Abstract
Benzoylformate decarboxylase (BFDC) and pyruvate decarboxylase (PDC) are thiamin diphosphate-dependent enzymes that share some structural and mechanistic similarities. Both enzymes catalyze the nonoxidative decarboxylation of 2-keto acids, yet differ considerably in their substrate specificity. In particular, the BFDC from P. putida exhibits very limited activity with pyruvate, whereas the PDCs from S. cerevisiae or from Z. mobilis show virtually no activity with benzoylformate (phenylglyoxylate). Previously, saturation mutagenesis was used to generate the BFDC T377L/A460Y variant, which exhibited a greater than 10,000-fold increase in pyruvate/benzoylformate substrate utilization ratio compared to that of wtBFDC. Much of this change could be attributed to an improvement in the Km value for pyruvate and, concomitantly, a decrease in the kcat value for benzoylformate. However, the steady-state data did not provide any details about changes in individual catalytic steps. To gain insight into the changes in conversion rates of pyruvate and benzoylformate to acetaldehyde and benzaldehyde, respectively, by the BFDC T377L/A460Y variant, reaction intermediates of both substrates were analyzed by NMR and microscopic rate constants for the elementary catalytic steps were calculated. Herein we also report the high resolution X-ray structure of the BFDC T377L/A460Y variant, which provides context for the observed changes in substrate specificity.
1. Introduction
The decarboxylases form the largest group of thiamin diphosphate (ThDP)-dependent enzymes [1]. The archetypal member of this group is pyruvate decarboxylase (PDC), which catalyzes the nonoxidative decarboxylation of pyruvate to yield acetaldehyde and carbon dioxide, a reaction critical to the fermentation pathway of several yeast and bacteria [2]. X-ray structures of PDCs from a variety of species show that, in addition to ThDP, the active site contains two ionizable acidic residues as well as two contiguous histidine residues that are located on an ordered loop [3,4,5,6]. The latter has been termed the HH-motif, and mutagenesis and kinetic studies have revealed that both histidines play significant roles in catalysis [7,8,9,10]. X-ray structures of several other ThDP-dependent decarboxylases reveal that, even though a variety of residues may line the substrate-binding pocket, thereby explaining the observed differences in substrate specificity, most possess the HH-motif and the two acidic residues [11,12,13].
Benzoylformate decarboxylase (BFDC) is the penultimate enzyme in the mandelate pathway, a secondary metabolic pathway that allows various pseudomonads to grow using R-mandelate as their only source of carbon [14,15,16]. Also a ThDP-dependent enzyme, BFDC catalyzes the nonoxidative decarboxylation of benzoylformate, generating benzaldehyde and carbon dioxide [14]. Given the similarity of their reactions, and their using the same cofactor, it may have been expected that the active sites of PDC and BFDC would be similar. However, this proved not to be the case, for the active site of BFDC was found to contain two leucine residues occupying positions equivalent to the HH-motif [17]. That said, the active site of BFDC does contain two histidine residues (His70 and His281) and mutagenesis studies suggest that they too are catalytically important [18,19]. Another difference was that the BFDC active site lacked the two acidic residues found in the HH-motif family. Instead, BFDC’s only other ionizable residue is a serine, Ser26, which has also been implicated in the catalytic mechanism [18].
Although their active sites and substrate preference may differ, the catalytic cycles of BFDC and PDC can still be broken into the same series of steps (Scheme 1) [20]. The first two steps are the deprotonation of the C2 carbon of ThDP to generate either an ylid [16,21,22] or a carbene [23], followed by non-covalent binding of the 2-keto substrate to provide the Michaelis complex. Addition of the C2 carbanion/carbene of ThDP to the carbonyl of the substrate gives rise to the first covalent tetrahedral intermediate, which subsequently loses carbon dioxide resulting in the formation of carbanion/enamine intermediate typical of all ThDP-dependent enzymes. In decarboxylases such as BFDC and PDC, protonation of this enamine generates a second tetrahedral intermediate, which finally breaks down, releasing the product aldehyde and regenerating ThDP.
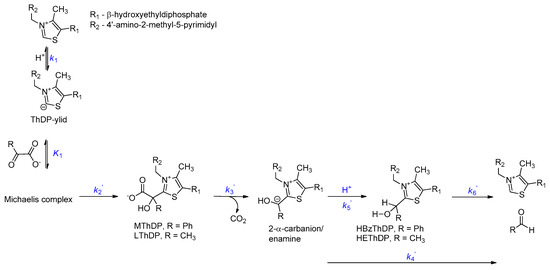
Scheme 1.
Mechanism of benzoylformate decarboxylase (BFDC) and pyruvate decarboxylase (PDC) with intermediates and net rate constants.
Over the past few years, the steady-state distribution of the covalent intermediates has been used to determine net rate constants for the elementary catalytic steps of several ThDP-dependent enzymes including PDC [24] and BFDC [25]. For the former, decarboxylation and product release were found to be partially rate-limiting, but formation of lactyl-ThDP (LThDP), the first covalent intermediate, was quite rapid [24]. Conversely, the rate-limiting step for BFDC was found to be the formation of the first covalent intermediate, mandelyl-ThDP (MThDP) [18,25]. Further, BFDC was found to decarboxylate its first covalent adduct an order of magnitude faster than any other ThDP-dependent enzyme studied to date [25].
As with many ThDP-dependent decarboxylases, BFDC and PDC are both are capable of stereoselective carboligation reactions leading to chiral α-hydroxy ketones [26]. These have been used as versatile building blocks in the pharmaceutical (and other) industries [26,27,28]. As a consequence there is considerable interest in expanding the capabilities of the decarboxylases to accept alternative substrates. While both BFDC and PDC exhibit some substrate promiscuity neither enzyme is particularly efficient at decarboxylating the other’s in vivo substrate [29]. Recently, as part of our ongoing efforts to understand the evolution of substrate specificity in ThDP-dependent enzymes, we used site-saturation mutagenesis in an attempt to convert a BFDC into a PDC [30]. The BFDC T377L/A460Y variant was observed during screening to have greatly enhanced PDC activity. Subsequently it was shown to have a 10,000-fold increase in its substrate utilization ratio between pyruvate and benzoylformate compared to that of wtBFDC [30].
Here we describe our efforts to identify the reasons for that change in specificity. In the first instance, we used NMR spectroscopy to analyze the intermediates obtained in the reaction of BFDC T377L/A460Y with both benzoylformate and pyruvate. The microscopic rate constants were calculated for the elementary catalytic steps in the reaction of both substrates. Further, in an attempt to understand the structural basis for the altered substrate specificity exhibited by the T377L/A460Y variant, its high resolution X-ray structure was determined.
2. Results and Discussion
2.1. Substrate Profile of T377L/A460Y
Our earlier study compared the reactions of wtBFDC, ScPDC and the T377L/A460Y variant only with benzoylformate and pyruvate. Here the comparison was extended to include a variety of straight- and branched-chain 2-keto acids, as well as phenylpyruvate (Table 1). The steady-state kinetic constants for reaction of wtBFDC with pyruvate could only be determined under V/K conditions, i.e., at substrate concentrations well below the Km value, which was estimated to be easily in excess of 100 mM [30]. By contrast the T377L/A460Y variant has a Km value for pyruvate of 4.6 mM, which is comparable to the pyruvate S0.5 value for ScPDC. In addition to improving utilization of pyruvate, the double variant was found to decarboxylate several other aliphatic 2-keto acids at a faster rate than wtBFDC. While it was most efficient with 2-ketohexanoate and 4-methylthio-2-ketobutanoate as substrates, there was only a 60-fold difference in kcat/Km values between the best substrate, 2-ketohexanoate, and its least efficient substrate, pyruvate. In many ways pyruvate is still the exception as its Km value was significantly higher than those of other aliphatic substrates. With regards to kcat, there is less than a 15-fold decrease between the substrate that is turned over the fastest, 3-methyl-2-ketopentanoate, and the substrate that the T377L/A460Y variant decarboxylates the slowest, 2-ketopentanoate. With the exception of benzoylformate, the T377L/A460Y variant was able to decarboxylate its substrates more efficiently than wtBFDC. Similarly, only pyruvate and its longer chain analogues, 2-ketobutanoate and 2-ketopentanoate were better substrates for ScPDC.

Table 1.
Kinetic characterization of wtBFDC, BFDC T377L/A460Y and ScPDC at pH 6.0 a.
Overall, the data in Table 1 clearly demonstrate that the T377L/A460Y variant is able to decarboxylate a wide range of aliphatic substrates and, in most cases, more efficiently that either wtBFDC or ScPDC. That said, it is also apparent that the ability of this variant to decarboxylate either benzoylformate or pyruvate is considerably less than that of wtBFDC or ScPDC, respectively. The similarity of substrate Km values suggests that substrate binding is not likely to be the issue, rather the changes in active site geometry are having a significant effect on one or more of the individual catalytic steps.
2.2. Intermediate Distribution Analysis for Reaction of T377L/A460Y with Benzoylformate
To explore this possibility the microscopic rate constants for the BFDC T377L/A460Y variant were determined for the decarboxylation of both benzoylformate and pyruvate. These have been determined previously for the reaction of wtBFDC with benzoylformate [25]. In that study it was shown that the rate-limiting step was the formation of MThDP by addition of the ThDP ylid to benzoylformate, and that BFDC carried out its decarboxylation step at least two orders of magnitude faster than any other ThDP-dependent enzyme [25].
All three major intermediates, unreacted ThDP, MThDP, and 2-hydroxybenzyl-ThDP (HBzThDP), can be detected at steady-state when T377L/A460Y reacts with benzoylformate (Figure 1). While the MThDP signal is close to the limits of resolution, it is clearly visible. Following correction for non-enzymatic decarboxylation of MThDP [25] analysis reveals that ~37% of the active sites contain C2-unsubstituted ThDP and 3% contain MThDP. The remaining 60% are occupied by HBzThDP. This is distinctly different to the result obtained with wtBFDC where a similar analysis provided values of 79%, 3% and 18% for ThDP, MThDP and HBzThDP, respectively [25]. A comparison of the derived kinetic constants for wtBFDC and T377L/A460Y can be seen in Table 2. In wtBFDC addition of the ThDP ylid to benzoylformate (i.e., C–C formation, k2′) is clearly rate limiting. Conversely, protonation of the enamine/elimination of benzaldehyde (k4′) has been reduced ~100-fold in the T377L/A460Y variant and has now become rate limiting. It is notable that for both variants decarboxylation of MThDP (k3′) is significantly faster than other steps in the reaction (Table 2). Further, even though the rate for T377L/A460Y has decreased considerably compared to that of the wild-type enzyme it is still faster than decarboxylation of LThDP by pyruvate-utilizing enzymes [20,31,32].
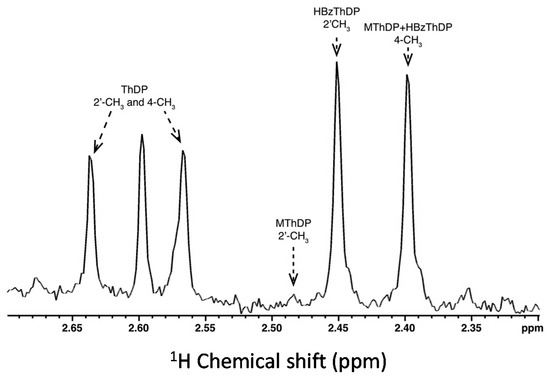
Figure 1.
The 2′-CH3 and 4-CH3 singlet signals of thiamin diphosphate (ThDP) and of benzoylformate-conjugated covalent intermediates were used for the assignment and quantitative analysis of the following intermediates: ThDP (2.64 and 2.57 ppm), mandelyl-ThDP (MThDP) (2.48 and 2.41 ppm), and 2-hydroxybenzyl-ThDP (HBzThDP) (2.45 and 2.40 ppm). Intermediates and cofactor were isolated from the protein by acid quench after a reaction time of 1 s and analyzed by 1H NMR at pH 0.75 at 25 °C as described in Materials and Methods. Relative concentrations of intermediates were determined from the relative integrals of the corresponding signals, which were used to calculate all microscopic rate constants of the catalytic cycle.

Table 2.
Net rate constants (s−1) for reaction of BFDC and PDC variants with benzoylformate and pyruvate.
2.3. Intermediate Distribution Analysis for Reaction of T377L/A460Y with Pyruvate
The microscopic rate constants for reaction of wtBFDC with pyruvate could not be determined as only trace amounts of the reaction intermediates were observable at steady-state using the NMR-based approach. This is not surprising as activity of wtBFDC with pyruvate could only be measured under V/K conditions [30]. The lack of any observable reaction intermediates, coupled with a pyruvate Km value in excess of 100 mM, indicates that the rate-limiting step is the initial formation of the Michaelis complex and/or the subsequent C–C bond formation (k2′) that provides LThDP.
For T377LA460Y the steady-state distribution of intermediates showed that, when pyruvate is used as substrate (Figure 2), all the key catalytic intermediates are present: ThDP, 2-lactyl-ThDP (LThDP) and 2-hydroxyethyl-ThDP (HEThDP). This finding indicates that all major steps of catalysis are on the same time scale. In addition to those anticipated intermediates, a minor fraction of 2-acetolactate-ThDP (ALThDP) was observed [31].
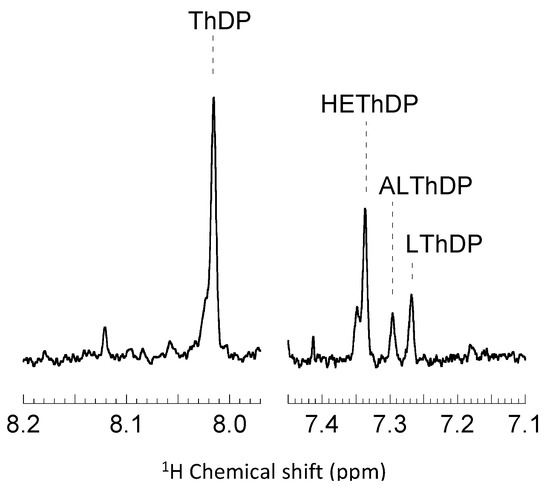
Figure 2.
The C6′-H singlet signals of ThDP (thiamin diphosphate) and of pyruvate-conjugated covalent intermediates were used for the assignment and quantitative analysis of the following intermediates: ThDP (8.01 ppm), LThDP (lactyl-ThDP) (7.27 ppm), HEThDP (2-hydroxyethyl-ThDP) (7.34 ppm) and ALThDP (2-acetolactate-ThDP) (7.30 ppm). Intermediates and cofactor were isolated by acid quench after a reaction time of 8 s and analyzed by 1H NMR at pH 0.75 at 25 °C as described in Materials and Methods. Relative concentrations of intermediates were determined from the relative integrals of the corresponding signals, which were used to calculate all microscopic rate constants of the catalytic cycle.
ALThDP arises during an off-pathway reaction in which a second molecule of pyruvate acts as the electron acceptor for the post decarboxylation carbanion intermediate. ALThDP has previously been observed during the determination of the microscopic rate constants for acetohydroxyacid acid synthase [34,35,36]. The presence of ALThDP during the reaction cycle of the BFDC T377L/A460Y variant suggests that the reaction may generate acetolactate. However, the only carboligation product detected was acetoin. While this seems at odds with the presence of ALThDP, Beigi et al. [37] recently described an α-hydroxy-β-keto acid rearrangement-decarboxylation reaction in which acetolactate spontaneously decarboxylates to yield acetoin. Currently, it is unclear if ALThDP decarboxylates to yield acetoin or if the decarboxylation of acetolactate occurs after it is released. Regardless, it would seem that the longer lifetime of the pyruvate-derived carbanion/enamine results in an increase in formation of a carboligation product.
If this off-pathway reaction is ignored, then the calculation of the forward net rate constants is possible. These are provided in Table 2. Examination of those rate constants shows that C–C bond formation (k2′) is the slowest step in the reaction of BFDC T377L/A460Y with pyruvate. Further, while the variant is clearly more efficient in binding pyruvate, the rate constant for the formation of the LThDP (6 s−1) is still 50- to 400-fold lower than those of the true PDCs. By comparison, decarboxylation of the LThDP intermediate (k3′) is ca. four-fold faster, and the subsequent steps involving enamine protonation/product release (k4′) are, at best, partially rate-limiting. These results are in sharp contrast with those found in previous studies with ZmPDC [24] and ScPDC [33] where formation of LThDP was the fast step and both decarboxylation and product release were appreciably slower (Table 2). That said, it is not without precedent, as initial C–C bond formation (k2′) is the slow step in the reaction of acetohydroxyacid synthase (AHAS) with pyruvate [32]. Finally, it is notable that decarboxylation of LThDP catalyzed by BFDC T377L/A460Y is the fast step in the overall process, and only approximately four-fold slower when compared to ScPDC. It seems that, even with a relatively poor substrate, the active site of BFDC seems to favor decarboxylation over the other reaction steps.
2.4. X-ray Structure of BFDC T377L/A460Y
In an attempt to understand how the BFDC T377L/A460Y variant resulted in a better binding site for pyruvate and, potentially, to explain the decrease in the rate of decarboxylation of benzoylformate, the X-ray structure for T377L/A460Y was determined. The structure was solved to a resolution of 1.56 Å and the coordinates have been deposited with the Protein Data Bank as PDB ID 4MZX. The refinement statistics are provided in Table 3.

Table 3.
Data, model, and crystallographic statistics for BFDC T377L/A460Y structure a.
The overall fold was found to be almost identical to that of wtBFDC holoenzyme, and both the space group and cell dimensions were also the same. There was a RMSD deviation of 0.28 Å between the BFDC T377L/A460Y variant and wtBFDC enzyme for the Cα atoms of a single monomer. As previously observed with BFDC structures, the region comprising residues 461–470 had the highest B-factors found in the protein.
During the initial rounds of refinement, positive electron density adjacent to the C2 atom of the thiazolium ring was observed when ThDP was modeled into the active site. This was indicative of the oxidation of ThDP to thiamin thiazolone diphosphate (TZD). Consequently TZD was modeled into the active site during subsequent rounds of refinement. This oxidation of ThDP to TZD has been observed previously in BFDC, as well as other ThDP-dependent enzymes, and has been attributed to radiation damage [38,39]. As seen in several earlier BFDC structures, the diphosphate tail of TZD is coordinated to Ca2+, rather than Mg2+. Presumably due to its high concentrations in the crystallization buffer, Mg2+ is often displaced by Ca2+ during the crystallization of BFDC [17,38]. In addition to the cofactors, electron density indicative of two glycerol molecules was also observed in the asymmetric unit. One glycerol was H-bonded to the side chains of Arg294 and Asp312, while the second was coordinated to the oxygen atoms of the backbone carbonyl of Asp477 and the side chain of Gln443. Finally, a sodium ion was found coordinated between the main chain carbonyl oxygens of Leu118, Arg120, and Asn117 from two separate monomers at the dimer–dimer interface. The BFDC T377L/A460Y structure retained the backbone conformation of the wtBFDC enzyme at the points of the mutations, but, not surprisingly, additional electron density was seen in the difference maps. This density corresponded to leucine and tyrosine residues at positions 377 and 460, respectively.
Comparison of the active sites of the wtBFDC and the T377L/A460Y variant showed that the side chain of His281 is displaced by more than 1 Å compared to the wtBFDC holoenzyme (Figure 3). The only other notable change was Leu110 being found in two rotamer conformations, one of which had not previously been observed in BFDC or its variants [17,18,25,38,40,41]. The net result was that the active site was reduced in volume by ~20%.
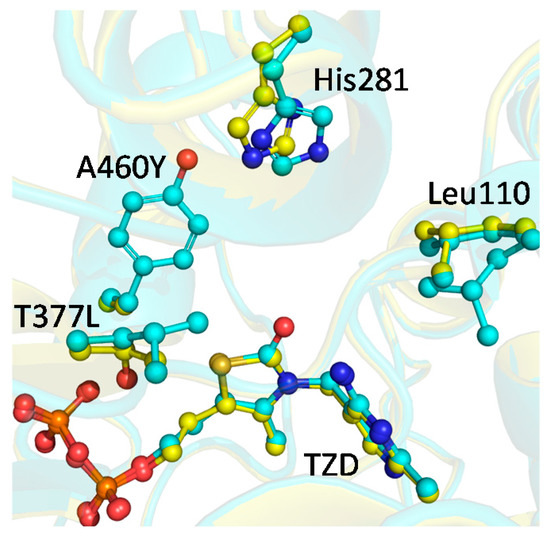
Figure 3.
Superposition of the active sites of wtBFDC (yellow) and the BFDC T377L/A460Y variant (cyan) reveals a shift in the position of His281 accompanied by multiple conformations of Leu110. In addition, ThDP was oxidized to thiamin thiazolone diphosphate (TZD). Figure prepared using PyMOL (Schrödinger, Inc., Portland, OR USA) using data from PDB ID 1BFD (wtBFDC) and 4MZX (BFDC T377L/A460Y).
The data in Table 1 show that the Km value of wtBFDC for pyruvate was in excess of 100 mM, and for the T377L/A460Y variant, the Km value was reduced to <5 mM. Clearly the latter must have a binding site much better able to accommodate pyruvate. To see how this was achieved we compared the binding of the LTHDP to PDC and to BFDC T377L/A460Y. Previously, the ZmPDC E473D structure in complex with LThDP was solved [42]. In this structure the methyl substituent of the lactyl moiety is within 5 Å of 11 atoms evenly distributed across seven different residues.
Superimposition of the LThDP intermediate on ThDP in the active site of wtBFDC revealed only three atoms within 5 Å of the methyl group (Figure 4A). By contrast, superimposition of LThDP on TZD in the T377L/A460Y structure (Figure 4B) shows that this variant now has 10 atoms contributing to the methyl-binding site. Thus the active site has become more like that of ZmPDC. There were some differences for, unlike the evenly distributed interactions which make up the ZmPDC methyl-binding pocket, 7 of the 10 potential interactions in the BFDC T377L/A460Y variant arise from the engineered residues, Leu377 and Tyr460. This suggests that changes in other active site residues may be able to further improve pyruvate binding and, potentially, in positioning the “new” substrate for decarboxylation and subsequent steps.
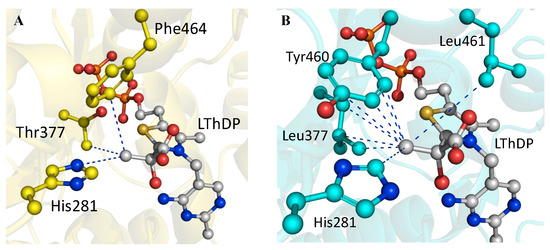
Figure 4.
(A) wtBFDC (PDB: 1BFD) with LThDP (gray) placed into the active site by superposition of the LThDP on the corresponding atoms of ThDP. In total, three atoms are within 5 Å of the methyl group of LThDP; (B) BFDC T377L/A460Y PDB 4MZX) with LThDP (gray) superimposed on TZD. In total 10 atoms, from four residues, are within 5 Å of the methyl group of LThDP. In both structures blue lines indicate distances of <5 Å. Figure prepared using PyMOL with LThDP data from PDB ID 3OE1 [42].
While this relatively simplistic modeling of LThDP offers an explanation for how the active site of the T377L/A460Y variant has improved pyruvate binding, there are still several issues to be addressed. For example, while the active site volume has been reduced by 20% from wtBFDC, the Km value for benzoylformate is only increased by a factor of two. Indeed, at 0.45 mM, it remains 10-fold lower than that of pyruvate (Table 2). To understand how this could be achieved, the tetrahedral intermediate from the reaction of wtBFDC with methyl benzoylphosphonate (MBP) [41] was superimposed on the TZD in the active site of T377L/A460Y. The MBP-ThDP intermediate is an analogue of MThDP, thus can be used to get an indication of potential changes in benzoylformate binding.
This simple model indicates a potentially severe steric clash between the phenyl group of the inhibitor and the engineered mutations (Figure 5). The γ-methyl of Leu377 is within <2.5 Å of several carbon atoms of the phenyl ring of MBP, whereas Tyr460 has a potential edge-to-edge contact with MBP that is also within <2.5 Å. While this appears to be at odds with the kinetic data indicating only a modest effect on Km value for benzoylformate, there is structural evidence that wtBFDC is able to accommodate unnaturally large substrates by slight rotations of active site residues [43]. Therefore it is not unreasonable to suggest that the side chains of Tyr460 and Leu377 could rotate to avoid or, at least, ameliorate unfavorable interactions with the MThDP. Equally plausible is that the phenyl group of the MThDP could rotate to prevent steric clash with these residues. Either scenario provides a rationale for the binding of benzoylformate being relatively unaffected.
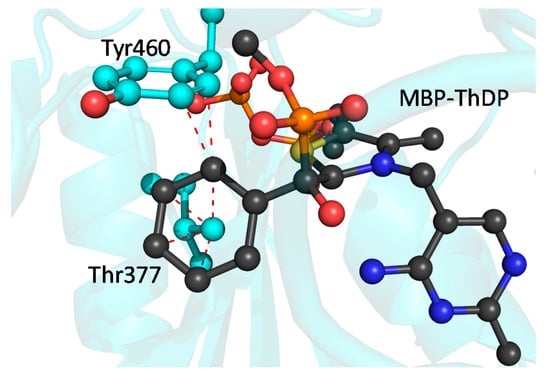
Figure 5.
BFDC T377L/A460Y with MBP-ThDP (black) placed into the active site. Red lines indicate distances of <2.5 Å. Superposition carried out with PyMOL as described in Materials and Methods using MBP-THDP data from PDB ID 3FSJ [41]
Figure 5 may also provide a partial explanation for the reduction in net rate constants for the decarboxylation of benzoylformate (Table 2). It is clear that some active site rearrangements will be necessary to avoid potential steric clashes and permit substrate binding. Conceivably, these may make it more difficult for the ThDP ylid to attack the carbonyl of benzoylformate, thereby explaining the observed 12-fold decrease in the rate of formation of the MThDP intermediate (k2′).
Figure 3 shows that the mutations have caused modest, but potentially significant changes in the orientation of Leu110 and His281. Together these changes provide some insight into the considerable decrease in the observed rate for the decarboxylation of the MThDP intermediate (k3′), as well as the release of benzaldehyde (Scheme 1). While its precise role in catalysis is not obvious, mutagenesis and crystallographic data suggest that Leu110 plays a vital role in the mechanism of BFDC. The L110A mutation results in greater than four-orders of magnitude decrease in kcat/Km, due to a simultaneous increase in Km and decrease in kcat values [44]. Analysis of the structure of wtBFDC in complex with MBP indicates that Leu110 may be responsible for positioning the glyoxylate moiety of benzoylformate [25,41]. In the T377L/A460Y variant, Leu110 adopts multiple conformations potentially reducing its ability to lock the glyoxylate moiety into the perpendicular arrangement of the carboxylate group to the thiazolium-C2α bond. This geometry is thought to promote decarboxylation by allowing maximum overlap of π electrons of the thiazolium ring and the p-orbital of the scissile bond [25,45]. The conformation has been observed in the X-ray structures of all the first tetrahedral intermediates, and intermediate analogues [25,42,46,47,48]. Further, Ser26 has been proposed to assist in the decarboxylation step by carrying out a nucleophilic attack on carboxylate group of MThDP [40]. In wtBFDC, not only does the perpendicular arrangement assure maximum orbital overlay, but also it positions the hydroxyl group of Ser26 within striking distance of the carboxylate group. Taken together, the alternative conformations of Leu110 observed may help explain the decrease in the rate of MThDP decarboxylation (k3′).
The net rate constant associated with product release (k4′) incorporates the protonation of the enamine/carbanion intermediate, as well as the breakdown of the resultant tetrahedral intermediate and subsequent release of benzaldehyde. The T377L/A460Y variant exhibits a 100-fold decrease in k4', i.e., the largest decrease of all net rate constants. Mutagenesis studies suggest that His281 is involved in the protonation of the enamine/carbanion intermediate [18], whereas His70 has been implicated in the abstraction of the proton from the HBzThDP intermediate, thereby facilitating product release [18,19]. Since these two steps of the mechanism are incorporated into this single net rate constant, it is difficult to say which step has been more affected in the T377L/A460Y variant. The X-ray structure shows that the position of His70 is largely unaffected by the mutations but that there is a 1 Å displacement of the imidazole ring of His281 (Figure 3). It is conceivable that this change could reduce proton transfer efficiency, resulting in an increase in the lifetime of the carbanion/enamine. It is well known that the BFDC H281A variant has been associated with an improvement in carboligation efficiency [49,50,51], which was attributed to the longer lifetime of the enamine. It is also notable that an increase in the lifetime of the carbanion/enamine in ZmPDC gave rise to formation of acetoin/acetolactate [52]. Given that the 1H NMR spectrum of the intermediates in the reaction of T377L/A460Y with pyruvate (Figure 2) showed the presence of 2-acetolactate-ThDP, it is reasonable to speculate that displacement of His281 is contributing to the decrease in k4'. That said, there is a clear increase in the steady-state concentration of HBzThDP, which certainly indicates that its breakdown and product release—i.e., k5' (Scheme 1)—also must be greatly reduced.
3. Materials and Methods
3.1. Materials
Plasmids containing genes for recombinant alcohol dehydrogenase from Equus caballus (HLADH) and C-terminally His6-tagged pyruvate decarboxylase from Saccharomyces cerevisiae were kind gifts from Bryce Plapp (University of Iowa) and Frank Jordan (Rutgers University), respectively. NADH, IPTG, yeast alcohol dehydrogenase (YADH), and the various 2-keto acids were purchased from Sigma-Aldrich (St Louis, MO, USA). Other assay reagents were purchased from either Sigma-Aldrich or Fisher Scientific (Waltham, MA, USA) and were of the highest commercially available grade.
3.2. Protein Expression and Purification
Expression and purification of wtBFDC and the BFDC T377L/A460Y variant was carried out as described in Yep et al. [30]. The protein was exchanged into storage buffer (100 mM KPO4, 1 mM MgSO4, 0.5 mM ThDP, pH 6.0, 10% glycerol v/v), and concentrated using EMD Millipore Amicon Ultra filters (Fisher Scientific). Expression and purification of ScPDC was carried out in a similar manner. In all cases purity was assessed by SDS-PAGE, and the protein concentration was determined by the Bradford method [53] using BSA as the standard.
3.3. Analysis of Substrate Spectrum
The activity assays with the purified enzymes have been described in detail elsewhere [30]. The assay contained the appropriate 2-keto acid substrate, NADH, and HLADH (YADH for short straight-chain aliphatic substrates) in 100 mM potassium phosphate buffer (pH 6.0) with a final volume of 1 mL. The reaction was carried out at 30 oC and was initiated by the addition of the enzyme. Kinetic data were fitted to the Michaelis-Menten equation using SigmaPlot 9.0.1.
3.4. Steady State Analysis of Reaction Intermediates by Acid Quench/NMR Spectroscopy
Prior to the chemical quench/NMR studies, excess ThDP was removed from the BFDC T377L/A460Y sample by dialysis overnight against 50 mM potassium phosphate buffer, pH 6.5, supplemented with 2.5 mM MgSO4. Enzyme concentrations were adjusted to 15 mg/mL and reacted with one equivalent volume of either 50 mM benzoylformate or 100 mM pyruvate prepared in the same buffer as the enzyme. After 1–10 seconds the reaction was quenched by manual mixing with the addition of 1 volume of an acidic solution comprising 65% (v/v) D2O, 25% (v/v) of trichloroacetic acid and 10% (v/v) concentrated HCl [24,25]. Samples were vortexed to facilitate complete quenching and protein denaturation. Samples were then centrifuged and the supernatant passed through a 0.22 µm filter. The filtrate, containing the intermediates, substrates and products, was analyzed by 1H NMR spectroscopy. NMR data collection, assignment of peaks, and calculation of net forward unimolecular rate constants was performed as previously described [24,25].
3.5. Crystallization of BFDC T377L/A460Y
Crystals of the BFDC T377L/A460Y variant were grown by the hanging drop diffusion method under the conditions used for the crystallization of wtBFDC [17]. Storage buffer was exchanged for crystallization buffer (0.1 mM MgCl2, 0.2 mM ThDP, 25 mM NaHEPES pH 7.0). The well solution consisted of 0.1 M Tris (pH 8.5), 0.15 M CaCl2 and 22% PEG 400 (v/v). Equal volumes of protein solution (10 mg/mL) and well solution were pipetted onto a silanized glass slide and mixed. A heavy precipitate quickly formed and single crystals emerged from the precipitate within days. Crystals were transferred to fresh crystallization buffer containing 36% glycerol (v/v) as a cryoprotectant and mounted on Hampton CryoLoops immediately prior to being flash-frozen in liquid nitrogen.
3.6. X-ray Data Collection
Diffraction experiments were carried out at 100 K on the 23-ID-B beamline administered by GM/CA-CAT at the Advanced Photon Source at Argonne National Laboratory. Datasets for the BFDC T377L/A460Y variant were scaled to the I222 space group. Data reduction and processing of dataset was done with HKL2000 software package and the CCP4 suite of programs [54]. Molecular replacement was performed using the search model wtBFDC (PDB 1BFD) with metals and waters removed. The asymmetric unit for each variant contained a single monomer.
3.7. Structure Solutions and Refinements
Refinement was performed using Phenix.refine [55]. After each round of refinement, the electron density was manually inspected and models were built using Coot [56]. Model refinement continued until the free R factor and the crystallographic R factor had converged. Validity of the model was checked using the Molprobity server [57,58]. All images were generated with PyMOL (Schrödinger Inc.).
3.8. Modeling of Reaction Analogues and Intermediates
Using PyMOL and the coordinates from PDB ID 3OE1, the lactyl-ThDP intermediate was placed into the active sites of both wtBFDC (PDB ID: 1BFD) and the T377L/A460Y structure. Further, using the coordinates from PDB ID 3FZN, the phosphonomandelyl-ThDP (MBP-ThDP) was also placed into the active site of the T377L/A460Y structure. In this process, the Cα atoms of the protein structures were first superimposed. Next, the atoms corresponding to the ThDP moiety of lactyl-ThDP and MBP-ThDP were aligned with the corresponding atoms of either ThDP, found in wtBFDC, or the thiamin thiazolone diphosphate (TZD) found in the BFDC T377L/A460Y structure. No energy minimization was carried out.
4. Conclusions
The BFDC variant T377L/A460Y effectively acts as a “pseudo” PDC. This is reflected in a broad substrate spectrum more akin to that of ScPDC than wtBFDC. NMR-based analysis of the steady-state reaction intermediates for the reaction of BFDC T377L/A460Y with pyruvate revealed that the rate-limiting step for this variant is the formation of the LThDP intermediate. This is in contrast to the reactions catalyzed by ZmPDC and ScPDC, where product release is rate-limiting. Comparing the methyl-binding pocket of T377L/A460Y to ZmPDC suggests that both are likely to have a similar number of interactions with the lactyl moiety. However, the contacts within the ZmPDC methyl-binding pocket are evenly distributed across seven residues compared to only four residues in T377L/A460Y. Further, the seven residues of ZmPDC are arranged to surround the methyl group thereby providing steric restraints from every direction. Presumably this arrangement would also allow for the maximum orbital overlap, thereby permitting attack of the ThDP ylid and facilititating the subsequent decarboxylation of the LThDP intermediate. On the other hand, the four residues comprising the T377L/A460Y methyl binding pocket are unevenly arranged and do not provide the precise substrate positioning seen in a native PDC. This likely accounts for the higher Km value for pyruvate, and the lower rate of formation of LThDP observed with the BFDC variant. Given that several aliphatic 2-keto acids are better substrates than pyruvate for T377L/A460Y, it is reasonable to predict that further evolution could well bring the properties of the “pseudo” PDC significantly closer to those of the native enzyme.
Over the years there have been many attempts to interconvert or otherwise alter substrate specificity in ThDP-dependent enzymes. This has been met with a resounding lack of success [1]. The results here provide some rationale for the difficulty. They highlight the fact that, in spite of the similarity in overall mechanism, ThDP-dependent enzymes often have different rate-determining steps. This means that the enzymes have evolved so that individual residues will provide an optimal alignment for (effectively) several substrates (intermediates) in the catalytic cycle. Thus, making a change that may be expected to improve binding for a new substrate may have a deleterious effect on another step in the mechanism. In the BFDC T377L/A460Y variant, for example, not only do we see a new ability to catalyze pyruvate decarboxylation, but it is done without greatly affecting its ability to bind benzoylformate. Rather, the decrease in kcat value for reaction with benzoylformate is due primarily to a change in rate-determining step. This result implies that structure-based redesign of ThDP-dependent enzymes will prove difficult. Rather, a more random approach will be required, probably involving saturation mutagenesis at a combination of sites identified on the basis of X-ray structures or homology models.
Acknowledgments
This work was supported by the National Science Foundation (CHE 1306877to M.J.M.) and the Deutsche Forschungsgemeinschaft (FOR1296 to K.T.). Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract DE-AC02-06CH11357. GM/CA @ APS has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104).
Author Contributions
F.H.A. and M.J.M conceived and designed the experiments; F.H.A and M.P.R carried out the enzyme kinetic experiments; F.H.A., C.W. and D.M. carried out the NMR experiments while K.T. analyzed the NMR data; F.H.A carried out the crystallography experiments and analyzed the data; F.H.A and M.J.M. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Andrews, F.H.; McLeish, M.J. Substrate specificity in thiamin diphosphate-dependent decarboxylases. Bioorg. Chem. 2012, 43, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Nichols, N.; Dien, B.; Bothast, R. Engineering lactic acid bacteria with pyruvate decarboxylase and alcohol dehydrogenase genes for ethanol production from Zymomonas mobilis. J. Ind. Microbiol. Biotechnol. 2003, 30, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Dyda, F.; Furey, W.; Swaminathan, S.; Sax, M.; Farrenkopf, B.; Jordan, F. Catalytic centers in the thiamin diphosphate dependent enzyme pyruvate decarboxylase at 2.4 Å resolution. Biochemistry 1993, 32, 6165–6170. [Google Scholar] [CrossRef] [PubMed]
- Dobritzsch, D.; König, S.; Schneider, G.; Lu, G. High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases. J. Biol. Chem. 1998, 273, 20196–20204. [Google Scholar] [CrossRef] [PubMed]
- Kutter, S.; Wille, G.; Relle, S.; Weiss, M.S.; Hübner, G.; König, S. The crystal structure of pyruvate decarboxylase from Kluyveromyces lactis. FEBS J. 2006, 273, 4199–4209. [Google Scholar] [CrossRef] [PubMed]
- Rother, D.; Kolter, G.; Gerhards, T.; Berthold, C.L.; Gauchenova, E.; Knoll, M.; Pleiss, J.; Müller, M.; Schneider, G.; Pohl, M. S-selective mixed carboligation by structure-based design of the pyruvate decarboxylase from Acetobacter pasteurianus. ChemCatChem 2011, 3, 1587–1596. [Google Scholar] [CrossRef]
- Liu, M.; Sergienko, E.A.; Guo, F.; Wang, J.; Tittmann, K.; Hübner, G.; Furey, W.; Jordan, F. Catalytic acid-base groups in yeast pyruvate decarboxylase. 1. Site-directed mutagenesis and steady-state kinetic studies on the enzyme with the D28A, H114F, H115F, and E477Q substitutions. Biochemistry 2001, 40, 7355–7368. [Google Scholar] [CrossRef] [PubMed]
- Sergienko, E.A.; Jordan, F. Catalytic acid-base groups in yeast pyruvate decarboxylase. 2. Insights into the specific roles of D28 and E477 from the rates and stereospecificity of formation of carboligase side products. Biochemistry 2001, 40, 7369–7381. [Google Scholar] [CrossRef] [PubMed]
- Sergienko, E.A.; Jordan, F. Catalytic acid-base groups in yeast pyruvate decarboxylase. 3. A steady-state kinetic model consistent with the behavior of both wild-type and variant enzymes at all relevant pH values. Biochemistry 2001, 40, 7382–7403. [Google Scholar] [CrossRef] [PubMed]
- Candy, J.M.; Duggleby, R.G. Structure and properties of pyruvate decarboxylase and site-directed mutagenesis of the Zymomonas mobilis enzyme. BBA Protein Struct. Mol. Enzymol. 1998, 1385, 323–338. [Google Scholar] [CrossRef]
- Schütz, A.; Sandalova, T.; Ricagno, S.; Hübner, G.; König, S.; Schneider, G. Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid. Eur. J. Biochem. 2003, 270, 2312–2321. [Google Scholar] [CrossRef] [PubMed]
- Berthold, C.L.; Gocke, D.; Wood, M.D.; Leeper, F.J.; Pohl, M.; Schneider, G. Structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis provides insights into the structural basis for the chemoselective and enantioselective carboligation reaction. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Versees, W.; Spaepen, S.; Vanderleyden, J.; Steyaert, J. The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 Å resolution. Implications for its catalytic and regulatory mechanism. FEBS J. 2007, 274, 2363–2375. [Google Scholar] [CrossRef] [PubMed]
- Hegeman, G.D. Benzoylformate decarboxylase (Pseudomonas putida). Methods Enzymol. 1970, 17, 674–678. [Google Scholar]
- Hegeman, G.D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida I. Synthesis of enzymes by the wild type. J. Bacteriol. 1966, 91, 1140–1154. [Google Scholar] [PubMed]
- Kluger, R.; Tittmann, K. Thiamin diphosphate catalysis: Enzymic and nonenzymic covalent intermediates. Chem. Rev. 2008, 108, 1797–1833. [Google Scholar] [CrossRef] [PubMed]
- Hasson, M.S.; Muscate, A.; McLeish, M.J.; Polovnikova, L.S.; Gerlt, J.A.; Kenyon, G.L.; Petsko, G.A.; Ringe, D. The crystal structure of benzoylformate decarboxylase at 1.6 Å resolution: Diversity of catalytic residues in thiamin diphosphate-dependent enzymes. Biochemistry 1998, 37, 9918–9930. [Google Scholar] [CrossRef] [PubMed]
- Polovnikova, E.S.; McLeish, M.J.; Sergienko, E.A.; Burgner, J.T.; Anderson, N.L.; Bera, A.K.; Jordan, F.; Kenyon, G.L.; Hasson, M.S. Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate decarboxylase. Biochemistry 2003, 42, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Sergienko, E.A.; Wang, J.; Polovnikova, L.; Hasson, M.S.; McLeish, M.J.; Kenyon, G.L.; Jordan, F. Spectroscopic detection of transient thiamin diphosphate-bound intermediates on benzoylformate decarboxylase. Biochemistry 2000, 39, 13862–13869. [Google Scholar] [CrossRef] [PubMed]
- Schütz, A.; Golbik, R.; König, S.; Hübner, G.; Tittmann, K. Intermediates and transition states in thiamin diphosphate-dependent decarboxylases. A kinetic and NMR study on wild-type indolepyruvate decarboxylase and variants using indolepyruvate, benzoylformate, and pyruvate as substrates. Biochemistry 2005, 44, 6164–6179. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.; Kern, G.; Neef, H.; Tittmann, K.; Killenberg-Jabs, M.; Wikner, C.; Schneider, G.; Hübner, G. How thiamin diphosphate is activated in enzymes. Science 1997, 275, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Frank, R.; Leeper, F.; Luisi, B. Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell Mol. Life Sci. 2007, 64, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Neumann, P.; Ficner, R.; Tittmann, K. Observation of a stable carbene at the active site of a thiamin enzyme. Nat. Chem. Biol. 2013, 9, 488–490. [Google Scholar] [CrossRef] [PubMed]
- Tittmann, K.; Golbik, R.; Uhlemann, K.; Khailova, L.; Schneider, G.; Patel, M.; Jordan, F.; Chipman, D.M.; Duggleby, R.G.; Hübner, G. NMR analysis of covalent intermediates in thiamin diphosphate enzymes. Biochemistry 2003, 42, 7885–7891. [Google Scholar] [CrossRef] [PubMed]
- Bruning, M.; Berheide, M.; Meyer, D.; Golbik, R.; Bartunik, H.; Liese, A.; Tittmann, K. Structural and kinetic studies on native intermediates and an intermediate analogue in benzoylformate decarboxylase reveal a least motion mechanism with an unprecedented short-lived predecarboxylation intermediate. Biochemistry 2009, 48, 3258–3268. [Google Scholar] [CrossRef] [PubMed]
- Pohl, M.; Lingen, B.; Müller, M. Thiamin-diphosphate-dependent enzymes: New aspects of asymmetric C–C bond formation. Chem. Eur. J. 2002, 8, 5288–5295. [Google Scholar] [CrossRef]
- Fuganti, C.; Grasselli, P. Synthesis of the C14 chromanyl moiety of natural α-tocopherol (vitamin E). J. Chem. Soc. Chem. Commun. 1982, 4, 205–206. [Google Scholar] [CrossRef]
- Gala, D.; DiBenedetto, D.J.; Clark, J.E.; Murphy, B.L.; Schumacher, D.P.; Steinman, M. Preparations of antifungal Sch 42427/SM 9164: Preparative chromatographic resolution, and total asymmetric synthesis via enzymic preparation of chiral α-hydroxy arylketones. Tetrahedron Lett. 1996, 37, 611–614. [Google Scholar] [CrossRef]
- Siegert, P.; McLeish, M.J.; Baumann, M.; Iding, H.; Kneen, M.M.; Kenyon, G.L.; Pohl, M. Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein Eng. Des. Sel. 2005, 18, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Yep, A.; McLeish, M.J. Engineering the substrate binding site of benzoylformate decarboxylase. Biochemistry 2009, 48, 8387–8395. [Google Scholar] [CrossRef] [PubMed]
- Tittmann, K.; Vyazmensky, M.; Hübner, G.; Barak, Z.; Chipman, D.M. The carboligation reaction of acetohydroxyacid synthase ii: Steady-state intermediate distributions in wild type and mutants by nmr. Proc Natl Acad Sci U S A 2005, 102, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Belenky, I.; Steinmetz, A.; Vyazmensky, M.; Barak, Z.; Tittmann, K.; Chipman, D.M. Many of the functional differences between acetohydroxyacid synthase (AHAS) isozyme I and other AHASs are a result of the rapid formation and breakdown of the covalent acetolactate-thiamin diphosphate adduct in AHAS I. FEBS J. 2012, 279, 1967–1979. [Google Scholar] [CrossRef] [PubMed]
- Joseph, E.; Wei, W.; Tittmann, K.; Jordan, F. Function of a conserved loop of the β-domain, not involved in thiamin diphosphate binding, in catalysis and substrate activation in yeast pyruvate decarboxylase. Biochemistry 2006, 45, 13517–13527. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, M.; Kaplun, A.; Vyazmensky, M.; Engel, S.; Golbik, R.; Tittmann, K.; Uhlemann, K.; Meshalkina, L.; Barak, Z.; Hübner, G.; et al. Monitoring the acetohydroxy acid synthase reaction and related carboligations by circular dichroism spectroscopy. Anal. Biochem. 2005, 342, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, A.; Vyazmensky, M.; Meyer, D.; Barak, Z.; Golbik, R.; Chipman, D.M.; Tittmann, K. Valine 375 and phenylalanine 109 confer affinity and specificity for pyruvate as donor substrate in acetohydroxy acid synthase isozyme II from Escherichia coli. Biochemistry 2010, 49, 5188–5199. [Google Scholar] [CrossRef] [PubMed]
- Vyazmensky, M.; Steinmetz, A.; Meyer, D.; Golbik, R.; Barak, Z.; Tittmann, K.; Chipman, D.M. Significant catalytic roles for Glu47 and Gln110 in all four of the C–C bond-making and -breaking steps of the reactions of acetohydroxyacid synthase II. Biochemistry 2011, 50, 3250–3260. [Google Scholar] [CrossRef] [PubMed]
- Beigi, M.; Loschonsky, S.; Lehwald, P.; Brecht, V.; Andrade, S.L.A.; Leeper, F.J.; Hummel, W.; Muller, M. α-hydroxy-β-keto acid rearrangement-decarboxylation: Impact on thiamine diphosphate-dependent enzymatic transformations. Org. Biomol. Chem. 2013, 11, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Andrews, F.H.; Tom, A.R.; Gunderman, P.R.; Novak, W.R.P.; McLeish, M.J. A bulky hydrophobic residue is not required to maintain the V-conformation of enzyme-bound thiamin diphosphate. Biochemistry 2013, 52, 3028–3030. [Google Scholar] [CrossRef] [PubMed]
- Berthold, C.L.; Moussatche, P.; Richards, N.G.; Lindqvist, Y. Structural basis for activation of the thiamin diphosphate-dependent enzyme oxalyl-coA decarboxylase by adenosine diphosphate. J. Biol. Chem. 2005, 280, 41645–41654. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.K.; Polovnikova, L.S.; Roestamadji, J.; Widlanski, T.S.; Kenyon, G.L.; McLeish, M.J.; Hasson, M.S. Mechanism-based inactivation of benzoylformate decarboxylase, a thiamin diphosphate-dependent enzyme. J. Am. Chem. Soc. 2007, 129, 4120–4121. [Google Scholar] [CrossRef] [PubMed]
- Brandt, G.S.; Kneen, M.M.; Chakraborty, S.; Baykal, A.T.; Nemeria, N.; Yep, A.; Ruby, D.I.; Petsko, G.A.; Kenyon, G.L.; McLeish, M.J.; et al. Snapshot of a reaction intermediate: Analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor. Biochemistry 2009, 48, 3247–3257. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Neumann, P.; Parthier, C.; Friedemann, R.; Nemeria, N.; Jordan, F.; Tittmann, K. Double duty for a conserved glutamate in pyruvate decarboxylase: Evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate. Biochemistry 2010, 49, 8197–8212. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Nemeria, N.S.; Balakrishnan, A.; Brandt, G.S.; Kneen, M.M.; Yep, A.; McLeish, M.J.; Kenyon, G.L.; Petsko, G.A.; Ringe, D.; et al. Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase. Biochemistry 2009, 48, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Andrews, F.H.; Rogers, M.P.; Paul, L.N.; McLeish, M.J. Perturbation of the monomer-monomer interfaces of the benzoylformate decarboxylase tetramer. Biochemistry 2014, 53, 4358–4367. [Google Scholar] [CrossRef] [PubMed]
- Turano, A.; Furey, W.; Pletcher, J.; Sax, M.; Pike, D.; Kluger, R. Synthesis and crystal structure of an analog of 2-(alpha-lactyl)thiamin, racemic methyl 2-hydroxy-2-(2-thiamin)ethylphosphonate chloride trihydrate. A conformation for a least-motion, maximum-overlap mechanism for thiamin catalysis. J. Am. Chem. Soc. 1982, 104, 3089–3095. [Google Scholar] [CrossRef]
- Wille, G.; Meyer, D.; Steinmetz, A.; Hinze, E.; Golbik, R.; Tittmann, K. The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography. Nat. Chem. Biol. 2006, 2, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.; Arjunan, P.; Furey, W.; Jordan, F. A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex e1 component modulates substrate utilization and chemical communication with the E2 component. J. Biol. Chem. 2007, 282, 28106–28116. [Google Scholar] [CrossRef] [PubMed]
- Arjunan, P.; Sax, M.; Brunskill, A.; Chandrasekhar, K.; Nemeria, N.; Zhang, S.; Jordan, F.; Furey, W. A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct. J. Biol. Chem. 2006, 281, 15296–15303. [Google Scholar] [CrossRef] [PubMed]
- Demir, A.S.; Dünnwald, T.; Iding, H.; Pohl, M.; Müller, M. Asymmetric benzoin reaction catalyzed by benzoylformate decarboxylase. Tetrahedron Asymmetry 1999, 10, 4769–4774. [Google Scholar] [CrossRef]
- Dünkelmann, P.; Kolter-Jung, D.; Nitsche, A.; Demir, A.S.; Siegert, P.; Lingen, B.; Baumann, M.; Pohl, M.; Müller, M. Development of a donor-acceptor concept for enzymatic cross-coupling reactions of aldehydes: The first asymmetric cross-benzoin condensation. J. Am. Chem. Soc. 2002, 124, 12084–12085. [Google Scholar] [CrossRef] [PubMed]
- Pohl, M.; Sprenger, G.A.; Müller, M. A new perspective on thiamine catalysis. Curr. Opin. Biotechnol. 2004, 15, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Walter, L.; Kolter, G.; Pohl, M.; Müller, M.; Tittmann, K. Conversion of pyruvate decarboxylase into an enantioselective carboligase with biosynthetic potential. J. Am. Chem. Soc. 2011, 133, 3609–3616. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bailey, S. The CCP4 suite—Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., 3rd; Snoeyink, J.; Richardson, J.S.; et al. Molprobity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. Molprobity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).