Abstract
Enzyme catalyzed reactions are complex reactions due to the interplay of the enzyme, the reactants, and the operating conditions. To handle this complexity systematically and make use of a design space without technical restrictions, we apply the model based approach of elementary process functions (EPF) for selecting the best process design for enzyme catalysis problems. As a representative case study, we consider the carboligation of propanal and benzaldehyde catalyzed by benzaldehyde lyase from Pseudomonas fluorescens (PfBAL) to produce (R)-2-hydroxy-1-phenylbutan-1-one, because of the substrate dependent reaction rates and the challenging substrate dependent PfBAL inactivation. The apparatus independent EPF concept optimizes the material fluxes influencing the enzyme catalyzed reaction for the given process intensification scenarios. The final product concentration is improved by 13% with the optimized feeding rates, and the optimization results are verified experimentally. In general, the rigorous model driven approach could lead to selecting the best existing reactor, designing novel reactors for enzyme catalysis, and combining protein engineering and process systems engineering concepts.
1. Introduction
The pharmaceutical industry is considering biocatalytic processes as a possible alternative to chemocatalytic processes [,,]. This is primarily due to the high stereoselectivity and specificity of biocatalytic processes, which lead to efficient production of high-quality active pharmaceutical ingredients (APIs) in fewer synthesis steps [,]. Less complex synthesis is of economic importance to the pharmaceutical industry as it could translate into cost savings and greener pharmaceutical processes []. In recent years, the design of biocatalysts for the efficient synthesis of APIs has witnessed significant advancements [].
To render enzyme processes economically viable, a high product concentration and low enzyme cost must be ensured []. To this end, the advancements in protein engineering [], appropriate reaction engineering concepts [], and process intensification strategies must be combined [,]. A traditional method for performing this optimization is by carrying out numerous experiments in the lab that are cost and time intensive. Therefore, computer aided model based approaches have been proposed to complement laboratory experiments [,]. As examples of the application of model based approaches to optimize enzyme catalyzed reactions, Stillger et al. developed an enzyme-membrane continuous stirred tank reactor (CSTR) for the carboligation of benzaldehyde and acetaldehyde catalyzed by benzaldehyde lyase from Pseudomonas fluorescens (PfBAL) to produce (R)-2-hydroxy-1-phenylpropanone (HPP) []. In their work, they performed simulations by using a kinetic model in combination with reactor models to select the best performing reactor. Parallel to this work, Hildebrand et al. [] investigated the production of HPP by PfBAL in a membrane CSTR using a kinetic model to simulate different reaction engineering strategies to select the best reactor design. In their work, however, PfBA inactivation was observed, but was not included in the kinetic model and, thus, could neither be predicted nor simulated []. Begemann et al. used a model based approach to analyze reactors and develop a control strategy for a two phase biocatalytic oxidoreduction system catalyzed by Candida parapsilosis carbonyl reductase 2 []. Based on simulations of their model, they showed that a fed-batch reactor performed better than a batch reactor for the chosen reaction. Their simulations also showed that controlling the pH could increase conversions and improve productivity. By performing simulations with coupled kinetic models and reactor design equations, Braun et al. minimized the enzyme costs for the biocatalytic production of 12-ketochenodeoxycholic acid in a batch reactor []. Marpani et al. [] utilized a kinetic model and simulation approach to determine the optimal operating conditions for the biocatalytic conversion of formaldehyde to methanol in a batch reactor. Although their work showed excellent agreement between the simulation and experimental results, it involved performing hundreds of simulations [].
Therefore, even simulation based approaches might be time consuming and could lead to sub-optimal results []. Furthermore, there is no guarantee that the design space explored by the simulations would lead to optimal results. Fortunately, model based process optimization methods have been established in process systems engineering, which could lead to (at least locally) guaranteed optimal reactor designs and operating conditions. Furthermore, most existing model based reactor design approaches for enzyme catalysis are based on comparing a limited subset of existing reactor types, which constrains the design space and limits the possibilities of designing novel intensified reactors for enzyme catalysis.
In the last decade, a model based reactor design approach that is based on the concept of elementary process functions (EPF) was proposed by Freund and Sundmacher. The basic premise of the EPF concept is to design reactors based on their essential reaction functions rather than optimizing predefined apparatuses or equipment [] and, therefore, to formulate the reactor design problem as a reaction function optimization problem. To describe and optimize processes with the EPF approach, a fluid element is considered traveling through the thermodynamic state space of a (bio)chemical reaction that is acted upon by internal and external mass and energy fluxes. These fluxes are then designed to drive the fluid element to the desired state in the thermodynamic state space, thus maximizing a particular objective function or performance metric, such as the final product concentration, space-time yield, or total enzyme turnover number. Material and energy balances of the fluid element are set up from first principles to model the fluid element. They are accompanied by detailed reaction kinetics and thermodynamic relations, which explain the key phenomena taking place within and outside the fluid element.
Given that the fluid element changes with time, a dynamic optimization problem is formulated by using the balances, reaction kinetic and thermodynamic equations, and constraints that are inherent to the particular process. The dynamic optimization problem is then solved for different intensification cases. Based on the corresponding simulations, the best intensification strategy is selected and then technically approximated by using an off-the-shelf reactor or by designing a novel reactor to approximate the optimal fluxes obtained []. The EPF approach has been applied extensively to select the best existing reactor or to design novel reactors for the optimal production of bulk chemicals [,,,,,]. Recently, we worked on extending the EPF approach to biotechnological processes [,]. In this paper, we build on our previous work by presenting a clear guideline for applying the EPF approach to enzyme catalysis.
The objective of this paper is to implement and experimentally demonstrate the applicability of the EPF approach for enzyme catalysis problems. As a case study, we chose the PfBAL catalyzed carboligation of propanal and benzaldehyde to form (R)-2-hydroxy-1-phenylbutan-1-one, which is representative of biocatalytic carboligations to optically pure 2-hydroxy ketones, key organic intermediates for producing a vast array of APIs [,,]. The PfBAL catalyzed conversion of benzaldehyde in the presence of propanal can, in theory, yield four different enantiopure compounds []: the two asymmetric products (R)-2-hydroxy-1-phenylbutan-1-one and (R)-1-hydroxy-1-phenylbutan-2-one, as well as the two symmetric products (R)-benzoin and (R)-propioin. However, in the case of PfBAL catalyzed carboligation, the aromatic substrate benzaldehyde is preferred as the donor molecule over the aliphatic aldehyde propanal, thus eliminating two of the possible products []. In Figure 1, we show the corresponding reaction system, adapted from Ohs et al. [].
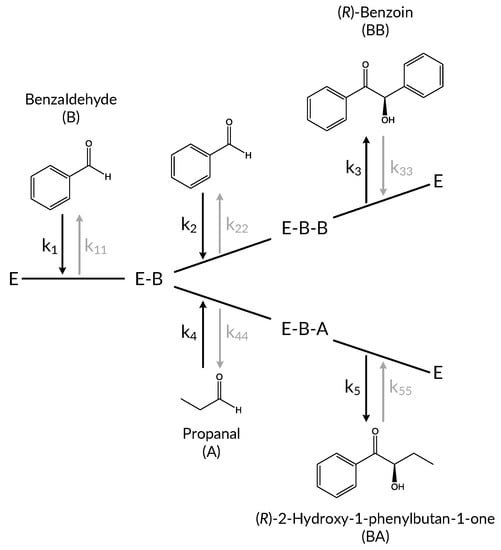
Figure 1.
Branched reaction scheme for benzaldehyde lyase from Pseudomonas fluorescens (PfBAL) catalyzed carboligations with benzaldehyde and propanal as substrates.
The simplified network can be described as three coupled reaction pathways. In the first step, benzaldehyde (B) forms a covalent bond with the ThDP cofactor in the PfBAL (E) active site. Starting from this first intermediate, the pathway branches into two separate reactions depending on the second substrate: self-carboligation occurs if a second benzaldehyde molecule binds to the enzyme, acting as an acceptor molecule. The resulting (R)-benzoin (BB) is regarded as a side product in this study. In the cross-carboligation, propanal (A) acts as the acceptor substrate, leading to the desired product (R)-2-hydroxy-1-phenylbutan-1-one (BA). The self-carboligation and the cross-carboligation are modeled as ordered bi-uni-reaction mechanisms. The third reaction pathway describes the transfer reaction between the two carboligation products and is modeled as a ping-pong-bi-bi mechanism.
Because the reaction rate and PfBAL inactivation depend on the substrate concentration [], the objective is to maximize the final concentration of (R)-2-hydroxy-1-phenylbutan-1-one by tuning the fluxes due to substrate feeding. To our knowledge, this work constitutes the first study that designs and experimentally reproduces an optimal enzyme catalyzed carboligation using the model based EPF approach with a kinetic considering enzyme inactivation.
2. Results and Discussion
Following the EPF strategy, three intensification cases were investigated systematically to ascertain the best process intensification scenario for the maximization of the final concentration of (R)-2-hydroxy-1-phenylbutan-1-one produced from the PfBAL catalyzed carboligation between propanal and benzaldehyde. A batch reactor was selected as the reference case because it is the most common reactor used for enzyme catalyzed reactions. Due to the rapid enzyme inactivation caused by the substrates (propanal and benzaldehyde), it was hypothesized that dosing concepts could be advantageous over the batch reactor resulting in the following case studies.
Case 1: Dosing of propanal. In this case, the propanal dosing flux was incorporated into the material balance and dynamically optimized. In addition to the dosing flux of propanal, the initial volume and the optimal initial concentrations of propanal and benzaldehyde required for the highest possible concentration of (R)-2-hydroxy-1-phenylbutan-1-one were degrees-of-freedom in the optimization.
Case 2: Dosing of benzaldehyde. In a similar fashion to Case 1, the dosing of only benzaldehyde along the reaction coordinate was investigated. The dosing rate of benzaldehyde, the initial volume, and initial concentrations of propanal and benzaldehyde were optimized with the same goal of maximizing the final concentration of the target product.
Case 3: Dosing of propanal and benzaldehyde. In Case 3, all tunable operating conditions were optimized for maximizing the cross-carboligation product concentration, namely the dynamic dosing fluxes of the reactants propanal and benzaldehyde, as well as the initial concentrations of the substrates and the enzyme.
In the subsequent sections, experimental data are presented for the reference case to confirm the validity of the kinetic model derived previously and for the predicted best intensification scenario. However, to carry out the model based optimization, a mathematical model of the underlying reaction phenomena that includes material and energy fluxes is required first. Note that in this study, energy balances are not considered, due to the mild conditions at which the PfBAL catalyzed carboligation is performed and the assumption that these conditions are not energy intensive [,]. Only material balances that follow the so-called Lagrangian formulation [] are considered. The essential terms in these balances are the reaction rate expressions, which are defined by a kinetic model in Section 3.1.2.
2.1. Reference Case: Batch Reactor
The concentration profiles of the optimized batch reactor are shown in Figure 2a-i, including the concentration progress of the respective experimental data. For better readability, concentrations of A and E are displayed on the second y-axis and indicated by arrows, and the concentration of active enzyme in the system has been shortened to “enzyme concentration”.
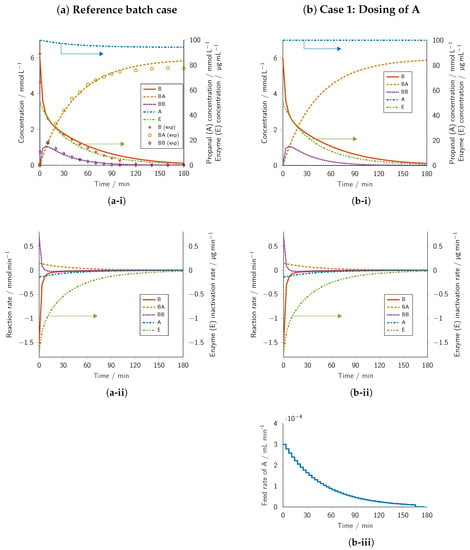
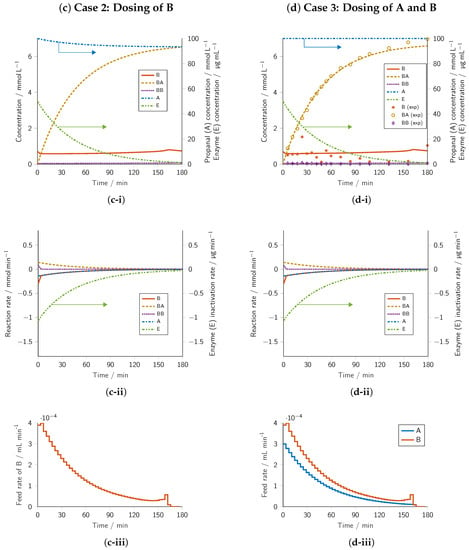
Figure 2.
Results of the dynamic optimization and their experimental verification. Columns: (a) Batch reference, (b) Intensification Case 1 involving the dosing of A, (c) Intensification Case 2 involving the dosing of B, and (d) Intensification Case 3 involving the dosing of A and B. Rows: (i) concentration profiles, (ii) reaction rate profiles, and (iii) volumetric flow rate profiles. Starting concentrations within the design space were optimized so that the concentration of BA at final time point was maximized. Lower concentration bounds were defined as 0 −1 and upper bounds as 100, 149.35, −1, and 50 / for A, B, BB, and E, respectively. Experiments were conducted at 30 in a reaction volume of 30 (70% v/v TEA buffer with cofactors and 30% v/v DMSO) at pH = 8.5. Batch reference: = −1, , Case 3: , .
The optimized initial concentrations for A and E of 100 and 50 /, respectively, are restricted by the upper bounds, whereas that of B of 5.94 is not (Table 1). The optimization result is because the BA product formation rate is proportional to E and approximately proportional to A and B (Equations (14)–(16)). Note that the enzyme is inactivated much more strongly by benzaldehyde than by propanal, corresponding to the 15 fold higher inactivation rate parameters (Table 2). Due to the increased reaction rate towards BA and the lower inactivation rate parameter of A, a large excess concentration of the aliphatic compound compared to benzaldehyde seemed beneficial for the investigated objective function. The simulated final concentration of the active enzyme reached /, indicating nearly full enzyme inactivation of % over the 180 reaction time. The simulated optimal final BA concentration of 5.83 together with the final propanal and benzaldehyde concentrations of 94.17 and 0.11 , respectively, showed that propanal and benzaldehyde were converted quantitatively, with a conversion of over 98% for the limiting substrate benzaldehyde and eliminating benzoin byproduct formation at the reaction endpoint after 180 .

Table 1.
Summary of the optimization results: initial concentrations , final concentrations , solution time for the reference case and the three process intensification scenarios, as well as the residual sum of squares (RSS) and mean squared error (MSE) for the experimental validations.

Table 2.
Kinetic parameters for the PfBAL kinetic model.
The initial decrease rate of B was higher than that of A for the first 7 of the reaction, which coincided with the rapid formation of byproduct benzoin (BB) (Figure 2a-ii). These initial rates could be attributed to the higher affinity of the enzyme at the acceptor position to benzaldehyde over propanal []. The byproduct benzoin (BB) concentration peak indicated that the net BB formation rate equaled zero and may be explained by the enzyme specificity in the donor position, which was highest for BB (PfBAL has been discovered as lyase []) followed by the aromatic benzaldehyde (B). In contrast, the aliphatic propanal was practically not accepted as the substrate in the presence of aromatic compounds. The rate of AB formation exactly mirrored that of A consumption. The experimental data successfully confirmed the predicted model optimum, although minor discrepancies could be observed; see Figure 2a-i. The experimentally determined initial concentrations of B amounted to 6.23 instead of the optimal value of 5.94 . The deviation in the initial concentrations, in turn, led to an initially higher concentration of the byproduct BB during the first 30 of the reaction time. Up to around 60 of the reaction time, the measured concentrations of substrate B and product BA were nearly superimposable with the simulation results, but dropped below the predicted values afterward. In addition, the mass balance with respect to B represented in the sum of B, BA, and two times BB was not closed over the reaction period, but instead declined slightly due to evaporation to finally reach 5.47 or a loss of 11%. The same evaporation loss also applied to Compound A, which, however, was assumed to have a negligible impact on the progress curves due to its high stoichiometric excess. However, the conversion of B to BA after the full reaction time matched the simulation. It is important to note the excellent quality of the optimization prediction to the experimental reproducibility run, in particular given that the kinetic model was obtained using a separate set of experimental data [] and partially obtained in a different lab using different instrumentation.
In summary, optimizing the batch case for a maximum concentration of BA led to nearly full conversion of B with selectivity to the target compound of over 98%, albeit at the cost of almost complete enzyme inactivation.
2.2. Case 1: Dosing of Propanal
The optimization of the dosing of propanal (A) along the reaction coordinate led to a slightly higher allowable initial concentration of B of 6 (those of E and A were again restricted by their upper bounds) and a slightly higher final BA concentration of 5.88 at comparable 98% conversion (Table 1). The concentration profile of reactant A remained at a constant value of 100 throughout the course of the reaction (Figure 2b-i), which was given by the upper bound defined in Equation (17j). The constant propanal concentration was achieved by a decreasing propanal dosing rate along the reaction coordinate (Figure 2b-iii), which followed its consumption rate (Figure 2b-ii). The maintained optimal concentration of A showed that an increased reaction rate toward BA outweighed the enzyme inactivation caused by A. Note that setting the upper bound to a higher value beyond the design space limit used in this study might further increase the concentration of BA. Except for the concentration of A, all other concentration and reaction rate profiles resembled those of the reference batch case (Figure 2a-i,a-ii). In summary, the only marginal increase in the final concentration of BA for this case was essentially due to the constant concentration of A, but suggested that the added experimental effort for the intensification was probably not warranted.
2.3. Case 2: Dosing of Benzaldehyde
In the case of dosing benzaldehyde (B) (Figure 2c-i), the optimal initial concentration of B was reduced by nearly an order of magnitude to 0.71 with A and E being restricted by their upper bounds. The concentration profile for A resembled that of the reference batch case. A final BA concentration of 6.52 was obtained (Table 1), which corresponded to a 12% increase over the batch reference.
In Figure 2c-i, we show that the concentration of B was maintained at a relatively constant low concentration. Similar to the propanal dosing case, the almost constant concentration of B was achieved by the dosing profile (Figure 2c-iii) mirroring the benzaldehyde consumption rate (Figure 2c-ii). However, the low B concentration drove the cross-carboligation pathway to maximize the formation of BA while the competing side product BB was formed at a relatively low concentration. In Figure 2c-ii, we highlight this effect, as the reaction rate of BB stays close to zero after a very short initial formation phase. The consumption rates of A and B nearly overlapped, which further supported the assumption that the optimal dosing of A shifted the reaction network significantly toward the conversion of A and B to form the target compound BA. However, the low concentration of B minimized the inactivation of the enzyme due to B, resulting in a significantly reduced enzyme inactivation rate (Figure 2c-ii), which was accompanied by a less steep enzyme activity loss than in the reference batch case (Figure 2a-i) and Case 1 (Figure 2b-i). As a result, a higher concentration of active enzyme was available for the production of BA at any time point. The low concentration of B at each time point, therefore, yielded the optimal balance between reaction kinetics toward BA and the conservation of PfBAL.
2.4. Case 3: Dosing of Propanal and Benzaldehyde
The results for the case of dosing propanal (A) and benzaldehyde (B) simultaneously are shown in Figure 2d-i. The optimal initial concentration of B of 0.70 was only slightly lower than on dosing of benzaldehyde and led to a final BA concentration of 6.59 (Table 1) with a yield of 88%. This product concentration corresponded to a 13% increase over the batch reference case and was only slightly higher than the value obtained for Case 2, thus confirming that dosing of B played an essential role in maximizing the final concentration of BA.
The dosing of A and B in Case 3 could be considered as a superposition of Case 1 and Case 2. Thus, the discussions for Cases 1 and 2 also apply here. Dosing of A and B simultaneously led to the highest final BA concentration from the intensification cases. Therefore, dosing of propanal (A) and benzaldehyde (B) was the best intensification strategy among all cases considered in this work and was selected for experimental reproducibility. The experimental setup was technically approximated as a fed-batch reactor.
In Figure 2d-i, we show the overlay of the simulated concentration progress for the optimized initial conditions and feeding profiles with the experimental data, which confirmed the optimization results. The measured concentration of BA nearly perfectly overlayed with the simulation for approximately 120 and then slightly diverged to higher values by up to 6%. As the measured initial concentrations were in line with the optimization case, but more benzaldehyde substrate was converted toward the end of the experiment compared to the model prediction, slower enzyme inactivation in the real experiment than in the predicted model seemed a plausible cause for this deviation. Measured concentrations of BB stayed constantly below 0.1 , which was in line with the simulation. The high ratio of BA to BB also confirmed the optimization result that keeping concentrations of B low during the experiment increased the selectivity toward BA and slowed down enzyme inactivation.
In contrast to the progress of BA and BB, the experimental concentration progress of B varied over time, presumably due to the manual sample withdrawal. Although the outlet of the dosing tube was submerged in the reaction solution, non-ideal mixing may have led to increased substrate concentrations near the tube outlet. For the products BA and BB, this issue did not apply as these compounds were homogeneously produced in the liquid body. An additional source of experimental error might exist due to HPLC sample storage in slightly variable vials, where the volatile substrate B may evaporate to the headspace, whereas BA and BB have low vapor pressures at ambient conditions.
In summary, the initial concentrations and feed rates obtained from the model based optimization applied to the laboratory setup could be experimentally reproduced and led to an enhanced final BA concentration, thus demonstrating successfully that the concentration of BA after a total runtime of 180 min could be increased by switching from the optimal batch scenario to an optimized fed-batch setup with time dependent feeding rates of Substrates A and B.
2.5. Data Summary
The optimized initial substrate and enzyme concentrations, together with the predicted final BA concentration and solution times for all cases, as well as the residual sum of squares and mean squared error for the experimental validations are summarized in Table 1. The solution times for all cases computed were between 1 and 4 and, thus, orders of magnitude faster than the time required to perform laboratory experiments. As laboratory experiments successfully validated the model based predictions, they could significantly reduce the time for the development of enzymatic processes.
The model based prediction quality surpassed the typical results of response-surface-model optimization using statistical experimental design. Thus, this study bolsters the importance and justifies the effort to identify the proper mechanistically based kinetic model.
3. Materials and Methods
3.1. Computational Methods
3.1.1. Material Balances
The dynamic model consisted of the material balances for Species A, B, BA, BB, and E. Instead of using the mole balances, as typically done in the EPF approach, we used a concentration basis because of numerical robustness during optimization. Furthermore, a perfectly mixed fluid element was assumed to avoid additional complexity introduced by the residence time distribution. The corresponding mass balances for the reactants A and B are as follows:
where:
and where and denote the concentrations of propanal (A) and benzaldehyde (B) in the reactor, respectively; j, q, and r the corresponding fluxes, feeding rates, and reaction rates, respectively; whereas and denote the inlet feed concentrations of pure liquid propanal and benzaldehyde of 13.946 M and 9.8 M, respectively. To avoid redundancy and provide a generalized representation of the material balances for all cases considered in this work, the binary terms and were introduced. That is, the reference batch case and dosing of either A and B were modeled via the binary decision variables, and . For example, the material balance equations for the reference batch case could be obtained by setting and . Moreover, note that the variables , , , , , , and V were all time varying, but for the sake of readability, the time varying argument was omitted. This also holds for variables , , and in the following paragraphs. Similarly, the material balances for the products (R)-2-hydroxy-1-phenylbutan-1-one (BA) and benzoin (BB), and the enzyme PfBAL (E), where no dosing occurs, are defined as:
Finally, the change in volume with time due to the amount of A and B added during the reaction is defined as follows:
3.1.2. Mechanistic Kinetic Model
The mechanistic kinetic model was based on the reaction mechanism shown in Figure 1 and was derived by solving the mass balances for substrates, products, and enzyme species under steady state conditions for the enzyme intermediates. Furthermore, the model considered the rate of inactivation of PfBAL, which was derived previously by combining progress curve analysis and optimal experimental design []. In this work, we additionally considered thermal deactivation of the enzyme, described by the reaction rate constant . The mechanistic kinetic model is given by:
For the ease of representation, the terms , , and D are defined as the following constitutive equations:
Note that the original parameter values in [] were not directly applicable as these parameters were derived for a kinetic model without thermal PfBAL inactivation. However, in this study, all kinetic parameters in Equations (9)–(16) were estimated using the methods and data presented in [] and are summarized in Table 2.
3.1.3. Dynamic Optimization Problem and Solution Strategy
The dynamic optimization problem aimed to maximize the final concentration of BA with respect to the time varying material balances, the volume change, and the reaction kinetic equations and is defined for the various intensification cases as:
The upper bounds on were set to 100 , which was the maximum pumping rate of the dosing pumps (Equation (17e)); those for and were set to , which was the maximum volume of the reactor used in the experimental setup (Equations (17f) and (17g)).
The inequality constraints (Equations (17h) and (17i)) represent the bounds for the initial and time varying concentrations and , respectively. The corresponding upper bounds were set to their respective maximum values used when estimating the kinetic parameters with of 100 and of 50 , with = 58919 (Equations (17j) and (17m)). The upper bounds for B and BB were set to their solubility limits (Equations (17k) and (17l)). Finally, the maximum reaction time was set to 180 min for the experimental duration time.
The dynamic optimization problem (Equation (17)) was solved by using the direct simultaneous approach, because of its ability to efficiently handle numerical instabilities and path constraints [,]. Specifically, the dynamic optimization problem for all cases was transcribed into algebraic equations by using the method of orthogonal collocation on finite elements []. The resulting system of algebraic equations falls into a class of optimization problems called nonlinear programming (NLP) []. The NLP was implemented in the MATLAB API for CasADi 3.4.0, a framework for automatic differentiation and numerical optimization []. Furthermore, 50 finite elements and three collocation points were used to discretize the dynamic optimization problems for all intensification cases considered. The number of 50 finite elements selected was found to provide a good trade-off between numerical accuracy and computational cost; while the three collocation points were chosen as recommended in []. The resulting NLP was solved by using IPOPT, an interior point solver designed for large scale NLPs [], in combination with the sparse symmetric linear solver MA57 [,]. All computations were performed on a Linux computer running a CentOS 7 operating system with an Intel(R) Core(TM) i7-4789 processor at 3.60 GHz and 16 GB RAM.
3.2. Experimental Setup
3.2.1. Materials
All chemicals used in this study were of analytical grade and used as purchased. Benzoin (racemic), propanal, thiamine diphosphate (ThDP), and triethanolamine (TEA) were purchased from Sigma Aldrich; acetonitrile (ACN), dimethyl sulfoxide (DMSO), and Bradford reagent from Carl Roth GmbH + Co. KG; 3,5-dimethoxy-benzaldehyde (DMBA), benzaldehyde, and bovine serum albumin (BSA) from Thermo Fisher Scientific; and trichloroacetic acid (TCA) from PanReac AppliChem. No further purification steps were performed before the experiment. Pure (R)-2-hydroxy-1-phenylbutan-1-one was provided by the Institute of Bio- and Geosciences at Forschungszentrum Jülich, Germany. PfBAL was kindly provided by the Institute of Bio- and Geosciences at Forschungszentrum Jülich as well; the His tagged enzyme was expressed in Escherichia coli SG13009, subsequently purified via immobilized metal affinity chromatography on a Ni-NTA-column, and finally desalted with size exclusion chromatography []. The pooled fractions were diluted to a concentration of 1 / with a desalting buffer (10 / TEA, / MgSO, / ThDP, adjusted to pH 7.5 with 1 HCl), lyophilized for several days, and then kept at in an airtight container.
3.2.2. Preparation of PfBAL Solution
Before each experiment, the enzyme solution and the reactant solution were freshly prepared in separate flasks. For the enzyme solution, the PfBAL lyophilizate was added to 10–20 mL of a prepared TEA buffer solution (50 TEA, adjusted to pH 8.5 with 1 HCl) containing cofactors (0.72 ThDP and 3.56 Mg). The PfBAL concentration in the stock solution was determined with a standard Bradford assay [] using a Tecan Spark photometer. The enzyme stock solution was appropriately diluted with 50 TEA buffer solution containing cofactors to the concentration desired for the specific reaction.
PfBAL activity was measured via the PfBAL catalyzed ligation of fluorescent DMBA to (R)-3,3,5,5-tetramethoxy-benzoin, causing a decrease in fluorescence over time []. Three different solutions with 70% v/v TEA buffer with cofactors and 30% v/v DMSO were prepared for the activity test. Solution I was obtained by diluting the enzyme stock solution with TEA buffer with cofactors and DMSO to a final PfBAL concentration of 15 /. For Solution II, 2 DMBA was dissolved, and Solution III remained without further additions. Four samples and four blanks were prepared in a 96 well microtiter plate by mixing 20 of Solution I with 180 of either Solution II or III. The plate was placed inside the preheated plate reader (Tecan Spark) at a temperature of 30 , and the emission intensity at 460 nm with an excitation at 362 nm was measured in intervals of 5 for a total time of 90 . The activity of PfBAL was calculated with an existing calibration curve for the fluorescence of DMBA at varying concentrations.
For the reactant solution, benzaldehyde and propanal were dissolved in DMSO. This step was done immediately before starting the experiment to avoid losses of any volatile substances.
3.2.3. Progress Curve Experiments
The results of dynamic optimization for the intensification cases were experimentally reproduced by performing progress curve experiments under the respective optimal conditions. The experiments were conducted in a magnetically stirred glass cylinder, which was placed in a water bath for temperature control at 30 . To minimize the evaporation of volatile Substances A and B during the experimental procedure, a PTFE lid was placed inside the glass cylinder right above the fluid level. Small holes in the PTFE lid gave access for the feeding tubes. The position of the lid was manually adjusted to correct for volume changes caused by sampling and, to a lesser extent, dosing.
The reaction was initiated by pouring both solutions (9 of reactant solution and 21 of enzyme solution) into the glass cylinder. The liquid was homogenized with the magnetic stirrer, and the lid was placed into position. After 10 s, a 100 zero sample was taken from the mixture and transferred to a high performance liquid chromatography (HPLC) vial containing 50 of % TCA solution to quench the enzymatic reaction immediately. About 950 of ACN were added to dilute the sample for the following analysis and to minimize the headspace inside the HPLC vial. Samples were taken at 6 to 10 min intervals for the first hour. Then, the sampling frequency was reduced to minimize the overall extraction of the reaction volume. All samples were prepared for analysis in HPLC on the same day or stored in a freezer at until further processing.
In the case of the fed-batch experiments, two glass syringes were filled with A and B, respectively, before the start of the experiment and positioned in high precision neMESYS 290N syringe pumps (Cetoni GmbH, Korbussen). PTFE tubes connected to the syringes were threaded through the holes of the PTFE lid into the reaction vessel and submerged in the reaction mixture. The feeding profile obtained from the dynamic optimizations for both substrates was individually programmed via the neMESYS UserInterface software. Dosings of A and B were initiated simultaneously with the zero sample.
3.2.4. HPLC Analysis
All samples from the progress curve experiments were filtered through PTFE syringe filters and were subsequently analyzed in reversed phase chromatography (RP-HPLC). About 30 of the sample volume were injected onto an RP8-column (LiChrospher 100 RP-8 (5 ), Merck KGaA) at 40 . The eluent was a mixture of ultrapure water and ACN with a flow rate of /. Initially, the eluent composition was 82% v/v ultrapure water and 18% v/v ACN. After , the composition was gradually changed within 2 to 100% ACN and maintained for 1 . Then, the composition was gradually changed back to 82% v/v ultrapure water and 18% v/v ACN within 2 min. The column was then flushed for 3 min to conclude the method, with a total runtime of min. Peaks of the individual compounds appeared after retention times of for B, for BA, and for BB. HPLC calibration was performed using standard samples of B, BB, and BA dissolved in 70% v/v TEA buffer solution and 30% v/v DMSO.
4. Conclusions
This contribution presents the elementary process functions (EPF) approach as a viable model based tool for designing optimal operating conditions for enzyme catalyzed reactions. For PfBAL catalyzed cross-carboligation of benzaldehyde and propanal, the final concentration of the product (R)-2-hydroxy-1-phenylbutan-1-one was chosen as the objective function to be optimized, and three process intensification scenarios were evaluated making use of the EPF concept.
The best final product concentration was obtained when dosing both aldehyde substrates, leading to a 13% increase compared to the optimized batch case that was used as the reference scenario. It was found that dosing propanal only served in replenishing the amount converted during the reaction so that the concentration constantly stayed at the upper limit specified in the design space. In contrast, dosing of benzaldehyde was found to be optimal when the concentration was set to a low initial value and never reached higher than −1. This strategy slowed down the PfBAL inactivation considerably, as well as the competing reaction to the byproduct benzoin. Although fed-batch processes are widely used in cases where substrates inhibit or even inactivate enzymes, determining time dependent dosing strategies is feasible only using model based approaches as presented. The value of the model based design becomes even more significant for optimization problems where the objective function is less intuitive than the product concentration. In the case of dosing propanal and benzaldehyde simultaneously and the derived optimal feeding profiles, experimental data confirmed the predicted results in the lab.
The apparatus independent EPF approach presented in this work could serve as a tool that enables process engineers to design novel processes and reactors systematically for enzyme catalyzed reaction systems in general. Additional reaction conditions, such as temperature, pH value, and composition of the reaction medium, might be incorporated into the kinetic model and open up the possibility for further improvements. Once the effects of protein engineering become tractable by appropriate structure–function relations, EPF based optimization of biocatalytic reactions may contribute to combining protein engineering and process systems engineering concepts [,].
Supplementary materials
Supplementary File 1Author Contributions
Conceptualization, V.N.E., D.H., R.S., and A.C.S.; investigation, D.H.; software, V.N.E.; writing, original draft preparation, V.N.E. and D.H.; writing, review and editing, R.S. and A.C.S.; visualization, V.N.E. and D.H.; supervision, R.S. and A.C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Research Foundation (DFG), Research Group for Diversity of Asymmetric Thiamine Catalysis, Grant/Award Number: 128900243.
Acknowledgments
We thank the Institute of Bio- and Geosciences at Forschungszentrum Jülich, especially Martina Pohl, for providing (R)-2-hydroxy-1-phenylbutan-1-one, as well as benzaldehyde lyase lyophilizate. The author V.N.E. is also affiliated with the International Max Planck Research School for Advanced Methods in Process and Systems Engineering (IMPRS-ProEng), Magdeburg, Germany. Moreover, we thank Ulrike Krewer for helpful comments on a previous version of the manuscript and Rüdiger Ohs for his support regarding experimental and modeling procedures. We also acknowledge support from the German Research Foundation and the Open Access Publication Funds of the Technische Universität Braunschweig.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; nor in the decision to publish the results.
Nomenclature
| Abbreviations | |
| molecular weight () | |
| E. coli | Escherichia coli |
| PfBAL | benzaldehyde lyase from Pseudomonas fluorescens |
| A | propanal |
| ACN | acetonitrile |
| B | benzaldehyde |
| BA | (R)-2-hydroxy-1-phenylbutan-1-one |
| BB | (R)-benzoin |
| CSTR | continuous stirred tank reactor |
| DMBA | 3,5-dimethoxy-benzaldehyde |
| DMSO | dimethyl sulfoxide |
| E | enzyme |
| EPF | elementary process functions |
| HPLC | high performance liquid chromatography |
| HPP | (R)-2-hydroxy-1-phenylpropanone |
| NLP | nonlinear programming |
| PTFE | polytetrafluoroethylene |
| RP-HPLC | reversed phase chromatography |
| TEA | triethanolamine |
| ThDP | thiamine diphosphate |
| Indices, subscripts, and superscripts | |
| f | final |
| i | species or component index |
| 0 | initial |
| in | inlet feed |
| UB | upper bound |
| Latin symbols | |
| C | concentration () |
| D | denominator term for the reaction rate |
| j | dosing flux () |
| k | reaction rate constant |
| N | numerator term for the reaction rate |
| q | feeding rate () |
| r | reaction rate () |
| s | binary variable (−) |
| t | reaction time () |
| V | volume () |
References
- Lalonde, J. Highly engineered biocatalysts for efficient small molecule pharmaceutical synthesis. Curr. Opin. Biotechnol. 2016, 42, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Pollard, D.J.; Woodley, J.M. Biocatalysis for pharmaceutical intermediates: The future is now. Trends Biotechnol. 2007, 25, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Woodley, J.M. New opportunities for biocatalysis: Making pharmaceutical processes greener. Trends Biotechnol. 2008, 26, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Ringborg, R.H.; Woodley, J.M. The application of reaction engineering to biocatalysis. React. Chem. Eng. 2016, 1, 10–22. [Google Scholar] [CrossRef]
- Kiss, A.A.; Grievink, J.; Rito-Palomares, M. A systems engineering perspective on process integration in industrial biotechnology. J. Chem. Technol. Biotechnol. 2015, 90, 349–355. [Google Scholar] [CrossRef]
- Woodley, J.M. Bioprocess intensification for the effective production of chemical products. Comput. Chem. Eng. 2017, 105, 297–307. [Google Scholar] [CrossRef]
- Braun, M.; Link, H.; Liu, L.; Schmid, R.D.; Weuster-Botz, D. Biocatalytic process optimization based on mechanistic modeling of cholic acid oxidation with cofactor regeneration. Biotechnol. Bioeng. 2011, 108, 1307–1317. [Google Scholar] [CrossRef]
- Schenkendorf, R. Supporting the shift towards continuous pharmaceutical manufacturing by condition monitoring. In Proceedings of the Conference on Control and Fault-Tolerant Systems, SysTol, Barcelona, Spain, 7–9 September 2016; pp. 593–598. [Google Scholar]
- Stillger, T.; Pohl, M.; Wandrey, C.; Liese, A. Reaction engineering of benzaldehyde lyase from Pseudomonas fluorescens catalyzing enantioselective C-C bond formation. Org. Process Res. Dev. 2006, 10, 1172–1177. [Google Scholar] [CrossRef]
- Hildebrand, F.; Kühl, S.; Pohl, M.; Vasic-Racki, D.; Müller, M.; Wandrey, C.; Lütz, S. The production of (R)-2-hydroxy-1-phenyl-propan-1-one derivatives by benzaldehyde lyase from Pseudomonas fluorescens in a continuously operated membrane reactor. Biotechnol. Bioeng. 2007, 96, 835–843. [Google Scholar] [CrossRef]
- Begemann, J.; Ohs, R.B.; Ogolong, A.B.; Eberhard, W.; Ansorge-Schumacher, M.B.; Spiess, A.C. Model based analysis of a reactor and control concept for oxidoreductions based on exhaust CO2-measurement. Process Biochem. 2016, 51, 1397–1405. [Google Scholar] [CrossRef]
- Marpani, F.; Sárossy, Z.; Pinelo, M.; Meyer, A.S. Kinetics based reaction optimization of enzyme catalyzed reduction of formaldehyde to methanol with synchronous cofactor regeneration. Biotechnol. Bioeng. 2017, 114, 2762–2770. [Google Scholar] [CrossRef] [PubMed]
- Emenike, V.N.; Schenkendorf, R.; Krewer, U. A systematic reactor design approach for the synthesis of active pharmaceutical ingredients. Eur. J. Pharm. Biopharm. 2018, 126, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Freund, H.; Sundmacher, K. Towards a methodology for the systematic analysis and design of efficient chemical processes. Chem. Eng. Process. Process Intensif. 2008, 47, 2051–2060. [Google Scholar] [CrossRef]
- Peschel, A.; Freund, H.; Sundmacher, K. Methodology for the design of optimal chemical reactors based on the concept of elementary process functions. Ind. Eng. Chem. Res. 2010, 49, 10535–10548. [Google Scholar] [CrossRef]
- Hentschel, B.; Peschel, A.; Freund, H.; Sundmacher, K. Simultaneous design of the optimal reaction and process concept for multiphase systems. Chem. Eng. Sci. 2014, 115, 69–87. [Google Scholar] [CrossRef]
- Jokiel, M.; Kaiser, N.M.; Kováts, P.; Mansour, M.; Zähringer, K.; Nigam, K.D.P.; Sundmacher, K. Helically coiled segmented flow tubular reactor for the hydroformylation of long-chain olefins in a thermomorphic multiphase system. Chem. Eng. J. 2019. [Google Scholar] [CrossRef]
- Kaiser, N.M.; Jokiel, M.; McBride, K.; Flassig, R.J.; Sundmacher, K. Optimal reactor design via flux profile analysis for an integrated hydroformylation process. Ind. Eng. Chem. Res. 2017, 56, 11507–11518. [Google Scholar] [CrossRef]
- Maußner, J.; Dreiser, C.; Wachsen, O.; Freund, H. Systematic model based design of tolerant chemical reactors. J. Adv. Manuf. Process. 2019. [Google Scholar] [CrossRef]
- Peschel, A.; Karst, F.; Freund, H.; Sundmacher, K. Analysis and optimal design of an ethylene oxide reactor. Chem. Eng. Sci. 2011, 66, 6453–6469. [Google Scholar] [CrossRef]
- Freund, H.; Maußner, J.; Kaiser, M.; Xie, M. Process intensification by model based design of tailor-made reactors. Curr. Opin. Chem. Eng. 2019, 26, 46–57. [Google Scholar] [CrossRef]
- Emenike, V.N.; Schenkendorf, R.; Krewer, U. Model based optimization of biopharmaceutical manufacturing in Pichia pastoris based on dynamic flux balance analysis. Comput. Chem. Eng. 2018, 118, 1–13. [Google Scholar] [CrossRef]
- Emenike, V.N.; Xie, X.; Schenkendorf, R.; Spiess, A.C.; Krewer, U. Robust dynamic optimization of enzyme catalyzed carboligation: A point estimate based back-off approach. Comput. Chem. Eng. 2019, 121, 232–247. [Google Scholar] [CrossRef]
- Müller, M.; Sprenger, G.A.; Pohl, M. C–C bond formation using ThDP dependent lyases. Curr. Opin. Chem. Biol. 2013, 17, 261–270. [Google Scholar] [CrossRef]
- Demir, A.S.; Pohl, M.; Janzen, E.; Müller, M. Enantioselective synthesis of hydroxy ketones through cleavage and formation of acyloin linkage. enzymatic kinetic resolution via C–C bond cleavage. J. Chem. Soc. Perkin Trans. 2001, 1, 633–635. [Google Scholar] [CrossRef]
- Pohl, M.; Gocke, D.; Müller, M. Thiamine based enzymes for biotransformations. In Handbook of Green Chemistry; Wiley-VCH: Weinheim, Germany, 2009. [Google Scholar]
- Ohs, R.; Fischer, K.; Schoepping, M.; Spiess, A.C. Derivation and identification of a mechanistic model for a branched enzyme catalyzed carboligation. Biotechnol. Prog. 2019, e2868. [Google Scholar] [CrossRef] [PubMed]
- González, B.; Vicuña, R. Benzaldehyde lyase, a novel thiamine PP-requiring enzyme, from Pseudomonas fluorescens biovar 1. J. Bacteriol. 1989, 171, 2401–2405. [Google Scholar] [CrossRef] [PubMed]
- Biegler, L.T. An overview of simultaneous strategies for dynamic optimization. Chem. Eng. Process. Process Intensif. 2007, 46, 1043–1053. [Google Scholar] [CrossRef]
- Biegler, L.T. Nonlinear Programming; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2010. [Google Scholar]
- Cuthrell, J.E.; Biegler, L.T. On the optimization of differential-algebraic process systems. AIChE J. 1987, 33, 1257–1270. [Google Scholar] [CrossRef]
- Andersson, J.A.E.; Gillis, J.; Horn, G.; Rawlings, J.B.; Diehl, M. CasADi: A software framework for nonlinear optimization and optimal control. Math. Program. Comput. 2019, 11, 1–36. [Google Scholar] [CrossRef]
- Wächter, A.; Biegler, L.T. On the implementation of an interior-point filter line-search algorithm for large-scale nonlinear programming. Math. Program. 2006, 106, 25–57. [Google Scholar] [CrossRef]
- Duff, I.S. MA57—A code for the solution of sparse symmetric definite and indefinite systems. ACM Trans. Math. Softw. 2004, 30, 118–144. [Google Scholar] [CrossRef]
- HSL. A Collection of Fortran Codes for Large Scale Scientific Computation. Available online: http://www.hsl.rl.ac.uk (accessed on 29 September 2017).
- Ohs, R.; Leipnitz, M.; Schöpping, M.; Spiess, A.C. Simultaneous identification of reaction and inactivation kinetics of an enzyme catalyzed carboligation. Biotechnol. Prog. 2018, 34, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zavrel, M.; Schmidt, T.; Michalik, C.; Ansorge-Schumacher, M.; Marquardt, W.; Büchs, J.; Spiess, A.C. Mechanistic kinetic model for symmetric carboligations using benzaldehyde lyase. Biotechnol. Bioeng. 2008, 101, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Woodley, J.M. Integrating protein engineering with process design for biocatalysis. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2018, 376. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).