Optimized Tandem Affinity Purification Strategy Enables High-Yield Isolation and Functional Characterization of Native COMPASS in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Results and Discussion
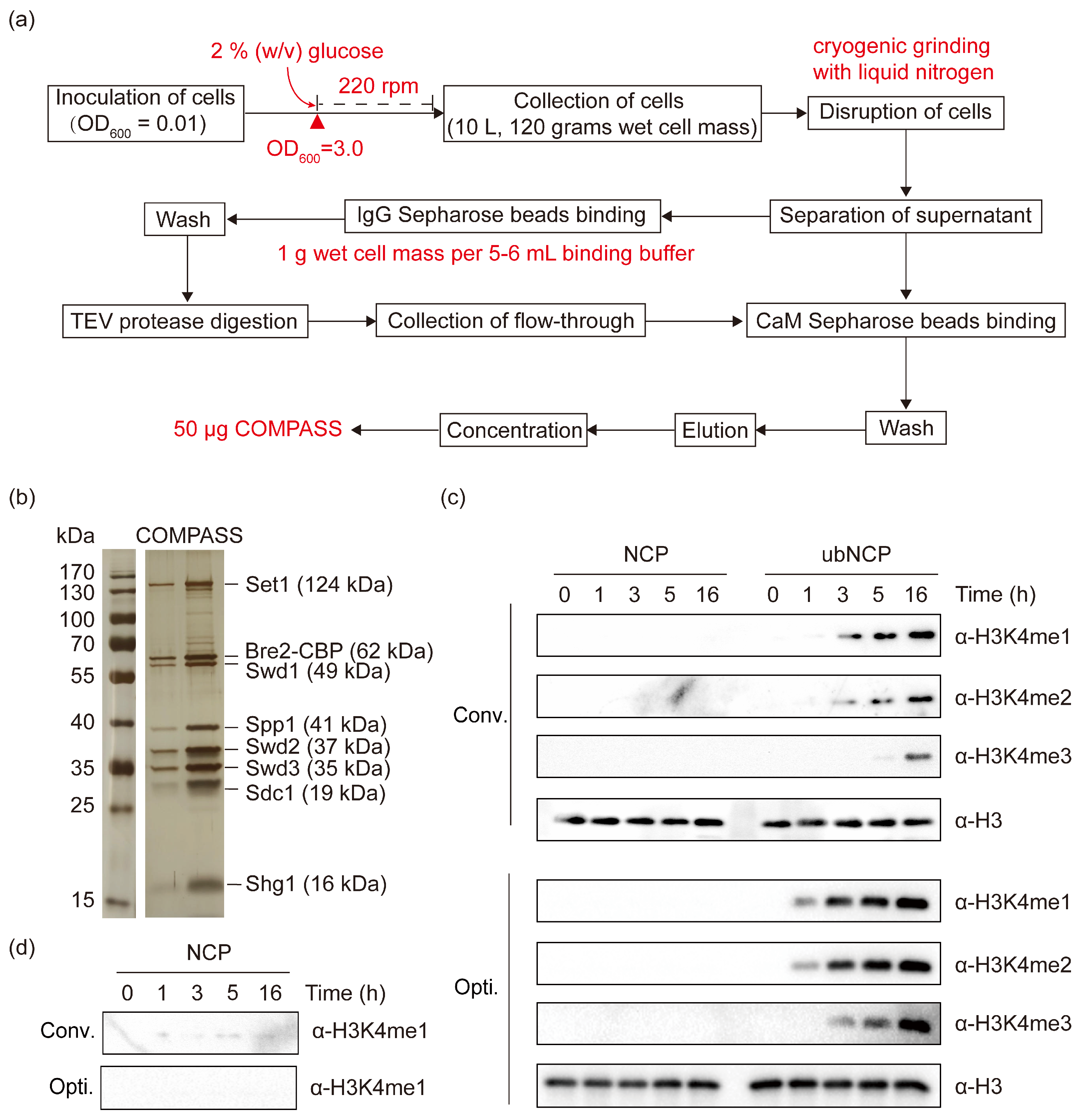
2.1. Conventional TAP-Based Purification Fails to Efficiently Isolate Endogenous COMPASS from Yeast
2.2. Optimizing Culture Conditions Leads to Enhanced Expression of COMPASS Core Subunit Set1
2.3. Optimized Cell Lysis and Affinity Purification Enable Efficient Preparation of High-Quality COMPASS
2.4. Endogenous COMPASS Purified Using the Optimized Procedure Exhibits Strict H2B Ubiquitination Dependency for Catalyzing H3K4 Methylation
3. Materials and Methods
3.1. Yeast Strains
3.2. Culture Conditions
3.3. Growth Curve of Yeast Under Different Culture Conditions
3.4. Conventional TAP-Tagged COMPASS Purification
3.5. Optimized Tandem Affinity Purification of Native COMPASS Complex
3.6. SDS-PAGE for Purity and Integrity Analysis
3.7. Western Blot Analysis of Purification Efficiency
3.8. In Vitro COMPASS Methyltransferase Assays
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COMPASS | Complex of proteins associated with Set1 |
| H3K4me | Histone 3 lysine 4 methylations |
| H3K4me1 | Histone 3 lysine 4 mono-methylation |
| H3K4me2 | Histone 3 lysine 4 di-methylation |
| H3K4me2 | Histone 3 lysine 4 trimethylated |
| AdoMet | S-adenosylmethionine |
| TAP | Tandem affinity purification |
| ProA | Protein A |
| TEV | Tobacco Etch Virus |
| CaM | Calmodulin |
| CBP | Calmodulin-binding peptide |
| OD600 | Optical density at 600 nm |
| SET1RC | SET1 mRNA-associated complex |
| H2Bub | H2B ubiquitination |
| NCPs | Nucleosome core particles |
| UbNCPs | H2Bub-modified NCPs |
| CM | Catalytic module |
| eCM | Extended CM |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
References
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Tolsma, T.O.; Hansen, J.C. Post-Translational Modifications and Chromatin Dynamics. Essays Biochem. 2019, 63, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Hu, X.; Wang, X. Crossing Epigenetic Frontiers: The Intersection of Novel Histone Modifications and Diseases. Signal Transduct. Target. Ther. 2024, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Helin, K. Roles of H3K4 Methylation in Biology and Disease. Trends Cell Biol. 2025, 35, 115–128. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Chong, S.Y.; Cutler, S.; Lin, J.-J.; Tsai, C.-H.; Tsai, H.-K.; Biggins, S.; Tsukiyama, T.; Lo, Y.-C.; Kao, C.-F. H3K4 methylation at active genes mitigates transcription-replication conflicts during replication stress. Nat. Commun. 2020, 11, 809. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and Predictive Chromatin Signatures of Transcriptional Promoters and Enhancers in the Human Genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Kim, T.; Buratowski, S. Dimethylation of H3K4 by Set1 Recruits the Set3 Histone Deacetylase Complex to 5′ Transcribed Regions. Cell 2009, 137, 259–272. [Google Scholar] [CrossRef]
- Yan, J.; Chen, S.-A.A.; Local, A.; Liu, T.; Qiu, Y.; Dorighi, K.M.; Preissl, S.; Rivera, C.M.; Wang, C.; Ye, Z.; et al. Histone H3 Lysine 4 Monomethylation Modulates Long-Range Chromatin Interactions at Enhancers. Cell Res. 2018, 28, 204–220. [Google Scholar] [CrossRef]
- Ruthenburg, A.J.; Allis, C.D.; Wysocka, J. Methylation of Lysine 4 on Histone H3: Intricacy of Writing and Reading a Single Epigenetic Mark. Mol. Cell 2007, 25, 15–30. [Google Scholar] [CrossRef]
- Deshpande, N.; Jordan, R.; Henderson Pozzi, M.; Bryk, M. Histone 3 Lysine 4 Monomethylation Supports Activation of Transcription in S. Cerevisiae During Nutrient Stress. Curr. Genet. 2022, 68, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Dehé, P.-M.; Dichtl, B.; Schaft, D.; Roguev, A.; Pamblanco, M.; Lebrun, R.; Rodríguez-Gil, A.; Mkandawire, M.; Landsberg, K.; Shevchenko, A.; et al. Protein Interactions within the Set1 Complex and Their Roles in the Regulation of Histone 3 Lysine 4 Methylation. J. Biol. Chem. 2006, 281, 35404–35412. [Google Scholar] [CrossRef] [PubMed]
- Nieborak, A.; Schneider, R. Metabolic Intermediates—Cellular Messengers Talking to Chromatin Modifiers. Mol. Metab. 2018, 14, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Al Temimi, A.H.K.; Amatdjais-Groenen, H.I.V.; Reddy, Y.V.; Blaauw, R.H.; Guo, H.; Qian, P.; Mecinović, J. The Nucleophilic Amino Group of Lysine Is Central for Histone Lysine Methyltransferase Catalysis. Commun. Chem. 2019, 2, 1–14. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Rey, F.E.; Denu, J.M. Chemical Signaling between Gut Microbiota and Host Chromatin: What Is Your Gut Really Saying? J. Biol. Chem. 2017, 292, 8582–8593. [Google Scholar] [CrossRef]
- Miller, T.; Krogan, N.J.; Dover, J.; Erdjument-Bromage, H.; Tempst, P.; Johnston, M.; Greenblatt, J.F.; Shilatifard, A. COMPASS: A Complex of Proteins Associated with a Trithorax-Related SET Domain Protein. Proc. Natl. Acad. Sci. USA 2001, 98, 12902–12907. [Google Scholar] [CrossRef]
- Li, T.; Kelly, W.G. A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Caenorhabditis elegans Germ Line. PLoS Genet. 2011, 7, e1001349. [Google Scholar] [CrossRef]
- Mersman, D.P.; Du, H.-N.; Fingerman, I.M.; South, P.F.; Briggs, S.D. Charge-Based Interaction Conserved within Histone H3 Lysine 4 (H3K4) Methyltransferase Complexes Is Needed for Protein Stability, Histone Methylation, and Gene Expression. J. Biol. Chem. 2012, 287, 2652–2665. [Google Scholar] [CrossRef]
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3K4me3 Instructive for Transcription Activation? BioEssays 2017, 39, 1–12. [Google Scholar] [CrossRef]
- Bae, H.J.; Dubarry, M.; Jeon, J.; Soares, L.M.; Dargemont, C.; Kim, J.; Geli, V.; Buratowski, S. The Set1 N-Terminal Domain and Swd2 Interact with RNA Polymerase II CTD to Recruit COMPASS. Nat. Commun. 2020, 11, 2181. [Google Scholar] [CrossRef]
- Dehé, P.-M.; Géli, V. The Multiple Faces of Set1. Biochem. Cell Biol. 2006, 84, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Quílez, J.; Roig-Soucase, S.; Rodríguez-Navarro, S. Sharing Marks: H3k4 Methylation and H2b Ubiquitination as Features of Meiotic Recombination and Transcription. Int. J. Mol. Sci. 2020, 21, 4510. [Google Scholar] [CrossRef] [PubMed]
- South, P.F.; Fingerman, I.M.; Mersman, D.P.; Du, H.-N.; Briggs, S.D. A Conserved Interaction between the SDI Domain of Bre2 and the Dpy-30 Domain of Sdc1 Is Required for Histone Methylation and Gene Expression. J. Biol. Chem. 2010, 285, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, K.; Li, T.; Cai, R.; Chen, Y. The Engagement of Histone Lysine Methyltransferases with Nucleosomes: Structural Basis, Regulatory Mechanisms, and Therapeutic Implications. Protein Cell 2023, 14, 165–179. [Google Scholar] [CrossRef]
- Xue, H.; Yao, T.; Cao, M.; Zhu, G.; Li, Y.; Yuan, G.; Chen, Y.; Lei, M.; Huang, J. Structural Basis of Nucleosome Recognition and Modification by MLL Methyltransferases. Nature 2019, 573, 445–449. [Google Scholar] [CrossRef]
- Hsu, P.L.; Shi, H.; Leonen, C.; Kang, J.; Chatterjee, C.; Zheng, N. Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS. Mol. Cell 2019, 76, 712–723.e4. [Google Scholar] [CrossRef]
- Han, J.; Li, T.; Li, Y.; Li, M.; Wang, X.; Peng, C.; Su, C.; Li, N.; Li, Y.; Xu, Y.; et al. The Internal Interaction in RBBP5 Regulates Assembly and Activity of MLL1 Methyltransferase Complex. Nucleic Acids Res. 2019, 47, 10426–10438. [Google Scholar] [CrossRef]
- Li, Y.; Han, J.; Zhang, Y.; Cao, F.; Liu, Z.; Li, S.; Wu, J.; Hu, C.; Wang, Y.; Shuai, J.; et al. Structural Basis for Activity Regulation of MLL Family Methyltransferases. Nature 2016, 530, 447–452. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, Z.; Liu, X.; Bao, Y.; Huang, M.; Wong, C.C.L.; Hong, X.; Cong, Y. Architecture and Subunit Arrangement of the Complete Saccharomyces Cerevisiae COMPASS Complex. Sci. Rep. 2018, 8, 17405. [Google Scholar] [CrossRef]
- D’Urso, A.; Takahashi, Y.-H.; Xiong, B.; Marone, J.; Coukos, R.; Randise-Hinchliff, C.; Wang, J.-P.; Shilatifard, A.; Brickner, J.H. Set1/COMPASS and Mediator Are Repurposed to Promote Epigenetic Transcriptional Memory. eLife 2016, 5, e16691. [Google Scholar] [CrossRef]
- Giaever, G.; Lissina, E.; Nislow, C. Network Dynamics of the Yeast Methyltransferome. Microb. Cell 2019, 6, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Janna, A.; Davarinejad, H.; Joshi, M.; Couture, J.-F. Structural Paradigms in the Recognition of the Nucleosome Core Particle by Histone Lysine Methyltransferases. Front. Cell Dev. Biol. 2020, 8, 600. [Google Scholar] [CrossRef] [PubMed]
- Worden, E.J.; Zhang, X.; Wolberger, C. Structural Basis for COMPASS Recognition of an H2B-Ubiquitinated Nucleosome. eLife 2020, 9, e53199. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Takahashi, Y.-H.; Yang, Y.; Hu, H.; Zhang, Y.; Brunzelle, J.S.; Couture, J.-F.; Shilatifard, A.; Skiniotis, G. Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex. Cell 2018, 174, 1117–1126.e12. [Google Scholar] [CrossRef]
- Hsu, P.L.; Li, H.; Lau, H.-T.; Leonen, C.; Dhall, A.; Ong, S.-E.; Chatterjee, C.; Zheng, N. Crystal Structure of the COMPASS H3K4 Methyltransferase Catalytic Module. Cell 2018, 174, 1106–1116.e9. [Google Scholar] [CrossRef]
- Cho, U.-S. The Core Complex of Yeast COMPASS and Human Mixed-Lineage Leukemia (MLL), Structure, Function, and Recognition of the Nucleosome. Subcell. Biochem. 2024, 104, 101–117. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.-A.; McGinty, R.K.; Nguyen, U.T.T.; Muir, T.W.; Allis, C.D.; Roeder, R.G. The N-SET Domain of Set1 Regulates H2B Ubiquitylation-Dependent H3K4 Methylation. Mol. Cell 2013, 49, 1121–1133. [Google Scholar] [CrossRef]
- Schneider, J.; Wood, A.; Lee, J.-S.; Schuster, R.; Dueker, J.; Maguire, C.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Shilatifard, A. Molecular Regulation of Histone H3 Trimethylation by COMPASS and the Regulation of Gene Expression. Mol. Cell 2005, 19, 849–856. [Google Scholar] [CrossRef]
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces Cerevisiae Proteome. Cell Syst. 2018, 6, 192–205.e3. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Lee, J.-S.; Shukla, A.; Schneider, J.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Bhaumik, S.R.; Shilatifard, A. Histone Crosstalk between H2B Monoubiquitination and H3 Methylation Mediated by COMPASS. Cell 2007, 131, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Bhojoo, U.; Biggar, K.K. Single-Step Purification of Intrinsic Protein Complexes in Saccharomyces Cerevisiae Using Regenerable Calmodulin Resin. MethodsX 2018, 5, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Schlichter, A.; Cairns, B.R. Histone Trimethylation by Set1 Is Coordinated by the RRM, Autoinhibitory, and Catalytic Domains. EMBO J. 2005, 24, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Puig, O.; Caspary, F.; Rigaut, G.; Rutz, B.; Bouveret, E.; Bragado-Nilsson, E.; Wilm, M.; Séraphin, B. The Tandem Affinity Purification (TAP) Method: A General Procedure of Protein Complex Purification. Methods 2001, 24, 218–229. [Google Scholar] [CrossRef]
- Li, Y. The Tandem Affinity Purification Technology: An Overview. Biotechnol. Lett. 2011, 33, 1487–1499. [Google Scholar] [CrossRef]
- Valero, M.L.; Sendra, R.; Pamblanco, M. Tandem Affinity Purification of Histones, Coupled to Mass Spectrometry, Identifies Associated Proteins and New Sites of Post-Translational Modification in Saccharomyces Cerevisiae. J. Proteom. 2016, 136, 183–192. [Google Scholar] [CrossRef]
- Roguev, A.; Schaft, D.; Shevchenko, A.; Pijnappel, W.W.M.P.; Wilm, M.; Aasland, R.; Stewart, A.F. The Saccharomyces Cerevisiae Set1 Complex Includes an Ash2 Homologue and Methylates Histone 3 Lysine 4. EMBO J. 2001, 20, 7137–7148. [Google Scholar] [CrossRef]
- Woo, H.; Oh, J.; Cho, Y.-J.; Oh, G.T.; Kim, S.-Y.; Dan, K.; Han, D.; Lee, J.-S.; Kim, T. N-Terminal Acetylation of Set1-COMPASS Fine-Tunes H3K4 Methylation Patterns. Sci. Adv. 2024, 10, eadl6280. [Google Scholar] [CrossRef]
- Tapia, H.; Morano, K.A. Hsp90 Nuclear Accumulation in Quiescence Is Linked to Chaperone Function and Spore Development in Yeast. Mol. Biol. Cell 2010, 21, 63–72. [Google Scholar] [CrossRef]
- Alberghina, L. The Warburg Effect Explained: Integration of Enhanced Glycolysis with Heterogeneous Mitochondria to Promote Cancer Cell Proliferation. Int. J. Mol. Sci. 2023, 24, 15787. [Google Scholar] [CrossRef]
- Torello Pianale, L.; Caputo, F.; Olsson, L. Four Ways of Implementing Robustness Quantification in Strain Characterisation. Biotechnol. Biofuels Bioprod. 2023, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Halbach, A.; Zhang, H.; Wengi, A.; Jablonska, Z.; Gruber, I.M.L.; Halbeisen, R.E.; Dehé, P.-M.; Kemmeren, P.; Holstege, F.; Géli, V.; et al. Cotranslational Assembly of the Yeast SET1C Histone Methyltransferase Complex. EMBO J. 2009, 28, 2959–2970. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, A.; Mukherjee, P.; Scheer, E.; Kamenova, I.; Antonova, S.; Mendoza Sanchez, P.K.; Yayli, G.; Morlet, B.; Timmers, H.T.M.; Tora, L. Hierarchical TAF1-Dependent Co-Translational Assembly of the Basal Transcription Factor TFIID. Nat. Struct. Mol. Biol. 2023, 30, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Urh, M.; Simpson, D.; Zhao, K. Chapter 26 Affinity Chromatography. General Methods. In Methods in Enzymology; Elsevier Inc.: New York City, NY, USA, 2009; Volume 463, pp. 417–438. [Google Scholar] [CrossRef]
- Singh, M.R.; Roy, S.; Bellare, J.R. Influence of Cryogenic Grinding on Release of Protein and DNA from Saccharomyces Cerevisiae. Int. J. Food Eng. 2009, 5, 1–23. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Quan, S.; Zheng, Y. Optimized Tandem Affinity Purification Strategy Enables High-Yield Isolation and Functional Characterization of Native COMPASS in Saccharomyces cerevisiae. Catalysts 2025, 15, 521. https://doi.org/10.3390/catal15060521
Li Y, Quan S, Zheng Y. Optimized Tandem Affinity Purification Strategy Enables High-Yield Isolation and Functional Characterization of Native COMPASS in Saccharomyces cerevisiae. Catalysts. 2025; 15(6):521. https://doi.org/10.3390/catal15060521
Chicago/Turabian StyleLi, Ya, Shu Quan, and Yongxin Zheng. 2025. "Optimized Tandem Affinity Purification Strategy Enables High-Yield Isolation and Functional Characterization of Native COMPASS in Saccharomyces cerevisiae" Catalysts 15, no. 6: 521. https://doi.org/10.3390/catal15060521
APA StyleLi, Y., Quan, S., & Zheng, Y. (2025). Optimized Tandem Affinity Purification Strategy Enables High-Yield Isolation and Functional Characterization of Native COMPASS in Saccharomyces cerevisiae. Catalysts, 15(6), 521. https://doi.org/10.3390/catal15060521






