of nitric oxide synthase by blocking AP-1 activation. J. Biomed. Sci. 2005, 12, 949–959. [Google Scholar] [CrossRef]
- Gorzalczany, S.; Moscatelli, V.; Ferraro, G. Artemisia copa aqueous extract as vasorelaxant and hypotensive agent. J. Ethnopharmacol. 2013, 148, 56–61. [Google Scholar] [CrossRef]
- Ramirez, G.; Zamipla, A.; Zavala, M.; Perez, J.; Morales, D.; Tortoriello, J. Chrysoeriol and other polyphenols from Tecoma stans with lipase inhibitory activity. J. Ethnopharmacol. 2016, 185, 1–8. [Google Scholar] [CrossRef]
- Baltz, R.H. Antimicrobials from actinomycetes: Back to the future. Microbe 2007, 2, 125. [Google Scholar]
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef]
- Hostetler, G.L.; Ralston, R.A.; Schwartz, S.J. Flavones: food sources, bioavailability, metabolism, and bioactivity. Adv. Nutr. 2017, 8, 423–435. [Google Scholar] [CrossRef]
- Parajuli, P.; Pandey, R.P.; Nguyen, T.T.H.; Shrestha, B.; Yamaguchi, T.; Sohng, J.K. Biosynthesis of natural and non-natural genistein glycosides. RSC Adv. 2017, 7, 16217–16231. [Google Scholar] [CrossRef]
- Parajuli, P.; Pandey, R.P.; Sohng, J.K. Regiospecific biosynthesis of tamarixetin derivatives in Escherichia coli. Biochem. Eng. J. 2018, 133, 113–121. [Google Scholar] [CrossRef]
- Park, Y.; Moon, B.H.; Yang, H.; Lee, Y.; Lee, E.; Lim, Y. Complete assignments of NMR data of 13 hydroxymethoxyflavones. Magn. Reson. Chem. 2007, 45, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Erenler, R.; Sen, O.; Yildiz, I.; Aydin, A. Antiproliferative activities of chemical constituents isolated from Thymus pracox subsp. Grossheimii (Ronniger) Jalas. Rec. Nat. Prod. 2016, 10, 766–770. [Google Scholar]
- Guha, R. On exploring structure-activity relationships. Methods Mol. Biol. 2013, 993, 81–94. [Google Scholar] [PubMed]
- Wang, T.; Wu, M.B.; Lin, J.P.; Yang, L.R. Quantitative structure-activity relationship: Promising advances in drug discovery platforms. Expert Opin. Drug Discov. 2015, 10, 1283–1300. [Google Scholar] [CrossRef] [PubMed]
- Pervaiz, I.; Ahmad, S.; Madni, M.A.; Ahmad, H.; Khaliq, F.H. Microbial biotransformation: A tool for drug designing. Appl. Biochem. Microbiol. 2013, 49, 437–450. [Google Scholar] [CrossRef]
- Bianchini, L.F.; Arruda, M.F.; Vieira, S.R.; Campelo, P.M.; Grégio, A.M.; Rosa, E.A. Microbial biotransformation to obtain new antifungals. Front. Microbiol. 2015, 6, 1433. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.P.; Parajuli, P.; Koffas, M.A.G.; Sohng, J.K. Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 2016, 34, 634–662. [Google Scholar] [CrossRef] [PubMed]
- Restaino, O.F.; Marseglia, M.; De Castro, C.; Diana, P.; Forni, P.; Parrilli, M.; De Rosa, M.; Schiraldi, C. Biotechnological transformation of hydrocortisone to 16α-hydroxy hydrocortisone by Streptomyces roseochromogenes. Appl. Microbiol. Biotechnol. 2014, 98, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Teffo, L.S.; Aderogba, M.A.; Eloff, J.N. Antibacterial and antioxidant activities of four kaempferol methyl ethers isolated from Dodonaea viscosa Jacq. var. angustifolia leaf extracts. S. Afr. J. Bot. 2010, 76, 25–29. [Google Scholar] [CrossRef]
- Choi, H.R.; Park, J.S.; Kim, K.M.; Kim, M.S.; Ko, K.W.; Hyun, C.G.; Ahn, J.W.; Seo, J.H.; Kim, S.Y. Enhancing the antimicrobial effect of genistein by biotransformation in microbial system. J. Ind. Eng. Chem. 2018, 63, 255–261. [Google Scholar] [CrossRef]
- Mori, A.; Nishino, C.; Enoki, N.; Tawata, S. Antibacterial activity and mode of action of plant flavonoids against Proteus vulgaris and Staphylococcus aureus. Phytochemistry 1987, 26, 2231–2234. [Google Scholar] [CrossRef]
- Ikigai, H.; Nakae, T.; Hara, Y.; Shimamura, T. Bactericidal catechins damage the lipid bilayer. Biochim. Biophys. Acta 1993, 1147, 132–136. [Google Scholar] [CrossRef]
- Babii, C.; Mihalache, G.; Bahrin, L.G.; Neagu, A.N.; Gostin, I.; Mihai, C.T.; Sârbu, L.G.; Birsa, M.; Stefan, M. A novel synthetic flavonoid with potent antibacterial properties: In vitro activity and proposed mode of action. PLoS ONE 2018, 4, e0194898. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Lamb, A.J. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Daglia, M. Polyphenols as antimicrobial agents. Biotechnology 2012, 23, 174–181. [Google Scholar] [CrossRef] [PubMed]

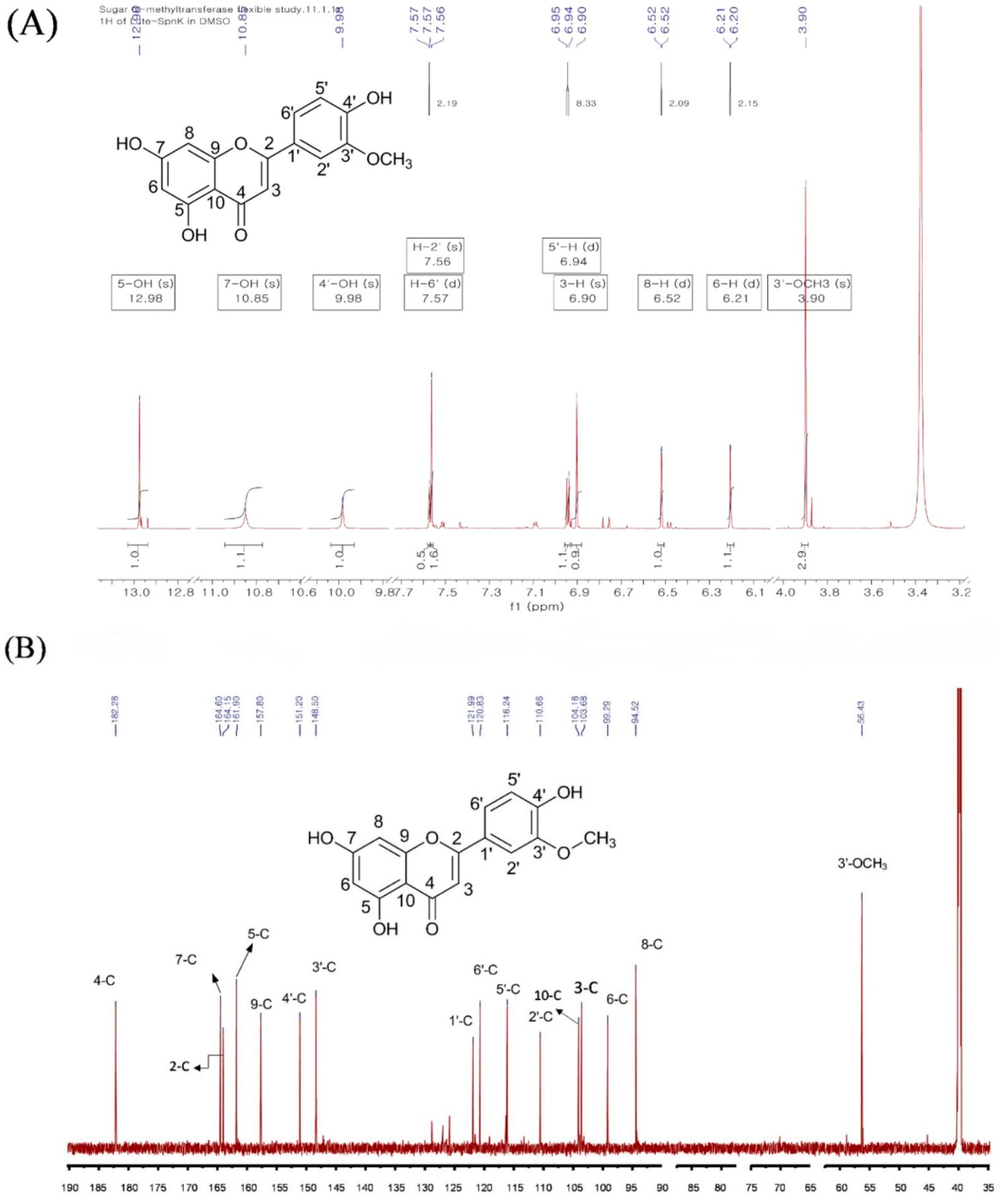
| 1H-NMR | 13C-NMR | ||
|---|---|---|---|
| Position of Protons | Position of Carbons | ||
| 5-OH | 12.96 | 1 | - |
| 7-OH | 10.85 | 2 | 164.15 |
| 4′-OH | 9.98 | 3 | 103.68 |
| 6′ | 7.57 (d, J = 2.19 Hz) | 4 | 182.28 |
| 2′ | 7.56 (s) | 5 | 161.90 |
| 5′-H | 6.94 (d, J = 8.33 Hz) | 6 | 99.29 |
| 3-H | 6.90 (s) | 7 | 164.60 |
| 8-H | 6.52 (d, J = 2.1 Hz) | 8 | 94.52 |
| 6-H | 6.21 (d, J = 2.15 Hz) | 9 | 157.80 |
| 3′-OCH3 3.90 (s) | 10 | 104.14 | |
| 1′ | 121.99 | ||
| 2′ | 110.66 | ||
| 3′ | 148.50 | ||
| 4′ | 151.20 | ||
| 5′ | 116.24 | ||
| 6′ | 120.83 | ||
| O-CH3 | 56.43 | ||
| Zone of Inhibition (mm) | |||
|---|---|---|---|
| - | Luteolin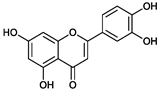 | Chrysoeriol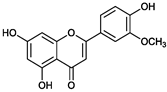 | Diosmetin |
| Gram-Positive Strains | |||
| S. aureus CCARM 3640 (MRSA) | 9 ± 0.2 | 15 ± 0.3 | ND |
| S. aureus CCARM 3089 (MRSA) | 9 ± 0.12 | 14 ± 0.35 | ND |
| S. aureus CCARM 33591(MRSA) | 9 ± 0.10 | ND | ND |
| S. aureus CCARM 0205 (MSSA) | 9 ± 0.2 | 9 ± 0.2 | ND |
| S. aureus CCARM 0204 (MSSA) | 10 ± 0.2 | 9 ± 0.12 | ND |
| S. aureus CCARM 0027 (MSSA) | ND | ND | ND |
| S. aureus CCARM 3090 (MRSA) | 12 ± 0.3 | 15 ± 0.21 | ND |
| S. aureus CCARM 3634 (MRSA) | 9 ± 0.1 | 14 ± 0.25 | ND |
| S. aureus CCARM 3635 (MRSA) | 11 ± 0.11 | 10 ± 0.15 | ND |
| Bacillus subtilis ATCC 6633 | 8 ± 0.1 | 9 ± 0.2 | ND |
| Enterococcus faecalis 19433 | ND | ND | ND |
| Enterococcus faecalis 19434 | ND | ND | ND |
| Kocuria rhizophilla NBRC 12708 | 8 ± 0.2 | ND | ND |
| Gram-Negative Strains | |||
| Salmonella enterica ATCC 14028 | 8 ± 0.2 | ND | ND |
| Klebsiella pneumonia ATCC 10031 | ND | ND | ND |
| Escherichia coli ATCC 25922 | ND | ND | ND |
| Proteus hauseri NBRC 3851 | ND | 14 ± 0.2 | ND |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashyal, P.; Parajuli, P.; Pandey, R.P.; Sohng, J.K. Microbial Biosynthesis of Antibacterial Chrysoeriol in Recombinant Escherichia coli and Bioactivity Assessment. Catalysts 2019, 9, 112. https://doi.org/10.3390/catal9020112
Bashyal P, Parajuli P, Pandey RP, Sohng JK. Microbial Biosynthesis of Antibacterial Chrysoeriol in Recombinant Escherichia coli and Bioactivity Assessment. Catalysts. 2019; 9(2):112. https://doi.org/10.3390/catal9020112
Chicago/Turabian StyleBashyal, Puspalata, Prakash Parajuli, Ramesh Prasad Pandey, and Jae Kyung Sohng. 2019. "Microbial Biosynthesis of Antibacterial Chrysoeriol in Recombinant Escherichia coli and Bioactivity Assessment" Catalysts 9, no. 2: 112. https://doi.org/10.3390/catal9020112
APA StyleBashyal, P., Parajuli, P., Pandey, R. P., & Sohng, J. K. (2019). Microbial Biosynthesis of Antibacterial Chrysoeriol in Recombinant Escherichia coli and Bioactivity Assessment. Catalysts, 9(2), 112. https://doi.org/10.3390/catal9020112






