Abstract
Methane is a promising carbon feedstock for industrial biomanufacturing because of its low price and high abundance. Recent advances in metabolic engineering and systems biology in methanotrophs have made it possible to produce a variety of value-added compounds from methane, including secondary metabolites. Isoprenoids are one of the largest family of secondary metabolites and have many useful industrial applications. In this review, we highlight the current efforts invested to methanotrophs for the production of isoprenoids and other secondary metabolites, including riboflavin and ectoine. The future outlook for improving secondary metabolites production (especially of isoprenoids) using metabolic engineering of methanotrophs is also discussed.
1. Introduction
Methane is the major component of natural gas and a huge amount of methane is available due to advances in natural and shale gas production technology [1]. Unfortunately, atmospheric methane is 20 times as potent as carbon dioxide as a greenhouse gas that needs to be mitigated. Of the total methane emissions, anthropogenic emissions (combustion of fossil fuels, livestock industry, landfills, and biomass burning) account for 63%, and natural sources (wetlands, oceans, rivers, lakes, and permafrost) account for 37%. Anthropogenic emissions have steadily increased, with an increase in methane gas production as relevant industry develops [2,3]. Currently, most methane is used to produce electricity in gas turbine boilers and is supplied to homes for heating and cooking [4].
One challenging technical issue on methane utilization is gas-to-liquid (GTL) conversion because methane gas is not easily transported [5]. Various chemical GTL conversion technologies have been developed to date, but there are some disadvantages. The chemical gas-to-liquid conversion technology consists of a methane reforming process to produce syngas, a Fischer-Tropsch process to convert syngas into hydrocarbons, and an upgrading and separation process [6]. Generally, these processes require high temperature up to 800 °C and high-pressure conditions. Therefore, chemical gas-to-liquid conversion requires high operating costs. Biological gas-to-liquid conversion technology is being developed to address these issues [5]. Biological gas-to-liquid conversion technology uses methanotrophs, which can use methane as a sole carbon source. This gas-to-liquid bioconversion using methanotrophs shows higher efficiency than the chemical conversion [7]. In addition, bioprocess proceeds under normal temperature and pressure conditions. By applying metabolic engineering and systems biology approaches, currently, engineered methanotrophs have been deployed for the production of several products, including secondary metabolites [8,9,10]. However, there are still some technical issues in the gas-to-liquid bioconversion using methanotrophs, such as limited mass transfer rates of methane, low volumetric productivity, and instability of recombinant strains [11].
Secondary metabolites produced by plants and microorganisms have many useful biological activities. Among the many secondary metabolites, isoprenoids are the largest family of natural products, with more than 50,000 members identified [12,13,14]. Many isoprenoids have been used in many applications such as pharmaceuticals, nutraceuticals, fragrances, as well as the chemical industry. Additionally, due to the methyl-branched and cyclized hydrocarbon alkene structure, some isoprenoids have attracted more attention for their potential use as advanced biofuels. Plants are the major sources of isoprenoids. However, there are still many limitations for the production of high quantities of natural isoprenoids, including slow growth and tissue-specific biosynthesis, and difficulties in harvesting and extraction [12,13,14]. In contrast, a promising alternative method for isoprenoid production is the reconstruction of isoprenoid biosynthesis pathways from plants into microbes. The application of this approach has been extensively studied on model organisms like Escherichia coli or Saccharomyces cerevisiae for a variety of isoprenoids [12,14]. E. coli and S. cerevisiae are promising hosts since they possess simple genetic backgrounds, high growth rates as well as well-developed genetic tools. For example, high production of isoprenoid-based biofuels has been achieved in the metabolic engineering of microbes via endogenous or heterologous biosynthetic pathways in these hosts. However, the vast majority of these microbe-based isoprenoid production cases rely on sugar-based metabolism, which is likely to increase in the price of isoprenoids produced in large quantity.
The use of alternative carbon sources is attractive in industrial biotechnology for isoprenoid production. Furthermore, different type of pathway regulation in different microbes utilizing unusual carbon substrate like methane can play a vital role in metabolic engineering of various microbes including model organisms. Thus, metabolic engineering of methane-utilizing methanotrophs has recently attracted much attention, due to not only cheaper price of methane but also the novelty and potential of secondary metabolites biosynthetic capability of methanotrophs. Some secondary metabolites were naturally produced in methanotrophs, such as carotenoids and ectoine, which are responsible for the pigmentation and osmotic stress in a high salt environment, respectively [8,10]. Recently, some rational metabolic engineering strategies have been used in methanotrophs as a microbial cell factory platform for the bioconversion of methane to a variety of value-added products [9]. Based on these backgrounds, in this study, the biosynthesis pathways and characteristics of isoprenoids and other secondary metabolites in methanotrophs are reviewed. The current state of the production of various isoprenoids, riboflavin, and ectoine using methanotrophs is overviewed. In later sections of this review, we discuss the potential of methanotrophs and strategies for improving the production of isoprenoids through metabolic engineering of methanotrophs.
2. Methane Metabolism in Methanotrophs
Methanotrophs play important roles in global methane cycling and in the degradation of harmful substances. They use methane as a sole carbon source and were isolated in several conditions where methane and oxygen are both present. Freshwater and sediments are sinks of atmospheric methane. A variety of different methanotrophs inhabit these environments, and Methylomonas, Methylobacter, Methylosarcina, Methylococcus, and Methylosoma are predominant. Landfills and rice field soils are major sources of atmospheric methane. Studies have reported that landfills and rice field soils contain a variety of methanotrophs, such as Methylomonas, Methylobacter, Methylosarcina, Methylomicrobium, Methylococcus, Methylocaldum, Methylocystis, and Methylosinus. The distribution and abundance of methanotrophs in these methane-generating environments are affected by oxygen availability. Methanotrophs are also found in various extreme environments such as hot springs, the Antarctic, peatbogs, and deep-sea environments [15].
Methanotrophs are bacteria that have the ability to assimilate methane. This has always been considered a very unique microbial function. Over the past decade, knowledge of methane metabolism has been broadened by the help of systems biology approaches [16]. However, it also highlighted a large amount of fundamental knowledge gaps of methanotrophs that must be addressed to exploit the full potential of methanotrophs [17]. Oxidation of methane is carried out by methane monooxygenase (MMO), a membrane-associated particulate methane monooxygenase (pMMO) and soluble methane monooxygenase (sMMO) [18,19,20]. After that, methanol dehydrogenase (MDH) oxidized methanol into formaldehyde, which is then assimilated through C1 metabolism [18,19,20]. While calcium-dependent MDH encoded by mxaFI has been well-investigated, a novel MDH (i.e., lanthanide-dependent MDH encoded by xoxF) has also been discovered [21]. The role of xoxF in methane and methanol oxidation and the divergence and wide distribution of these enzymes among bacterial taxa demonstrated the importance of xoxF enzymes environmentally, and furthermore, xoxF may be ancestral to mxaF.
There are two types of methanotrophs—γ-proteobacteria (Type I) and α-proteobacteria (Type II). The distinction between these groups is based on the process of assimilating formaldehyde, the organization and arrangement of cell membranes, and related characteristics such as cell morphology [22,23]. Typically, type I methanotrophs assimilate formaldehyde through the RuMP pathway (including Type X Methylocaldum and Methylococcus). They also form a flat, disc-shaped inner cytoplasm and have a TCA cycle lacking α-ketoglurarate dehydrogenase activity [24]. However, there have been many novel findings related to the methane metabolism of this group that have been reported recently. For example, 13C carbon tracings were used to discover a complete oxidative TCA cycle operating during methanotrophic growth in Methylomicrobium buryatense 5GB1 [25]. Others demonstrated that the Embden Meyerhof pathway (EMP) is the main route for C1 assimilation in Methylomicrobium alcaliphilum 20Z and Methylomonas sp. LW13 [26,27].
The assimilation of formaldehyde in type II methanotroph is through the serine pathway. First, formaldehyde is oxidized to formate, subsequently incorporating into biomass via tetrahydrofolate-linked pathways and the serine cycle. The serine cycle first converts formaldehyde and CO2 into C3 and C4 compounds, converts them to acetyl-CoA, and sends them to the TCA cycle (carbon dioxide accounts for about 50% of the total carbon) [18]. It has been hypothesized that an efficient pathway for C1 assimilation requires the coupling of the serine cycle with a glyoxylate regeneration pathway such as a glyoxylate shunt or an ethylmalonyl-coA (EMC) pathway. A flux analysis based on 13C metabolomics technology was used to identify that the EMC pathway is essential for glyoxylate regeneration in M. trichosporium OB3b [28]. Unlike type I methanotrophs, Type II methanotrophs possess α-ketoglurarate dehydrogenase activity, but the TCA cycle does not work perfectly because of its low activity. Type II methanotrophs also forms inner cytoplasm, which is similar to cytoplasm, except in Methylocella [22,23].
3. Genetic Tool Development for Methanotrophs
Genetic tool development is basic requiment for enabling the potential of industrial methane biocatalysis. To date, many attempts for the development of genetic tools for methantrophs have been conducted [8,29]. The introduction of replicable plasmids containing RK2/RP4 origin into methanotrophs via conjugation using Escherichia coli S17-1 as a donor strain was the most common method for foreign gene transfer and expression in methanotrophs [8,17,18,29]. In addition, integration and deletion vectors were also used in wide range of methanotrophs by marker exchange via homologous recombination methods [8,28]. Along with this, counter-selectable marker-based techniques for mutagenesis was developed for methanotrophs in which sacB or pheS gene were employed as the most common markers [9,30].
Electroporation-based genetic manipulation methods have been developed in both type I and type II methanotrophs [9,31]. Recently, one research group from the National Renewable Energy Laboratory has successfully developed a dual-plasmid, broad-host-range CRISPR/Cas9 gene-editing system for Methylococcus capsulatus (Bath) [32]. The DNA targeting and double-strand DNA cleavage by the deployment of heterologous Cas9 endonuclease and synthetic single-guide RNA were applied to the manipulation of cell vitality, fluorescent protein conversion, and protein function disruption by point mutation. This is a proof-of-concept study of novel genetic toolbox development for methanotrophic genetic manipulation. The advances in genetic toolbox development will provide efficient methods to study metabolic features of secondary metabolites in methanotrophs and enable genetic editing for methanotrophs for production of high value-added secondary metabolites.
4. Isoprenoid Production in Methanotrophs
4.1. Isoprenoid Biosynthesis Via MEP Pathway
The basic isoprenoid building blocks isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) are synthesized through one of two distinct biosynthetic routes—the mevalonic acid (MVA) pathway or the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. Over the last few decades, the MVA pathway was considered to be the unique pathway for IPP and DMAPP biosynthesis [33]. However, in the early 1990s, an alternative pathway named the MEP pathway was discovered in bacteria for the biosynthesis of IPP and DMAPP (Figure 1). While archaea and most eukaryotes use the MVA pathway for IPP and DMAPP synthesis, the bacteria (including key pathogens such as all Gram-negative bacteria and mycobacteria) use the MEP pathway [34].
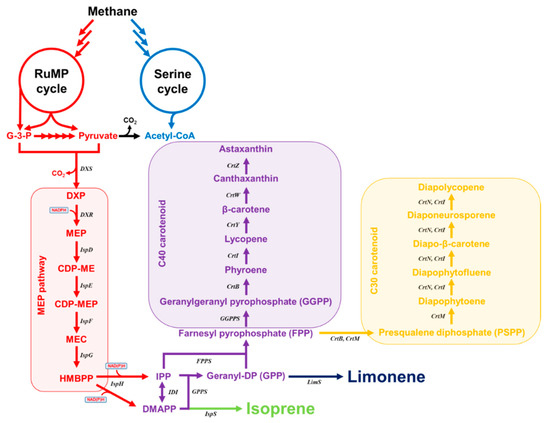
Figure 1.
Biosynthetic pathway of isoprenoids in methanotrophs.
The MEP pathway consists of seven enzymatic steps which initiate with the condensation of pyruvate and d-glyceraldehyde 3-phosphate (G3P) to form 1-deoxy-d-xylulose 5-phosphate (DXP) by 1-deoxy-d-xylulose-5-phosphate synthase (DXS). Next, DXP is converted to MEP by 1-deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) and is subsequently converted to methylerythritol 2,4-cyclodiphosphate (MEcDP) through three consecutive enzymatic steps, cytidylation (CTP dependent), phosphorylation (ATP-dependent), and cyclization encoded by ispD, ispE, and ispF. In the next step, MEcDP is converted to hydroxymethylbutenyl diphosphate (HMBDP) catalyzed by HMBDP synthase (HDS) encoded by the ispG gene. In the final step, HMDBP is reduced to IPP and DMAPP by the action of the ispH gene product, HMBDP reductase (HDR). IPP and DMAPP are isomerized by isopentenyl diphosphate isomerase (IDI) [35]. The regulatory network of the MEP pathway consists of different regulatory mechanisms involving intermediate metabolites and enzymes. Methylerythritol cyclodiphosphate (MEcDP), the fifth intermediate of this pathway, was recently reported as a key metabolite and may play a critical role in MEP pathway regulation. MEcDP may connect the MEP pathway with other cellular metabolism in making precursors for isoprenoids [35]. An efficient fermentation-based process for isoprenoid production has been achieved in engineered E. coli by the heterologous expression of the MVA pathway or upregulation of the native MEP pathway. Among them, the expression of the heterologous MVA pathway is the superior route for enhancing isoprenoid production compared to the MEP pathway. However, the MEP pathway possesses many advantages for isoprenoid production, including higher theoretical yield and better balance than that of the MVA pathway [36]. Specifically, the theoretical maximum IPP yield on glucose via the MEP pathway is 0.83 C-mol IPP/C-mol glucose and is higher than that via the MVA pathway, which is 0.56 C-mol IPP/C-mol glucose IPP [37].
Methanotrophs currently play an important role in a wide range of biotechnological applications with their ability to utilize single carbon (C1) feedstock, i.e., methane and methanol, to produce various high-value compounds. Our genome analysis indicated the existence of a set of genes encoding the methylerythritol 4-phosphate (MEP) pathway in both type I and type II methanotrophs [24] (Table 1). Interestingly, many strains belong to type I methanotrophs such as Methylomonas sp. DH-1, M. alcaliphilum 20Z, and M. buryatense 5GB1, and possess two genes encoding DXS, which catalyzes the first step of the MEP pathway. On the other hand, some strains belonging to type II methanotrophs such as M. trichosporium OB3b consisted of two copies of DXR, which converted DXP to MEP in the second step of MEP pathway (Table 1). In contrast to E. coli and Bacillus subtilis, IDI, which isomerized IPP and DMAPP in the last step of MEP, did not exist in either type I or type II methanotrophs. These results suggest that the regulatory mechanism of the MEP pathway in methanotrophs might be different compared to other microbes, which need to be elucidated in further studies. In agreement with this suggestion, our comparative transcriptome analysis of Methylomonas sp. DH-1 showed the extreme upregulation of MEP pathway encoding genes when shifting substrates from methane to methanol [38]. In addition, the pools of pyruvate and G3P, precursors for the MEP pathway, were approximately 150 and 30-fold higher than that of the acetyl-CoA pool, suggesting that the MEP pathway in methanotrophs is a suitable route to produce isoprenoid-related products [39].

Table 1.
The existence of genes encoding the methylerythritol 4-phosphate (MEP) pathway in methanotrophs and other microbes.
4.2. C5: Isoprene
Isoprene, a volatile 5-carbon hydrocarbon, is an important platform chemical for the production of polyisoprene (for tires and the rubber industry), elastomers (for sporting goods), and isoprenoids (for adhesives and pharmaceuticals) [40,41]. Isoprene is produced by direct separation from the oil cracking fraction and dehydration of C5 isoalkanes and isoalkenes. However, seven gallons of crude oil are needed to make one gallon of isoprene. In other words, demand is higher than supply, and studies are being conducted to biologically synthesize isoprene to solve this problem. Biologically, isoprene is produced by various organisms such as microorganisms, plants, and animals through the MVA and MEP pathways [42]. Currently, algae and cyanobacteria are used as biomass for biological isoprene production. These have the advantage of being renewable resources, but more than half of the mass is composed of oxygen, which results in low conversion efficiency. This is because the oxygen content of the isoprene is lower than the oxygen content of the cellulosic biomass, so oxygen must be removed as waste [43]. Therefore, it is still not suitable for commercial production.
To solve this problem, studies are being conducted to synthesize isoprene from methane gas through methanotrophs. Methanotrophs have genes encoding the enzymatic components of the MEP pathway. Therefore, if methanotrophs have an isoprene synthase, it can synthesize isoprene from both methane and methanol (Figure 1). Recently, recombinant technology of methanotrophs has been developed, and the isoprene synthase (ispS) derived from sweet potato was expressed in M. alcaliphilum 20Z to synthesize 50 μg/mL of isoprene from methanol (Table 2) [44].

Table 2.
Production processes and productivity of secondary metabolites by methanotrophs.
4.3. C10: Limonene
Among the components contained in the epidermis of citrus fruits such as mandarins and oranges, limonene is an essential component of plants. Its chemical formula is C10H16, and it is naturally produced from monoterpene. (S)-limonene is an (S)-menthol precursor, which is the main component of mint. (R)-limonene is widely used in antimicrobials, deodorants, food, pharmaceuticals, and cosmetics and has recently been used as an alternative detergent. In recent studies, the efficacy of (R)-limonene’s anticancer properties has been determined, and research has been actively conducted in the medical field [51]. This limonene is produced by modifying the precursor geranyl pyrophosphate (GPP) by limonene synthase to form aromatic rings. Therefore, methanotrophs with the MEP pathway, which is the pathway for producing GPP, also have the potential to produce limonene (Figure 1). The heterologous expression of Mentha spicata limonene synthase in Methylomonas sp. 16a resulted in limonene production with a titer of 0.5 ppm (Table 2) [45].
4.4. C30 and C40: Carotenoids
Carotenoids are isoprenoid compounds that are often used as feed or food additives because of their unique color. In addition, since they have a long double bond chain and a ketone group or a hydroxyl group, they have strong antioxidative effects [52,53]. Currently, most carotenoids are produced through chemical synthesis, but there is a growing interest in natural synthesis as well as in the use of medicines and health foods.
Carotenoids are classified into C30 and C40 carotenoids depending on the number of carbon atoms. The most commonly known C40 carotenoids are lycopene, β-carotene, canthaxanthin, and astaxanthin. Many studies have been conducted on ways to biosynthesize these compounds. Studies have also been carried out to biosynthesize C40 carotenoids in methanotrophs (Figure 1). Farnesyl pyrophosphate, which is a precursor of carotenoids, was prepared through the MEP pathway of methanotrophs, and the gene of the subsequent process was expressed. Rick et al. synthesized canthaxanthin, adonixanthin, and astaxanthin by expressing crtE, crtB, crtI, crtY, crtW, and crtZ genes in Methylomonas sp. 16a. The content of astaxanthin was increased by controlling the number of copies of crtW and crtZ genes to synthesize up to 2.4 mg/g DCW (Table 2) [46,47]. Additionally, lycopene was naturally produced in Methylomonas sp. ZR1, as confirmed by HPLC-DAD and HPLC-MS/MS analysis methods [54].
C30 carotenoids are less studied than C40 carotenoids. However, it is known that C30 carotenoids such as diapolycopene, diapolycopen-dial, and diaponeurosporene have higher antioxidative effects than tocopherol [55]. Therefore, C30 carotenoids can sufficiently replace C40 carotenoids. C30 carotenoid biosynthesis related genes crtN, ald, and crtNb are contained in Methylomonas sp. [24]. It was confirmed that C30 carotenoid was synthesized via mutagenesis analysis. A colorless mutant was isolated through deletion of a promoter upstream of the C30 encoding gene cluster [46]. Methanotrophs can naturally synthesize C30 carotenoids so that high productivity could be achieved by applying the recent studies on metabolic engineering and systems biology of methanotrophs.
4.5. Advances in Engineering Central Carbon Metabolism to Improve Isoprenoid Production in Methanotrophs
As noted above, the MEP pathway serves as a good candidate to enhance isoprenoid production in methanotrophs by metabolic engineering. The MEP pathway uses glyceraldehyde 3-phosphate and pyruvate in the same molar ratio. However, the EMP pathway that is the main route for C1 assimilation in type I methanotrophs with 75% pyruvate produces an imbalanced ratio of G3P and pyruvate, which is unfavorable for the efficiency of the MEP pathway. Engineering the feeding module of these precursors via the Entner-Doudoroff (ED) pathway suggests the possibility to achieve the balancing of precursor pool, which subsequently improves the isoprenoid titer. The isoprene titer was increased 3.1-fold compared to the strain that used the EMP pathway only by overexpressing the ED and the pentose phosphate pathways [56]. Reconstruction of the MVA pathway in type I methanotrophs is another promising strategy for isoprenoid production if the intracellular concentration of acetyl-CoA (which is a critical branch-point metabolite in central metabolism) can be enhanced. An effective approach for metabolic engineering to improve acetyl-coA pools in methanotrophs could increase carbon flux through the phosphoketolase (PKT) pathway (Figure 2). The overexpression of this pathway in type I methanotrophs has been successfully used to increase the yield of acetyl-CoA. High carbon flux through the PKT pathway was achieved by the overexpression of two PKT isoforms in M. buryatense; among them, ptkB overexpression showed a two-fold increase in intracellular acetyl-CoA [57]. Therefore, the integration of PKT and MVA pathways in type I methanotrophs could improve isoprenoid production in type I methanotrophs. Furthermore, since glycogen and extracellular polymeric substance are the main carbon sink in Group I strains, the elimination of these processes could also increase the pool of secondary metabolites.
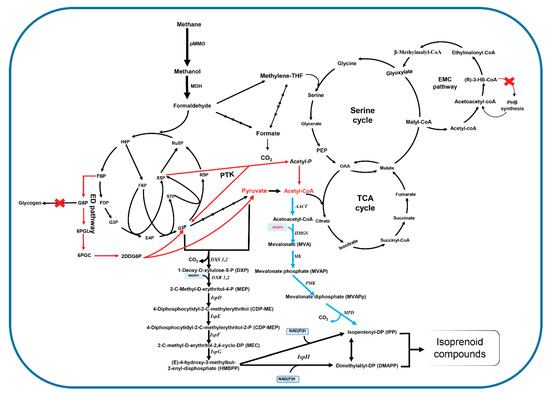
Figure 2.
Central metabolism in methanotrophs and proposed engineering strategies for isoprenoid production. Red arrows: predicted overexpression targets to improve carbon conversion efficiency for production of isoprenoid via MEP and mevalonic acid (MVA) pathway; red X: suggested knockout targets for improving flux into isoprenoids; blue arrows: heterologous expression of MVA pathway in methanotrophs.
The serine cycle is the main route for the C1 assimilation pathway in type II methanotrophs, in which formaldehyde was converted to formate and further incorporated into the serine cycle to produce acetyl-CoA. It should be noted that the production of acetyl-CoA from the serine cycle enhances the overall carbon conversion efficiency compared to RuMP-based C1 assimilation without loss of CO2 [17]. Type II methanotrophs possess higher flux to acetyl-coA production than type I methanotrophs and could be increased by eliminating polyhydroxybutyrate accumulation. Therefore, isoprenoids could also be produced by using both the heterologous expression of the MVA pathway and the endogenous MEP pathway. During initiation of isoprenoid biosynthesis via the MVA pathway, two molecules of acetyl-CoA are condensed to form acetoacetyl-CoA by an acetoacetyl-CoA thiolase. However, the heterologous expression of this enzyme has often been difficult to express in non-native hosts. Interestingly, this enzyme is also already presented in the primary metabolism of type II methanotrophs via the EMC pathway. This feature could be an advantage for engineering isoprenoid biosynthesis in type II methanotrophs (Figure 2). By applying this approach, reconstruction of the MVA pathway in a model methylotroph, Methylobacterium extorquens AM1, resulted in humulene concentrations of up to 1.65 g/L from methanol [58].
5. Non-Isoprenoid Secondary Metabolite Production from Methane Using Methanotrophs
5.1. Vitamin B2 (Riboflavin)
Riboflavin, a water-soluble vitamin, is biosynthesized by various microorganisms and most plants. However, it is not biosynthesized in vertebrates, including humans, and should be replenished by external food. It is a precursor to flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), the coenzyme necessary for the oxidation-reduction in all organisms and an essential nutrient for humans [59]. Riboflavin is produced in an annual capacity of 9000 tons and can be produced by chemical synthesis and biological synthesis. The chemical synthesis method is accomplished through a complicated process from glucose via ribose, rivamine, etc. Also, the production cost is high since the ribose is used as a precursor. Biological synthesis methods have been developed to replace the chemical method. The biological synthesis method involving the isolation of the microorganisms that naturally produce riboflavin is promising. There is also a method of producing riboflavin that involves improving the microorganism by a genetic engineering method or a chemical-physical method. The microorganisms that produce riboflavin include yeasts belonging to the genus Saccharomyces sp. and Candida sp., bacteria belonging to the genus Clostridium, Bacillus, and Corynebacterium, and fungi belonging to the genus Eremothecium and Ashbya [59,60].
Methanotrophs also biosynthesize riboflavin and release it in the form of riboflavin and flavin mononucleotide (FMN) (Figure 3). Both Type I and II methanotrophs can release the flavin to the medium. Methylocystis sp. strain M releases the flavin to the medium at 0.05 μM in the exponential phase, 0.056 μM in the late exponential phase, and 0.102 μM in the stationary phase. In the case of iron deficiency, up to 0.145 μM of flavin is released into the medium (Table 2). It is predicted that the flavin mononucleotide (FMN) will chelate the iron and act as an iron shuttle for cell use and respiration [48].
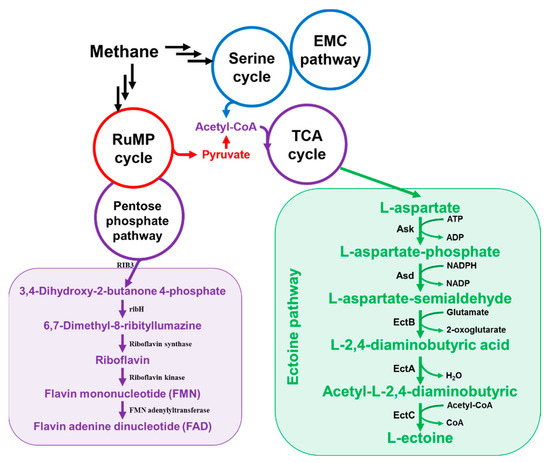
Figure 3.
Biosynthetic pathway of riboflavin and ectoine in methanotrophs.
5.2. Ectoine
Ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidine carboxylic acid) is a substance produced by microorganisms to withstand osmotic stress in a high salt environment. It prevents water loss, regulates cell collapse, and does not interfere with cell metabolism. Ectoine can be used for cosmetic and medical purposes. At present, ectoine has a demand of 15,000 tonnes per year, the market price is $1000 USD per kg, and it is commercially produced using Halomonas elongate [61].
The ectoine biosynthetic pathways have been investigated in a variety of halophilic bacteria. First, the aspartate commonly used in the synthesis of lysine, threonine and methionine is phosphorylated by aspartokinase (ask) to synthesize aspartyl phosphate. It is then converted to aspartate semialdehyde by aspartate semialdehyde dehydrogenase, which is used as a precursor to the ectoine synthesis pathway. Ectoine is synthesized by three enzymes, diaminobutyric acid transaminase (EctB), diaminobutyric acid acetyltransferase (EctA), and ectoine synthase (EctC). The synthesis of ectoine starts with conversion of aspartate semialdehyde to l-2,4-diaminobutyric acid (DABA) by EctB. l-2,4-diaminobutyric acid (DABA) is converted to acetyl-l-2,4-diaminobutyric acid by EctA. Acetyl-l-2,4-diaminobutyric acid is synthesized as ectoine by EctC. The genes coding this synthetic pathway have been studied in several halophilic bacteria and have been named ectABC, which is an ectoine synthesis cluster [62].
Studies on the synthesis route of ectoine were also conducted for methanotrophs. M. alcaliphilum 20Z, which is known to be a halophilic methanotroph, has an ectoine synthesis pathway. This pathway includes ask, l-2,4-diaminobutyric acid transaminase (ectB), L-2,4-diaminobutyric acid acetyltransferase (ectA) and l-ectoine synthase (ectC) (Figure 3), and it exists as one operon. Studies have been conducted to find factors that regulate the ectoine biosynthesis of M. alcaliphilum 20Z. As a result, the promoters ectAp1 and ectAp2 of the ectABAask operon were identified. This is similar to the σ70-dependent promoters of E. coli. In addition, a MarR-like transcriptional regulator, EctR1, was identified above the gene cluster, which acts as a negative regulator of the ectABCask operon. This confirmed that the purified EctR1 specifically binds to the ectAp1 promoter [63,64].
Studies on the production of ectoine through M. alcaliphilum 20Z are also underway. According to Cantera et al., the production of ectoine by M. alcaliphilum 20Z in stirred tank reactors (non-sterile conditions) varies depending on the NaCl concentration and the stirring rates [49]. When the concentration of NaCl in the medium increased from 3% to 6%, the intracellular ectoine content increased to 37.4 mg/g cell from 16.5 mg/g cell (Table 2). A high agitation rate stresses the cells and lowers the yield of ectoine. In addition, when the concentration of copper ion in the medium was increased from 0.05 mM to 25 mM, the amount of ectoine released from the cell increased by about 20%, although it did not affect the production of the intracellular ectoine [49].
A bio-milking process for the commercial production of ectoine was used to increase the production of ectoine from M. alcaliphilum 20Z. This is a method of continuously producing ectoine by sequentially in the culture medium. M. alcaliphilum 20Z was cultured in 6% NaCl and then alternately cultured in 0% controlling the NaCl concentration NaCl. As a result, it was observed that M. alcaliphilum 20Z released the accumulated ectoine at a low osmotic pressure and absorbed the released ectoine at high osmotic pressure. It was confirmed that the ectoine concentration in the cell was maintained at 70.4 mg/g cell in 6% NaCl and that 70% of the ectoine was released in 0% NaCl (Table 2). This shows that it is possible to continuously produce ectoine from methane using methanotrophs [50].
6. Future Aspects for Secondary Metabolite Production from Methane Using Methanotrophs
Methanotrophs might have advantages for secondary metabolite production, especially isoprenoid production, because the tolerance and the storage capacity are major limiting steps for isoprenoid production in other microbes. That is, one of the potentials of methanotrophs is the ability to accumulate higher amounts of isoprenoid on the basis of cell mass because methanotrophs need isoprenoids as a modulator of the oxidative stress or reactive oxygen species generated from oxygen split for C–H bond activation during methane oxidation. In addition, the carbon conversion efficiency (CCE) and yield of the secondary metabolites produced from methane are much higher than those of other substrates, highlighting the potential of secondary metabolite production in methanotrophs. For example, in comparison the theoretical yield for E. coli vs. methanotroph–based catalysts for isoprene production, the theoretical yield in methanotrophs is 71% (w/w) and is much higher than that of E. coli, which has a theoretical yield of 38% [18].
However, some critical issues need to be overcome to improve the productivity of both secondary metabolites and other target compounds. First, transferring metabolic engineering strategies of E. coli and yeast for increased isoprenoid production can be successfully applied to methanotrophs, although productivity enhancement of isoprenoids in methanotrophs is still limited by an incomplete understanding of flux control in the isoprenoid biosynthetic pathway. Isoprenoid production can be increased by optimal gene combination and dosage in methanotrophs. The DXS, DXR, and HDR are expected to act as the major flux control in MEP pathway, thus overexpression of these genes could be possible solution to increase isoprenoids productivity in methanotrophs.
For an efficient production of isoprenoids using E. coli, MVA pathway has been reconstructed. In addition, in order to circumvent energetically expensive MVA pathway consuming three ATP for the biosynthesis of an IPP molecule, the archaeal MVA pathway has been reconstructed in E. coli to save one mole ATP. However, MVA pathway reconstruction in methanotrophs might be not a good approach because the cellular amount of acetyl-CoA in methanotrophs is very low compared to pyruvate and GAP. Thus, metabolic engineering of the natural MEP pathway might be right option for constructing methanotrophic cell factory for isoprenoid production.
To provide potential metabolic engineering strategies to improve productivity of isoprenoids and other secondary metabolites in methanotrophs, a more deeper understanding on relevant secondary metabolism and biosynthetic regulatory network in methanotrophs is absolutely needed. For example, multi-omics studies together with construction of genome-scale model could be applied to achieve a system-level understanding of cellular behavior for accumulation of isoprenoids and other secondary metabolites, including specific and global gene expression regulation and control of their carbon flux. In particular, a systems biology–based understanding of the interrelationship between methane oxidation process and isoprenoids biosynthesis will be one of the key issues on isoprenoids productivity enhancement.
M. alcaliphilum 20Z strain, one of the platform strains used in methane bioconversion, is capable of growing under high salt conditions of up to 8.7%. Thus, M. alcaliphilum 20Z is expected to regulate the synthesis of relevant secondary metabolite, for example, ectoine, with high salt-induced stress. Thus, in-depth understanding of stress-regulatory network can play a key role in the isoprenoid production from methane using methanotrophs. Of course, the first step for this approach will be identification of appropriate stress factor.
The biosynthetic pathways and characteristics of isoprenoids in methanotrophs have not yet fully discovered. This means there might be novel genes, enzymes, and biosynthetic route in methanotrophs that can be used for the generation of new classes of isoprenoids. However, the amounts of secondary metabolites are low—not enough to be isolated efficiently for structure analysis and function characterization. Therefore, the development of precursor-overproducing platform methanotrophic strain—for example, MEP-related intermediates-overproducing strains—can be a promising approach for the production of novel isoprenoids that is too low to be isolated and characterized.
Funding
This research was supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2015M3D3A1A01064882).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hwang, I.Y.; Hur, D.H.; Lee, J.H.; Park, C.; Chang, I.S.; Lee, J.W.; Lee, E.Y. Batch conversion of methane to methanol using Methylosinus trichosporium OB3b as biocatalyst. J. Microbiol. Biotechnol 2015, 25, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, T.; Tauseef, S.; Abbasi, S. Anaerobic digestion for global warming control and energy generation-An overview. Renew. Sustain. Energy Rev. 2012, 16, 3228–3242. [Google Scholar] [CrossRef]
- Kirschke, S.; Bousquet, P.; Ciais, P.; Saunois, M.; Canadell, J.G.; Dlugokencky, E.J.; Bergamaschi, P.; Bergmann, D.; Blake, D.R.; Bruhwiler, L. Three decades of global methane sources and sinks. Nat. Geosci. 2013, 6, 813. [Google Scholar] [CrossRef]
- Nwaoha, C.; Wood, D.A. A review of the utilization and monetization of Nigeria’s natural gas resources: Current realities. J. Nat. Gas Sci. Eng. 2014, 18, 412–432. [Google Scholar] [CrossRef]
- Conrado, R.J.; Gonzalez, R. Chemistry. Envisioning the bioconversion of methane to liquid fuels. Science 2014, 343, 621–623. [Google Scholar] [CrossRef]
- Haynes, C.A.; Gonzalez, R. Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 2014, 10, 331. [Google Scholar] [CrossRef]
- Han, J.; Ahn, C.; Mahanty, B.; Kim, C. Partial oxidative conversion of methane to methanol through selective inhibition of methanol dehydrogenase in methanotrophic consortium from landfill cover soil. Appl. Biochem. Biotechnol. 2013, 171, 1487–1499. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Nguyen, A.D.; Nguyen, T.T.; Nguyen, L.T.; Lee, O.K.; Lee, E.Y. Biological conversion of methane to chemicals and fuels: Technical challenges and issues. Appl. Microbiol. Biotechnol. 2018, 102, 3071–3080. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Hwang, I.Y.; Lee, O.K.; Kim, D.; Kalyuzhnaya, M.G.; Mariyana, R.; Hadiyati, S.; Kim, M.S.; Lee, E.Y. Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2, 3-butanediol production from methane. Metab. Eng. 2018, 47, 323–333. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Rozova, N.; But, C.Y.; Mustakhimov, I.I.; Reshetnikov, A.S.; Beschastnyi, A.P.; Trotsenko, Y.A. Biosynthesis of secondary metabolites in methanotrophs: biochemical and genetic aspects (review). Appl. Biochem. Microbiol. 2015, 51, 140–150. [Google Scholar] [CrossRef]
- Fei, Q.; Guarnieri, M.T.; Tao, L.; Laurens, L.M.; Dowe, N.; Pienkos, P.T. Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol Adv 2014, 32, 596–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zada, B.; Wei, G.; Kim, S.W. Metabolic engineering and synthetic biology approaches driving isoprenoid production in Escherichia coli. Bioresour. Technol. 2017, 241, 430–438. [Google Scholar] [CrossRef]
- Tippmann, S.; Chen, Y.; Siewers, V.; Nielsen, J. From flavors and pharmaceuticals to advanced biofuels: production of isoprenoids in Saccharomyces cerevisiae. Biotechnol. J. 2013, 8, 1435–1444. [Google Scholar] [CrossRef]
- Li, Y.; Pfeifer, B.A. Heterologous production of plant-derived isoprenoid products in microbes and the application of metabolic engineering and synthetic biology. Curr. Opin. Plant Biol. 2014, 19, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Murrell, J.C.; McDonald, I.R.; Bourne, D.G. Molecular methods for the study of methanotroph ecology. FEMS Microbiol. Ecol. 1998, 27, 103–114. [Google Scholar] [CrossRef]
- Akberdin, I.R.; Thompson, M.; Kalyuzhnaya, M.G. Systems Biology and Metabolic Modeling of C1-Metabolism. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Cham, Switzerland, 2018; pp. 99–115. [Google Scholar]
- Kalyuzhnaya, M.G.; Puri, A.W.; Lidstrom, M.E. Metabolic engineering in methanotrophic bacteria. Metab. Eng. 2015, 29, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.K.; Hur, D.H.; Nguyen, D.T.N.; Lee, E.Y. Metabolic engineering of methanotrophs and its application to production of chemicals and biofuels from methane. Biofuel Bioprod. Biorefin. 2016, 10, 848–863. [Google Scholar] [CrossRef]
- Hakemian, A.S.; Rosenzweig, A.C. The biochemistry of methane oxidation. Annu. Rev. Biochem. 2007, 76, 223–241. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar]
- Chistoserdova, L.; Kalyuzhnaya, M.G. Current trends in methylotrophy. Trends Microbiol. 2018, 26, 703–714. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Dunfield, P.F. Facultative and obligate methanotrophs: how to identify and differentiate them. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 495, pp. 31–44. [Google Scholar]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy? Adv. Appl. Microbiol. 2011, 63, 183. [Google Scholar]
- Nguyen, A.D.; Hwang, I.Y.; Lee, O.K.; Hur, D.H.; Jeon, Y.C.; Hadiyati, S.; Kim, M.; Yoon, S.H.; Jeong, H.; Lee, E.Y. Functional analysis of Methylomonas sp. DH-1 genome as a promising biocatalyst for bioconversion of methane to valuable chemicals. Catalysts 2018, 8, 117. [Google Scholar]
- Fu, Y.; Li, Y.; Lidstrom, M. The oxidative TCA cycle operates during methanotrophic growth of the Type I methanotroph Methylomicrobium buryatense 5GB1. Metab. Eng. 2017, 42, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Yang, S.; Rozova, O.N.; Smalley, N.E.; Clubb, J.; Lamb, A.; Gowda, G.N.; Raftery, D.; Fu, Y.; Bringel, F. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 2013, 4, 2785. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.D.; Park, J.Y.; Hwang, I.Y.; Hamilton, R.; Kalyuzhnaya, M.G.; Kim, D.; Lee, E.Y. Genome-scale evaluation of core one-carbon metabolism in gammaproteobacterial methanotrophs grown on methane and methanol. Metab. Eng. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhanaya, M.G.; Yang, S.; Matsen, J.B.; Konopka, M.; Green-Saxena, A.; Clubb, J.; Sadilek, M.; Orphan, V.J.; Beck, D. Global molecular analyses of methane metabolism in methanotrophic alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. Metabolomics and 13C-labeling study. Front. Microbiol. 2013, 4, 70. [Google Scholar]
- Lee, O.K.; Nguyen, D.T.; Lee, E.Y. Metabolic Engineering of Methanotrophs for the Production of Chemicals and Fuels. In Methanotrophs; Springer: Cham, Switzerland, 2019; pp. 163–203. [Google Scholar]
- Ishikawa, M.; Yokoe, S.; Kato, S.; Hori, K. Efficient counterselection for Methylococcus capsulatus (Bath) by using a mutated pheS gene. Appl. Environ. Microbiol. 2018, 84, 1875. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.Y.; Rosenzweig, A.C. Recent advances in the genetic manipulation of Methylosinus trichosporium OB3b. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 605, pp. 335–349. [Google Scholar]
- Tapscott, T.; Guarnieri, M.T.; Henard, C.A. Development of a CRISPR/Cas9 system for Methylococcus capsulatus in vivo gene editing. Appl Environ Microbiol 2019, 85, 340. [Google Scholar] [CrossRef]
- Zhao, L.; Chang, W.; Xiao, Y.; Liu, H.; Liu, P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu. Rev. Biochem. 2013, 82, 497–530. [Google Scholar] [CrossRef]
- Odom, A.R. Five questions about non-mevalonate isoprenoid biosynthesis. PLoS Pathog. 2011, 7, e1002323. [Google Scholar] [CrossRef]
- Banerjee, A.; Sharkey, T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 2014, 31, 1043–1055. [Google Scholar] [CrossRef]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef]
- Niu, F.; Lu, Q.; Bu, Y.; Liu, J. Metabolic engineering for the microbial production of isoprenoids: Carotenoids and isoprenoid-based biofuels. Synth. Syst. Biotechnol. 2017, 2, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.D.; Kim, D.; Lee, E.Y. A comparative transcriptome analysis of the novel obligate methanotroph Methylomonas sp. DH-1 reveals key differences in transcriptional responses in C1 and secondary metabolite pathways during growth on methane and methanol. BMC Genomics 2019, 20, 130. [Google Scholar]
- Akberdin, I.R.; Thompson, M.; Hamilton, R.; Desai, N.; Alexander, D.; Henard, C.A.; Guarnieri, M.T.; Kalyuzhnaya, M.G. Methane utilization in Methylomicrobium alcaliphilum 20Z R: A systems approach. Sci. Rep. 2018, 8, 2512. [Google Scholar] [CrossRef] [PubMed]
- Kuzuyama, T. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 2002, 66, 1619–1627. [Google Scholar] [CrossRef]
- Clement, N.D.; Routaboul, L.; Grotevendt, A.; Jackstell, R.; Beller, M. Development of palladium–carbene catalysts for telomerization and dimerization of 1, 3-dienes: From basic research to industrial applications. Chem. Eur. J. 2008, 14, 7408–7420. [Google Scholar]
- Julsing, M.K.; Rijpkema, M.; Woerdenbag, H.J.; Quax, W.J.; Kayser, O. Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2007, 75, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Whited, G.M.; Feher, F.J.; Benko, D.A.; Cervin, M.A.; Chotani, G.K.; McAuliffe, J.C.; LaDuca, R.J.; Ben-Shoshan, E.; Sanford, K.J. Technology update: Development of a gas-phase bioprocess for isoprene-monomer production using metabolic pathway engineering. Ind. Biotechnol. 2010, 6, 152–163. [Google Scholar] [CrossRef]
- Song, J.; Cho, K.K.; Lee, K.S.; La, Y.H.; Kalyuzhnaya, M. Method for Producing Isoprene Using Recombinant Halophilic Methanotroph. U.S. Patent US20170211100A1, 27 July 2017. [Google Scholar]
- Dicosimo, D.J.; Koffas, M.; Odom, J.M.; Wang, S. Production of cyclic terpenoids. U.S. Patent US6818424B2, 2004. [Google Scholar]
- Rick, W.Y.; Yao, H.; Stead, K.; Wang, T.; Tao, L.; Cheng, Q.; Sharpe, P.L.; Suh, W.; Nagel, E.; Arcilla, D. Construction of the astaxanthin biosynthetic pathway in a methanotrophic bacterium Methylomonas sp. strain 16a. J. Ind. Microbiol. Biotechnol. 2007, 34, 289. [Google Scholar]
- Sharpe, P.L.; DiCosimo, D.; Bosak, M.D.; Knoke, K.; Tao, L.; Cheng, Q.; Rick, W.Y. Use of transposon promoter-probe vectors in the metabolic engineering of the obligate methanotroph Methylomonas sp. strain 16a for enhanced C40 carotenoid synthesis. Appl. Environ. Microbiol. 2007, 73, 1721–1728. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Levinson, B.T.; Rosenzweig, A.C. Secretion of flavins by three species of methanotrophic bacteria. Appl. Environ. Microbiol. 2010, 76, 7356–7358. [Google Scholar] [CrossRef]
- Cantera, S.; Lebrero, R.; Rodríguez, E.; García-Encina, P.A.; Muñoz, R. Continuous abatement of methane coupled with ectoine production by Methylomicrobium alcaliphilum 20Z in stirred tank reactors: A step further towards greenhouse gas biorefineries. J. Clean. Prod. 2017, 152, 134–141. [Google Scholar] [CrossRef]
- Cantera Ruiz de Pellón, S.; Lebrero Fernández, R.; Rodríguez, S.; García Encina, P.A.; Muñoz Torre, R. Ectoine bio-milking in methanotrophs: A step further towards methane-based bio-refineries into high added-value products. Chem. Eng. J. 2017, 328, 44–48. [Google Scholar]
- Sun, J. D-Limonene: Safety and clinical applications. Altern. Med. Rev. 2007, 12, 259. [Google Scholar] [PubMed]
- Noviendri, D.; Hasrini, R.F.; Octavianti, F. Carotenoids: Sources, medicinal properties and their application in food and nutraceutical industry. J. Med. Plant. Res. 2011, 5, 7119–7131. [Google Scholar]
- Umeno, D.; Tobias, A.V.; Arnold, F.H. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 2005, 69, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Li, D.; He, R.; Wu, M.; Chen, W.; Gao, F.; Zhang, Z.; Yao, Y.; Yu, L.; Chen, S. Synthesizing value-added products from methane by a new Methylomonas. J. Appl. Microbiol. 2017, 123, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, M.S.; Lee, B.Y.; Lee, P.C. Generation of structurally novel short carotenoids and study of their biological activity. Sci. Rep. 2016, 6, 21987. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, Y.; Ramos, K.R.M.; Nisola, G.M.; Valdehuesa, K.N.G.; Lee, W.; Park, S.J.; Chung, W. Combination of Entner-Doudoroff pathway with MEP increases isoprene production in engineered Escherichia coli. PLoS ONE 2013, 8, e83290. [Google Scholar] [CrossRef]
- Henard, C.A.; Smith, H.K.; Guarnieri, M.T. Phosphoketolase overexpression increases biomass and lipid yield from methane in an obligate methanotrophic biocatalyst. Metab. Eng. 2017, 41, 152–158. [Google Scholar] [CrossRef]
- Sonntag, F.; Kroner, C.; Lubuta, P.; Peyraud, R.; Horst, A.; Buchhaupt, M.; Schrader, J. Engineering Methylobacterium extorquens for de novo synthesis of the sesquiterpenoid α-humulene from methanol. Metab. Eng. 2015, 32, 82–94. [Google Scholar] [CrossRef]
- Schwechheimer, S.K.; Park, E.Y.; Revuelta, J.L.; Becker, J.; Wittmann, C. Biotechnology of riboflavin. Appl. Microbiol. Biotechnol. 2016, 100, 2107–2119. [Google Scholar] [CrossRef]
- Kato, T.; Park, E.Y. Riboflavin production by Ashbya gossypii. Biotechnol. Lett. 2012, 34, 611–618. [Google Scholar] [CrossRef]
- Strong, P.; Kalyuzhnaya, M.; Silverman, J.; Clarke, W. A methanotroph-based biorefinery: Potential scenarios for generating multiple products from a single fermentation. Bioresour. Technol. 2016, 215, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, A.U.; Bremer, E. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 2002, 68, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, A.S.; Khmelenina, V.N.; Trotsenko, Y.A. Characterization of the ectoine biosynthesis genes of haloalkalotolerant obligate methanotroph “Methylomicrobium alcaliphilum 20Z”. Arch. Microbiol. 2006, 184, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Mustakhimov, I.I.; Reshetnikov, A.S.; Glukhov, A.S.; Khmelenina, V.N.; Kalyuzhnaya, M.G.; Trotsenko, Y.A. Identification and characterization of EctR1, a new transcriptional regulator of the ectoine biosynthesis genes in the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 2010, 192, 410–417. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).