1. Introduction
Psychrophilic enzymes obtained from psychrophilic bacteria have been widely exploited because of their high activities at very low temperatures [
1,
2]. Due to their unique characteristics, these cold-active enzymes offer economic and ecological advantages in biotechnological applications over extremophiles, which operate at higher temperatures [
3]. Three major enzymes that have been studied in industrial applications are proteases, lipases, and amylases. However, lipases have attracted special attention from researchers since 1930 [
4]. In general, a lipase is a triacylglycerol hydrolase that catalyzes the hydrolysis of triglycerides to free fatty acids and glycerol at the oil–water interface [
5]. It also catalyzes bioconversion reactions in non-aqueous media and hydrolyzes organic carbonates in the absence of co-factors [
6,
7,
8,
9]. Cold-active lipases cover a broad spectrum of biotechnological applications, such as additives in detergents (cold washing), an additive in food industries (fermentation, cheese manufacture, bakery, meat tenderizing), bioremediations (Digesters, composting, oil, or xenobiotic biology applications), and molecular biology applications [
10,
11,
12].
Several conventional purification methods for cold-adapted lipases have been applied for many years in industrial and laboratory procedures [
13,
14]. For example, the lipase KE38 from
Pseudomonas fluorescens KE38 was successfully purified using (NH
4)
2SO
4 precipitation, Sephadex G-100 and, lastly, ultrafiltration with a recovery of 54.99% and a 41.13-fold purification [
15]. Likewise, three purification steps were used to purify extracellular LipP from
Pseudomonas sp. strain BII-I by using 20–65% (NH
4)
2SO
4 precipitation followed by a diethylaminoethyl (DEAE)-cellulose and graphite column with 38-fold purification and a recovery of 17% of the lipase activity [
13]. Although a good yield of purified enzyme was obtained from these chromatographic techniques, the production and purification of large amounts of enzyme are often laborious and expensive, which hinders applications for these enzymes [
16]. Therefore, a standard method that enables the purification of large amounts of highly pure, active, and stable enzymes is urgently needed. Reverse micellar extraction is one of the alternative purification methods under consideration for continuous downstream processing of bulk enzymes.
A reverse micelle (RM) is a surfactant (hydrophilic head group and hydrophobic tail) that forms a water microemulsion in a nonpolar liquid [
17]. It is an ideal model system for confined liquids. This approach has been extensively explored because of their resemblance to biological membranes and their capability for solubilizing enzymes and catalyzing biochemical reactions. The most common applications of RMs are in enzyme catalysis [
18], drug delivery [
19], nanocluster synthesis [
20], and materials development. As an alternative separation practice, RM offers several benefits, such as its ease of operation, low energy requirements, large interfacial area, low cost, and single-stage operation that combines extraction and stripping [
21]. Generally, Triton X-114 (a polyoxyethylene detergent, C
14EO
6) and members of the alkyl polyoxyethylene family (C
mEO
n), such as
n-decyltetraethylene oxide (C
10EO
4), are used as phase-forming surfactants [
22]. The formation of a large interface between the aqueous and nonpolar phases permits reactants that prefer different phases to be conveyed together. Most enzymes are hydrophilic molecules, while substrates may be soluble in nonpolar solutions. The interaction of these molecules occurs when there is an adequate interface between the phases [
23]. This partitioning behavior is governed by repulsive, steric, and excluded-volume interactions between the two components [
22]. As a result, the interactions between the lipase and surfactant-solvent complexes can be studied with atomic-level detail using molecular dynamics (MD) simulations.
Family 1.3 lipase is a hydrolytic enzyme. It is a bacterial lipase subfamily in which the lipase is mostly produced by
Pseudomonas and
Serratia sp. [
24]. Previously, mesophilic counterparts of the AMS8 lipase, the closed conformation of 1.3 lipases from
Pseudomonas sp. MIS38 (PML), were tested with substrate micelles (octane and trilaurin) using MD simulations. Lid 1 of PML was reported to be fully opened in the octane micelles compared to the trilaurin micelles and was followed by the opening of lid 2 in the presence of calcium ions [
25]. In addition, interfacial solubilized enzymes, such as lipase and horseradish peroxidase, show high activities at the mixed reverse micellar interface [
26]. Cytochrome c was successfully extracted into the Span 60/hexane reverse micelles [
27]. Therefore, it is important to verify this experimentally to determine the purified AMS8 and the mode of action of the lipase in the reverse micelles, which may subsequently lead to a higher recovery of activity and degree of purification.
In this work, nonionic surfactant Triton X-100/toluene reverse micelles were used to extract the cold-adapted lipase AMS8 and the procedure was optimized to gain a better production yield. The solvation dynamics (via in silico study) were investigated to visualize the structures and to understand the interaction between the lipase AMS8 and the TX-100/toluene reverse micelles at the molecular level.
2. Results and Discussion
There are many conventional strategies to extract proteins and gain purified enzymes. The cold-adapted lipase AMS8 has been previously purified using affinity and gel filtration chromatography with a final production yield of 23% and 9.7-fold purification [
28]. Since the lipase recovery was low, attempts have been made to extract lipases using liquid–liquid extraction, especially RME. The most widely reported examples of RME use the ionic surfactant di-2-ethylhexyl sodium sulfosuccinate (AOT) and cetyl trimethyl ammonium bromide (CTAB) for the protein extraction [
29,
30,
31]. Only a few publications report the application of nonionic surfactants (Triton X-100, Span 60, Tween 85, and Tween 20) [
27,
32]. Most studies have reported that nonionic surfactants lack the driving force to extract large molecular weight proteins [
33]. Affinity RME using cibacron blue (CB)-modified lecithin failed to extract bovine serum albumin (BSA) for this reason [
34]. However, one of the valuable properties of nonionic surfactant RME is that, unlike ionic surfactants, it does not denature or alter the structure of the solute during the extraction [
33]. This is very important for cold-adapted lipases since these proteins are very conformationally flexible and are easily affected by strong force interactions [
10]. Therefore, this study of nonionic surfactant RME for protein extraction should be helpful for understanding the liquid–liquid reverse micellar extraction process. In this study, we purify this cold-adapted enzyme by using an aqueous two-phase system in which reverse micellar extraction has been introduced.
2.1. Effect of Initial Aqueous Phase pH
The initial aqueous phase pH plays a major role in governing the behavior of the protein extraction in the reverse micellar system. The effect of the initial aqueous pH on the forward extraction of the lipase is shown in
Table 1. The concentration chosen was over the critical micellar concentration (0.25–0.30 mM), which is 0.05 M Triton X-100 in toluene. NaCl was added to all the initial pH phases at a concentration of 0.1 M. The recovery of the activity was obtained in both organic phase and aqueous phase at pH 7 (3.18% and 58.84%, respectively). In the backward extraction, 50 mM Tris-HCl and 1 M KCl at pH 9 were selected as the stripping solution (aqueous phase). This is because purified AMS8 lipase has optimum activity at alkaline solution, pH 9. This buffer was used for all sets of conditions during the optimizations of the forward extraction steps. Thus, the maximum activity recovery for the backward extraction for the aqueous solution was 58.84%. No recovery of the activity was observed at pH 8 to 9. In the ionic reverse micellar system, the protein extraction was significantly affected by changing the pH of the solutions. This is due to the electrostatic interaction between the polar head group of the surfactant and the protein surface [
35,
36,
37]. There is no net charge present in the nonionic surfactant system, but it has a surface charge (positive or negative) at neutral pH. This indicates that the protein extraction was favoured by a weak electrostatic interaction between the surfactant and the solute [
38]. Previously, larger proteins have needed a larger number of charged residues on their surface in order to be transferred into RMs, so the larger the protein, the further the pH of maximal transfer is from the isoelectric point [
39]. This theory holds true in the case of AMS8 lipase extraction as its optimal pH of maximum transfer is 7, which is different from the pI of AMS8 lipase (4.6).
The reaction mixture (refolded lipase, 0.05 M Triton X-100 in toluene) was incubated at 25 °C for 15 min under constant stirring (601× g). The organic phase was separated by centrifugation at 3607× g for 30 min at 25 °C. The lipase recovery was carried out by backward extraction using 1 M KCl in 50 mM Tris-Cl buffer, pH 9 (volume ratio 1:1), at 25 °C for 30 min. The initial total activity of refolded AMS8 lipase (10 mL) at different pHs (pH 5: 1039.5 U; pH 6: 7586.2 U; pH 7: 5301.7 U; pH 8: 892.8 U; pH 9: 666.22 U). The lipase was refolded in the different pHs and their recoveries were set as 100%.
2.2. Effect of Surfactant Concentration
Table 2 shows the recovery of activity as a function of the surfactant concentration in the range of 0.01–0.09 M. The initial crude pH of AMS8 lipase was maintained at pH 7 with 0.1 M NaCl. As the surfactant concentration increased from 0.01 to 0.03 M, there was a decline in the recovery of the activity from 118.81 to 77.05%. AMS8 lipase was unsuccessfully extracted at 0.09 M Triton X-100. This may be due to the deassembling of the reverse micelles in the organic phase due to intermicellar collision and a limitation on the solute diffusion due to surfactant aggregates at high surfactant concentrations [
33]. However, a small amount of recovery of the lipase activity of 20.75% was obtained at 0.07 M. As AMS8 lipase was assumed to be fully transported into the stripping solution, it was stated that the maximum activity recovery was 14.45%. However, about 6% of the activity was lost during the phase transfer process from the forward to the backward extraction. Increasing turbidity or cloudiness of the phases with increasing surfactant concentration was observed during the experiment.
The reaction mixture (refolded lipase, Triton X-100 in toluene) was incubated at 25 °C for 15 min under constant stirring (601× g). The organic phase was separated by centrifugation at 3607× g for 10 min at 25 °C. The lipase recovery was carried out by backward extraction using 1 M KCl in 50 mM Tris-Cl buffer, pH 9, for 30 min. The activity recovery of refolded lipase (control) was set as 100% (12,026 U).
2.3. Effect of Ionic Strength
The effect of the aqueous phase’s ionic strength on the enzyme extraction by the organic phase was investigated by varying the concentration of NaCl from 0.075 to 0.175 M (
Table 3). An insignificant decrease in the lipase recovery of organic phase from 85.46 to 81.90% was observed for 0.075 to 0.125 M NaCl. Then, it keeps decreasing to 52.92% at 0.175 M NaCl. In the case of the backward extraction, the highest recovery of activity was obtained at 0.125 M NaCl (39.47%). Some studies have shown that a minimum salt concentration enhanced the interactions between the biomolecule and surfactant head groups and reduced the repulsive forces between them [
21]. Others have shown that, as the ionic strength increased, the protein uptake capacity of the reverse micelles decreased. The electrostatic effects between the biomolecule and the surfactant existed as varying salt concentrations with the activity recovery [
29]. An increased ionic strength led to a reduction of the reverse micellar size, and thus the protein was easily expelled (the squeezing out effect). Decreases in the Debye length might have caused the lower activity recovery at higher salt concentrations [
33].
The reaction mixture (refolded lipase, 0.07 M Triton X-100 in toluene) was incubated at 25 °C for 15 min under constant stirring (601× g). The organic phase was separated by centrifugation at 3607× g for 10 min at 25 °C. Lipase recovery was carried out by backward extraction using 1 M KCl in 50 mM Tris-Cl buffer, pH 9, for 30 min. The activity recovery of refolded lipase (control) was set as 100% (11,193 U).
2.4. Effect of Temperature
The temperature is a key factor in the formation of nonionic surfactant reverse micelles as compared to the ionic surfactant. Generally, the size of nonionic polyoxyethylene micelles increases with temperature and the growth is more marked at a temperature close to the cloud point [
40]. As the micellar size of polyoxyethylene increases, the protein solubility in the organic phase increases to fill the micelles. Luisi et al. (1979) managed to increase the R-chymotrypsin and glucagon transfer yields into an NH
4+/chloroform phase to 50 and 100%, respectively [
41].
Table 4 shows the effect of temperature on the forward extraction. The optimum forward extraction temperature is 10 °C with 55.88% activity recovery. After 10 °C, the lipase activity recovery decreased. Since the cold-adapted lipase AMS8 has high activity at low temperature, at the desired temperature, the enzyme–surfactant interaction increased. Singh and Kishore (2006) have reported that the binding of Triton X-100 to BSA contributes to an increase in the K
1 value (by about a factor of 5) with an increase in temperature from 15 to 30 °C. At higher temperatures, proteins tend to lose their structures, leading to modification of the binding site and greater interaction between Triton X-100 and BSA [
42]. The initial lipase amount was set at about 30 mg, but lipase activity varies (10,000–30,000 U). This affects the activity recovery of the lipase released from the reverse micelles. Even with the same amount released, but it could be active or inactive. Even though the protein concentration increased with temperature, the activity recovery decreased. This is motivated by the high flexibility of the lipase AMS8 at low temperature.
The reaction mixture (0.125 M NaCl in crude lipase, 0.07 M Triton X-100 in toluene) was incubated for 15 min under constant stirring (500 rpm). The organic phase was separated by centrifugation at 3000 rpm for 10 min. The lipase recovery was carried out by backward extraction using 1 M KCl in 50 mM Tris-Cl buffer, pH 9, for 30 min. The activity recovery of the refolded lipase (control) was set as 100% (27,928 U)
2.5. Effect of the Triton X-100/Toluene Reverse Micelles on the Secondary Structure of AMS8
MD simulations have been extensively used for mimicking surfactant aggregation in micelles [
38]. YASARA is a powerful software tool for molecular graphics, modeling, and simulations to perform true interactive real-time simulations with highly accurate force fields on standard personal computers (PCs). It is well-known for its ease of manipulating complex protein-solvent parameters. Studying the surfactant-solvent interfacial structure with biomolecules provides insights into the structural changes enzymes undergo in the micelles. Since the cold-adapted lipase AMS8 has no crystal structure as yet, the closed conformation of the predicted model was used as an initial structure for the MD simulations. The interactions of the protein–reverse micelle complex structures were performed in 10 ns MD simulations at different temperatures (5, 10, 20, 25, 30, and 40 °C). To activate the enzyme, a transition from the closed to the open conformation has to occur. This interfacially activated lipase may give insight into how the lid domains act as a door to expose the catalytic triad for substrate accessibility (open conformation) in the presence of Triton X-100/toluene micelles with a temperature effect. Previously, this time-dependent simulation has shown that the second lid (residues 210–222) together with the first lid of the
Pseudomonas aeruginosa lipase act as a double door, protecting the hydrophobic catalytic site from the polar solvent [
43]. The tolerance of the AMS8 lipase has already been tested with several polar solvents (methanol, ethanol, propanol, and dimethyl sulfoxide—DMSO) and nonpolar solvents (hexane and toluene) in molecular dynamic simulations. Interestingly, the cold-adapted lipase AMS8 strongly interacts with toluene, which is responsible for lid 2 opening. This indicates that the conformational change in the AMS8 lipase from closed to open is influenced by toluene for substrate accessibility [
44].
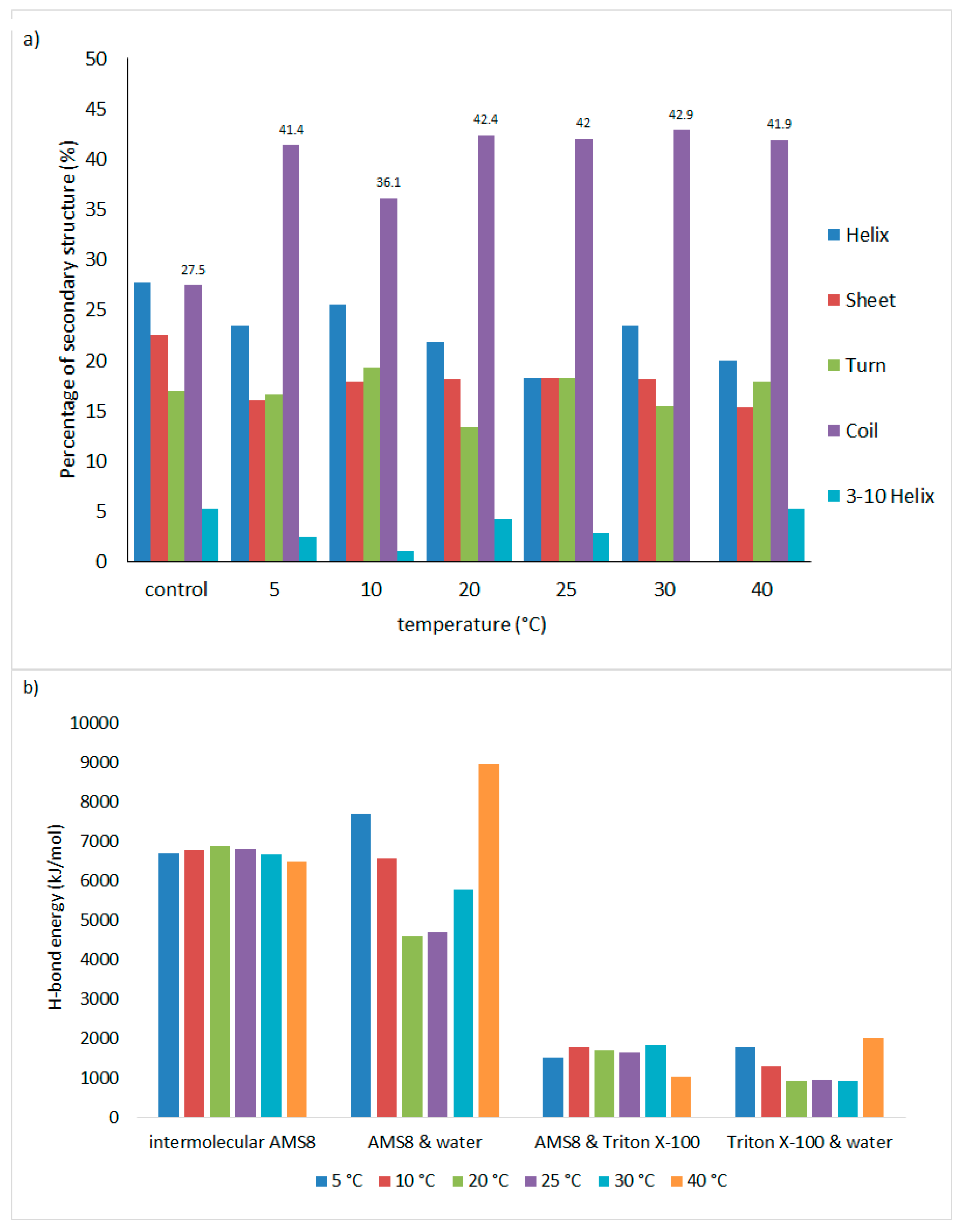
The influence of temperature on the secondary structure of AMS8 lipase in the Triton X-100/toluene reverse micelles was investigated (
Figure 1a). The secondary structures (α-helix, β-sheet, turn, and coil) were still conserved in the confined liquids. The α-helix structure significantly decreases at 25 and 40 °C compared to the control, with differences of 9.4 and 7.7, respectively. The β-sheet structure decreased up to 6.5 and 7.2 at 25 °C and 40 °C, respectively. A slight increase in the turn structure of about 2.3 and 1.3 at 10 and 25 °C and a sharp increase in the coil conformation at 30 and 20°C with differences of 15.4 and 14.9 from the control, respectively, were observed. This may be due to the adaptation of the AMS8 lipase to be more flexible in the confined liquids entrapped in the reverse micelles. Furthermore, these fluctuations were also enhanced by the intrinsic conformational preferences of the different amino acids, which are secondary structure tendencies that redirected the random coil state and the native protein structure association relationship [
45]. Therefore, enhanced stability of the protein secondary structure was prompted by hydrogen bonds to the α-helices and β-sheets. From
Figure 1b, the total energy required to form hydrogen bonds in the intramolecular AMS8 lipase slightly increased at 25 °C compared to other temperatures. Since the difference was small, the main residues involved in maintaining the structure, specifically in the catalytic triad and lid conformation (lid 1 and lid 2), were investigated. Unfortunately, the hydrogen bonds of the lid 1 nonpolar residues (Ala-51, Leu-53, Val-54, and Leu-57) had not been maintained from 10 to 30 °C. Throughout the simulations of these temperatures, the interacting lid 2 residues (Asp-157, Val-159, Ser-160, Asp-161, Leu-162, Ala-164, Ala-165, Leu-166, and Gly-167) were maintained after 10 ns of simulation.
Commonly, the residues involved in hydrogen bond formation are bonded to water molecules that mediate ligand interactions with protein polar and charged groups [
46]. Surprisingly, in these simulations, nonpolar residues also interacted with water molecules to stabilize the enzyme structure and reverse micelles. As the AMS8 lipase interacted with the surrounding water molecules, the lowest total binding energy for the hydrogen bonds (4585.17 kJ/mol) was found at 20 °C. Hence, for 20 °C, the selected residues in the active site (Asp-255) and lid 2 conformation (Glu-148, Leu-150, Asp-153, Ile-155, Asp-157, and Asp-161) interacted significantly with water molecules. For the main residue of lid 2, the O-atom of Leu-150 accepts a bond from the O-atom of H
2O-1599 with the lowest binding energy (17 kJ/mol) and a distance of 1.8 Å. Then, the OE2-atom of Glu-148 accepts a bond from the O-atom of H
2O-1577 with a shortest distance from the actual position of 1.53 Å. For the active site, the O-atom of H
2O-1523 donates a bond to the O-atom of Asp-255 with a distance below 2.0 Å. In conclusion, the energy of hydrogen bonds is required for enzyme stability at low temperatures for the AMS8 lipase to remain active in the Triton X-100/toluene reverse micelles.
2.6. Stability and Flexibility Analysis of AMS8 in Triton X-100/Toluene Reverse Micelles
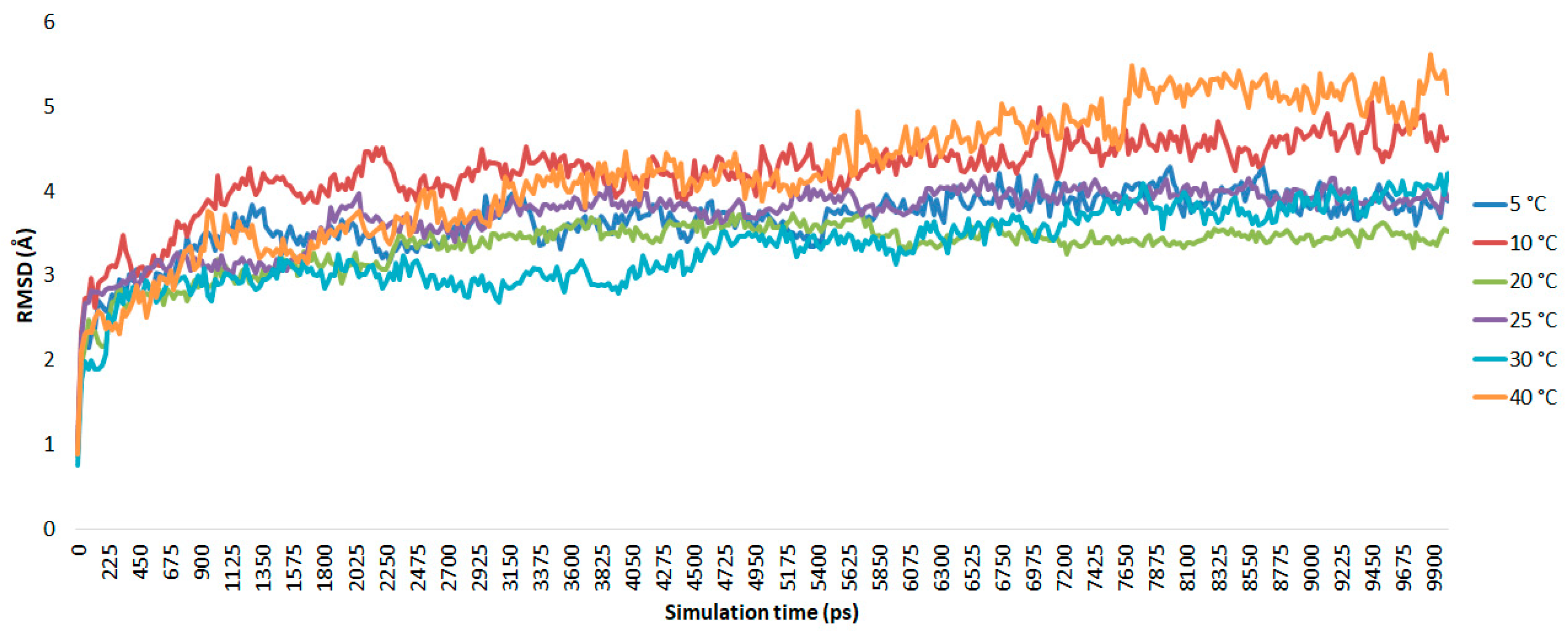
As shown in
Figure 2, from 5 to 40 °C, during the beginning of the trajectory the AMS8 lipase simulated at 5, 20, and 25 °C showed a similar pattern until the end of the simulation (t = 10 ns). An abrupt oscillation from the beginning (0.948 Å) until 3 ns (4.34 Å) was observed, which suggests that at this point equilibration and thermalization occurred at 10 °C. The root mean square deviations (RMSds) of the AMS8 lipase show minor conformational changes after 3 ns of simulation. However, at 30 °C, a gradual fluctuation at 2.6 to 4.1 ns occurred with an RMSd value of 2.85 to 2.99 Å and further slightly increased until the end of the simulation (4.2 Å). In contrast, at a higher temperature (40 °C), the deviation value from the beginning of the simulation continued to grow from 0.886 Å to 5.147 Å. This suggests that the conformation of the AMS8 lipase remained stable in the Triton X-100/toluene reverse micelles at low to moderate temperatures (5, 20, and 25 °C) as signified by the small deviation in RMSd value between them.
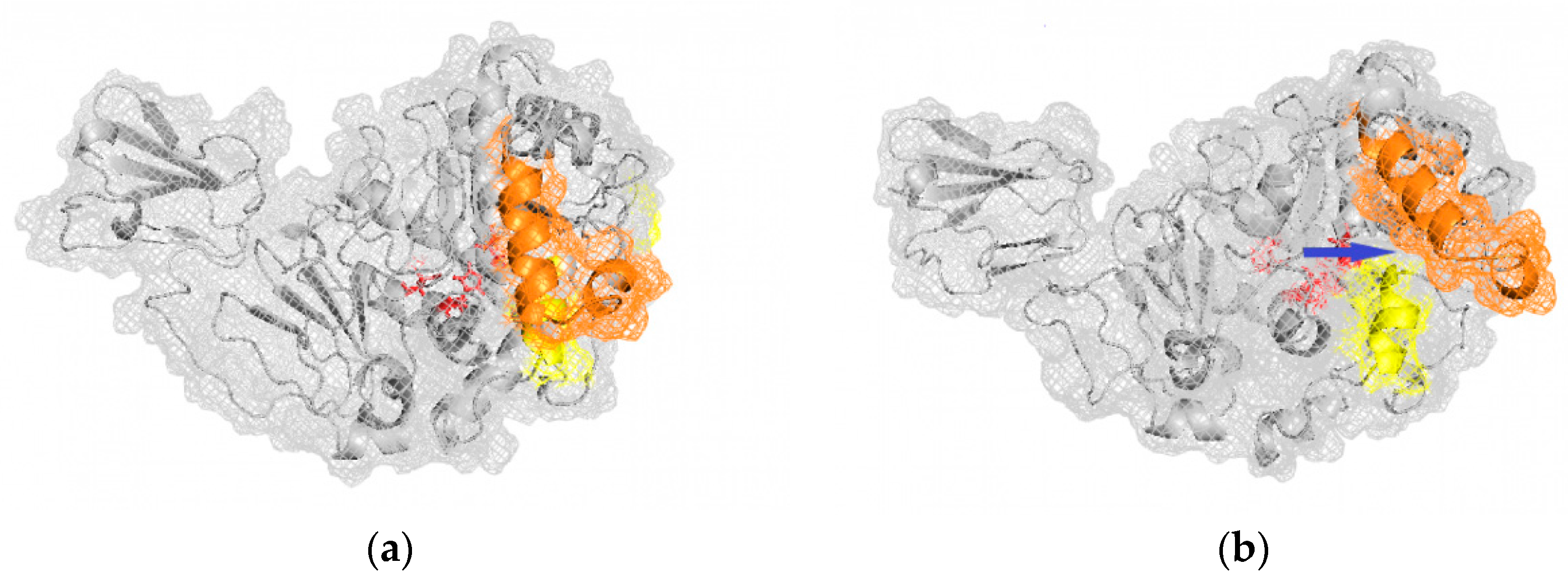
In the experimental conditions, the AMS8 lipase maintained 43% of its lipase activity at 10 °C, while at 5 °C and 20 °C, it remained 36% and 33% active, respectively. Thus, the AMS8 lipase was expected to be stable at these temperatures. However, at 10 °C, it has the highest stagnant RMSd values, probably due to difference from the actual experimental conditions, including the short incubation time and the Triton X-100 concentration. A superposition of the AMS8 lipase simulated at the optimum temperature in the Triton X-100/toluene reverse micelle is shown in
Figure 3. A movement of lid 2 from the catalytic site was observed, as the hydrophobic residues disrupt the strong hydrophobic interaction that usually conserves the large hydrophobic cavity, burying the catalytic site.
For comparison, the AMS8 lipase simulated in water [
32] exhibited a stable native-like structure at 0 and 5 °C. At higher temperatures (≥25 °C), the globular protein started to lose its native conformation to adapt to changes in temperature and the water density (solvent). For the AMS8 lipase simulated in toluene at 25 °C [
44], the RMSd values were as high as 6.76 Å at 10 ns of simulation. On the other hand, about 55.47% of the RMSd values were reduced in the Triton X-100/toluene reverse micelles. This shows that the AMS8 lipase is more stable in the reverse micelles than in water and toluene itself. It can be deduced that the cold-adapted protein tends to unfold and denature due to structural elements that confer high motion at a higher temperature (≥30 °C).
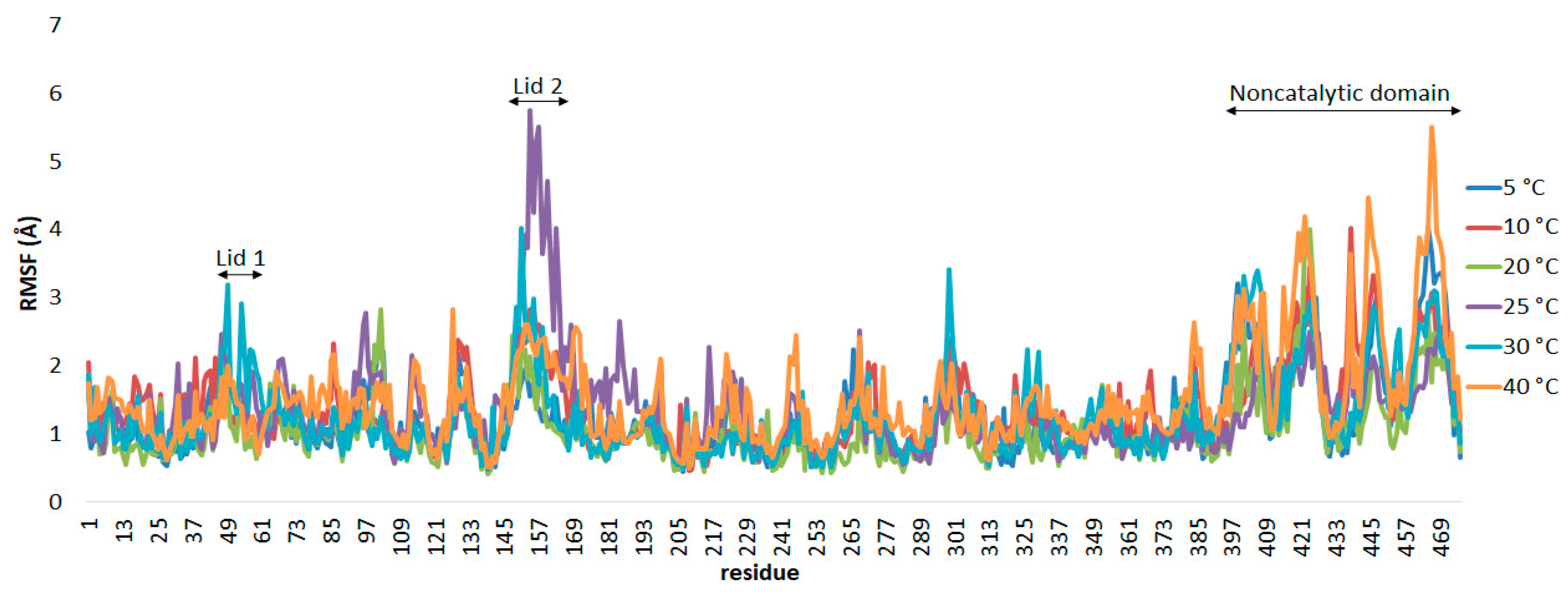
The flexibility of the enzyme was calculated using the root mean square fluctuation (RMSf) values for each residue throughout the whole trajectories.
Figure 4 shows the RMSf per residue for the AMS8 lipase simulated in Triton X-100/toluene reverse micelles at temperatures from 5 to 40 °C. The movement of lid 1 (residues 51–58) can only be seen in the simulation at 30 °C with the high deviation value of the amino acid Leu49 (3.19 Å). The lid 1 displacement was not greatly influenced by the temperature switch mechanism with the presence of reverse micelles.
The conformational transition of lid 2 (residues 148–167) upon contact with the Triton X-100/toluene reverse micelles was clearly perceived during the simulation, as indicated by the RMSf values, which were significantly larger than that of the entire protein. At temperatures of 10 and 20 °C, lid 2 was displaced via the polar residue Ser154, with deviations of 2.832 Å and 2.141 Å, respectively. At 25 °C, an Ile151–Thr152 peptide bond was broken, which leads to a higher RMSf value of 3.133–3.918 Å. The whole protein changed significantly beyond the optimum temperature, from 25 to 40 °C, as the heat energy absorbed disrupted protein bonds.
The terminal region is considered to be more flexible than other components of the protein [
47]. This can be clearly observed in the water simulation, where the AMS8 lipase gave high RMSf values, particularly in the C-terminal region and the coil conformation (residues 390–476). However, at 25 °C, this region has been significantly reduced. Instead, the lid 2 domain gave the highest RMSf values. In this region, the presence of high glycine residues of repeats-in-toxin (RTX) repeats contributes to the flexibility of the protein structure [
48]. The flexibility of the noncatalytic domain in the reverse micelles was reduced by about half as compared to the reverse micelle-free system, particularly at 25 °C and above. This showed that the stability of the non-catalytic domain improved in the reverse micelles.
In addition, the cold-adapted organic solvent tolerant AT2 lipase and a mutant demonstrated high RMSf values in the N-terminal and C-terminal moieties in water simulations at 25 and 45 °C [
49]. The same trend was found for the thermostable ARM lipase and a mutant, indicating high mobility in the N-terminal region at high temperature [
50]. For enzymes simulated in an organic solvent alone, more fluctuations were observed compared to in the reverse micelles. Residues 342–358 and residues 399–424 in the C-terminal region, and residues 78–93 in the N-terminal region of the AMS8 lipase, have lower RMSf values in the reverse micelles than in toluene alone. This shows that the AMS8 lipase is more stable in the reverse micelles than in the organic solvent alone.
3. Materials and Methods
3.1. Materials
Polyethylene glycol p-(1,1,3,3-tetramethylbutyl)-phenyl ether (Triton X-100) and toluene were purchased from R&M Chemicals (Essex, UK). Sodium citrate (C6H5Na3O7·2H2O), Tris buffer, disodium hydrogen phosphate (Na2HPO4), sodium dihydrogen phosphate (NaH2PO4), sodium chloride (NaCl), potassium chloride (KCl), calcium chloride (CaCl2), sodium hydroxide (NaOH), copper(II) sulphate (Cu2SO4), urea, p-Nitrophenol (pNP), and p-nitrophenol palmitate (pNPP) were obtained from Sigma-Aldrich (St. Louis, MO, USA).
DMSO and Arabic gum were sourced from Friendemann Schmidt (Oseterode am harz, Germany). Luria-Bertani (LB) broth and Folin–Ciocalteu reagent were obtained from Merck (Darmstadt, Germany). Isopropylthio-β-galactoside (IPTG) and ampicillin were purchased from Calbiochem (Billerica, MA, USA). The homogenizer was obtained from Cole-Parmer (Vernon Hills, IL, USA).
3.2. Preparation of the Recombinant Cold-Adapted Lipase AMS8
The stock culture of
Escherichia coli strain BL21 (DE3) harboring the recombinant plasmid pET32b/AMS8 was stored in 80% (
v/
v) glycerol and kept at −80°C. The culture was streaked onto LB agar containing ampicillin and incubated at 37 °C for 12 h. A single colony of
E. coli was selected and grown in LB broth containing ampicillin (50 µg/mL) at 37 °C for 16–18 h with agitation at 240×
g. The production of the recombinant AMS8 lipase was confirmed by streaking on a Tributyrin LB agar plate containing ampicillin. The plate was incubated at 37 °C overnight and later the plate was later transferred to 4 °C for 1–3 days. A single colony was grown in LB broth (50 mL) containing ampicillin (50 µg/mL) for the expression. IPTG (100 mM) was added to initiate the expression and incubated for another 16–18 h. The bacterial cells were harvested into a 50-mL Falcon tube and centrifuged at 6654×
g, using a centrifuge at 4 °C (Sigma 3–18K Centrifuge, Washington, DC, USA), for 30 min. The cell pellets were resuspended in 50 mM sodium phosphate buffer with 100 mM NaCl at pH 7 and lysed by sonication (Branson Sonifier 450) for 6 min. The disrupted cell suspension was centrifuged at 6654×
g for 30 min to separate the supernatant from the pellet. The pellet was collected and underwent treatment with 8 M urea, 100 mM NaCl, and 10 mM CaCl
2 for solubilization at 4 °C for 1.5 h. Then, the sample was centrifuged at at 6654×
g for 30 min and the supernatant was subjected to subsequent renaturation by using the 10-fold dilution method [
28].
3.3. Reverse Micellar Extraction (RME)
The extraction of the AMS8 was performed using the reverse micellar extraction method reported by Thenmozhi and Basheer (2013) with some modifications [
51]. RME consists of a two-step procedure which is forward extraction and backward extraction. Forward extraction involves transferring target biomolecules from an aqueous solution (refolded lipase) to a reverse micellar phase (organic phase) and a subsequent step (backward extraction) involves releasing biomolecules from reversed micelles to the aqueous phase (stripping solution) [
22] The organic phase is a mixture of the nonionic surfactant (Triton X-100) and an organic solvent (toluene). The refolded AMS8 lipase was transferred into the organic phase as a forward extraction. The total protein of refolded AMS8 lipase was standardized at about 30 mg. Then, 10 mL of refolded AMS8 lipase (starting sample) was injected into the organic phase (volume ratio 1:1) and homogenized thoroughly for 15 min at 601×
g. The mixture underwent phase separation by centrifugation for 30 min at 3607×
g. In the backward extraction, a fresh stripping solution, 50 mM Tris-HCl containing 1 M KCl at pH 9, was injected into the emulsion, stirred for 30 min at 601×
g, and subjected to the centrifugation for 45 min at 3607×
g.
Several parameters of the RME were optimized to obtain a better lipase recovery. In the forward extraction, the initial aqueous phase (pH 5–9), surfactant concentration (0.01–0.09 M), salt concentration (0.075–0.175 M), and temperature (10–40 °C) were optimized. The lipase activity and the protein content were estimated.
3.4. Enzyme Assay
A spectrophotometric method was used to measure the lipase activity using pNPP as a substrate [
52]. With some modifications, 400 µL of the 23.96 M pNPP solution was added dropwisely into 3 mL of 0.05 M sodium phosphate buffer (pH 7), 0.1 g Arabic gum, and 0.4 g Triton X-100 with stirring. The lipase extract (50 µL) was added to the reaction mixture and assayed at 20 °C for 5 min. A 10% (
w/
v) SDS solution was used to demulsify the reaction mixture from cloudiness. Liberation of pNP was measured at 410 nm using a Biochrom WPA UV/visible spectrophotometer (Cambridge, UK). The control absorbance was deducted from the sample absorbance. One unit (U) of lipase activity was defined as 1.0 nmol of pNP released per min under the conditions stated above.
3.5. Estimate of Protein Content
The estimate of protein content was adapted from Chandra et al. (1975) using bovine serum albumin (BSA) as a standard [
53]. A BSA standard curve was plotted using a protein solution in the range of 0–2000 µg of protein. For the sample, 1 mL was added to 5 mL of the alkaline copper reagent and vortexed to mix well. After 10 min, 0.5 mL of 1 N Folin–Ciocalteu reagent was mixed and left for detergent precipitation for at least 20 min. Then, it was centrifuged at 1202×
g for 10 min, and the absorbance was measured at 700 nm against a reagent blank.
3.6. Activity Recovery Estimation
The activity recovery of the extracted lipase was calculated according to the following equation:
3.7. Computational Analysis
3.7.1. Protein Sequence and Predicted Model of the AMS8 Lipase
The three-dimensional (3D) structure of cold-adapted lipase isolated from
Pseudomonas fluorescens strain AMS8 (GenBank accession No: ADM87309.1) was used in the molecular dynamic simulations. The predicted model of the AMS8 lipase was acquired by superimposing the crystal structures of the Serratia marcescens lipase (PDB ID: 2qua) and the
Pseudomonas sp. MIS38 lipase (PDB ID: 2z8x) due to their similar sequence identity [
54].
3.7.2. Simulation Parameters
The MD simulations were performed on the predicted model of the AMS8 lipase in the closed conformation with the calcium ions and zinc ion intact. The AMS8 lipase was used as the starting structure to analyze conformational changes in AMS8 in the presence of hydrocarbon (toluene and Triton X-100) micelles and water. The PDB coordinates for toluene and Triton X-100 were generated from PUBCHEM (
https://pubchem.ncbi.nlm.nih.gov/compound/). A varied simulation box (X = 77.49–95.05 Å, Y = 70.98–75.48 Å, Z = 66.30–66.78 Å) was filled with the toluene density appropriate to the temperature and Triton X-100 (100 molecules) was added randomly around AMS8 together with water and NaCl. The MD simulations were completed with the YASARA Structure (version 11.3.22, Krieger, Vienna, Austria) software [
55,
56]. Energy minimizations were performed using the steepest descent method for about ~2000 steps before the simulation started with the fixed AMS8 lipase model. The protein-solvent-surfactant simulation systems were executed at 278–313 K and 1 bar pressure under the steepest descent parameters. The total system size was ~45,000 atoms with all the molecules and AMS8 lipase filled in the simulation box. The AMBER03 force field with a cut off of 10.846 Å was used in the simulation. The lipase was studied using 400 saved simulation steps, with 40 picoseconds, representing 1 nanosecond. The MD simulations were permitted to run until 10 ns.
3.7.3. Simulation Analysis
The stability and flexibility of a residue in the enzyme at a given simulated time were obtained from the root mean square deviation (RMSd) and the root mean square fluctuation (RMSf). These revealed deviations from the native enzyme structure, showing an interruption of the molecular forces of the molecules. The α-helix and β-sheet, as well as the distance between the lid domains and the catalytic residues, were obtained from the simulation. The protein simulation snapshots in the Triton X-100/toluene reverse micelles obtained from various timelines were aligned in a parallel arrangement (by superimposing the structures), showing the movement of the lid, using the open-source PyMOL Molecular Graphics System Version 1.1 (2009) software developed by Schrödinger, Inc (
https://pymol.org/2/).