Structural Features Underlying the Mismatch Between Catalytic and Cytostatic Properties in L-Asparaginase from Rhodospirillum rubrum
Abstract
1. Introduction
- Type I, constitutively expressed cytoplasmic enzymes with high KM (~1–8 mM) for L-asparagine, generally lacking significant anti-tumor activity (e.g., Bacillus subtilis, Methanococcus jannaschii, and Pyrococcus horikoshii);
- Type II, periplasmic enzymes with low KM (~10–20 µM) for L-asparagine, demonstrating antiproliferative activity and including those used clinically (EcA and ErA), along with others such as Yersinia pseudotuberculosis, Helicobacter pylori, and Wolinella succinogenes.
2. Results and Discussion
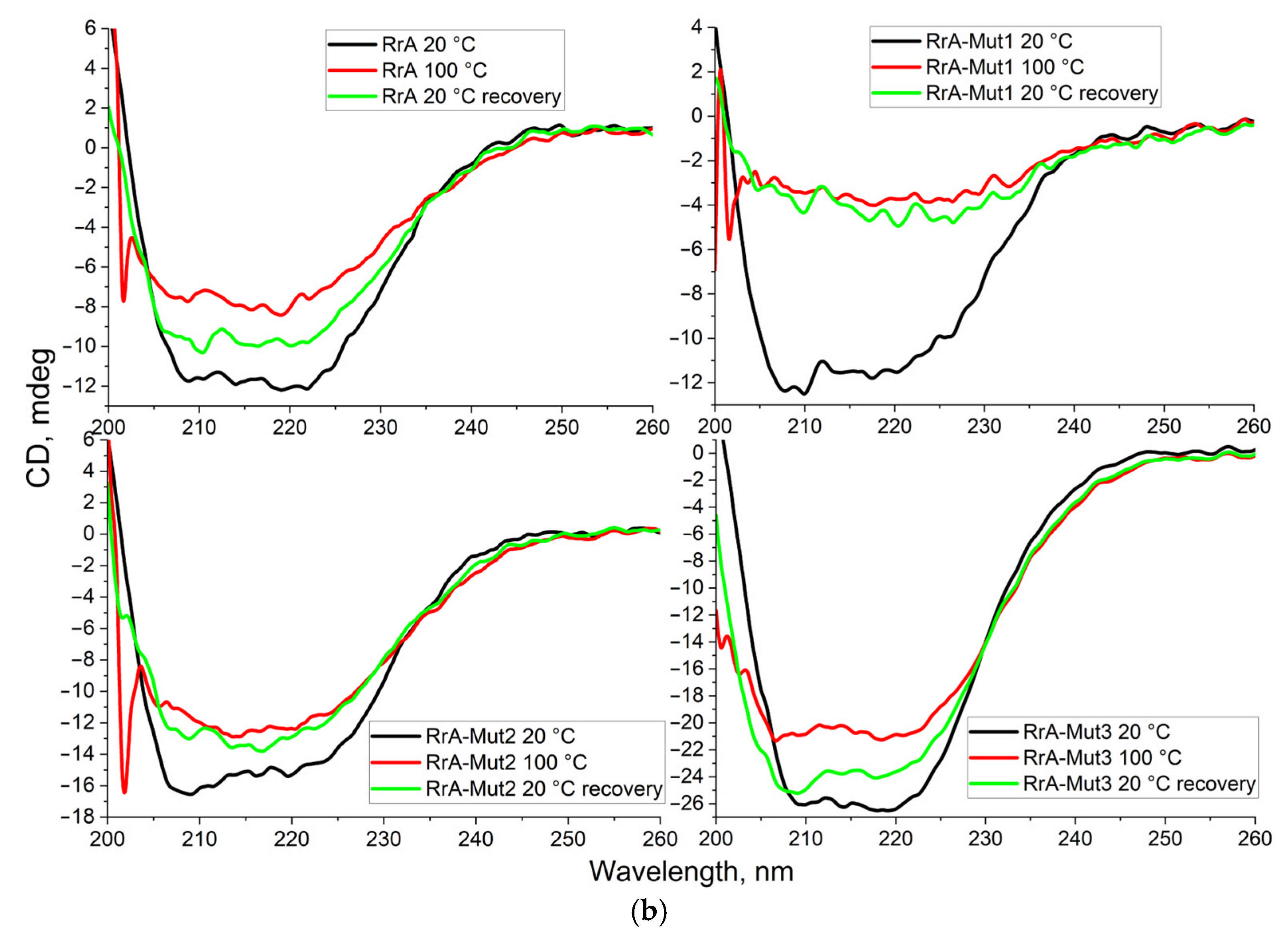
2.1. FTIR Spectroscopy for Characterization of Native and Mutant RrA L-Asparaginases and Comparison with Pharmaceutical (EcA) Preparations
2.2. Determination of the Catalytic Parameters of L-Asparaginase RrA and Its Mutant Forms
2.2.1. 3D Structures of RrA and Mutant Forms to Visualize How Amino Acid Substitutions Could Affect the Conformation Stability of L-Asparaginase
2.2.2. Kinetics Curves
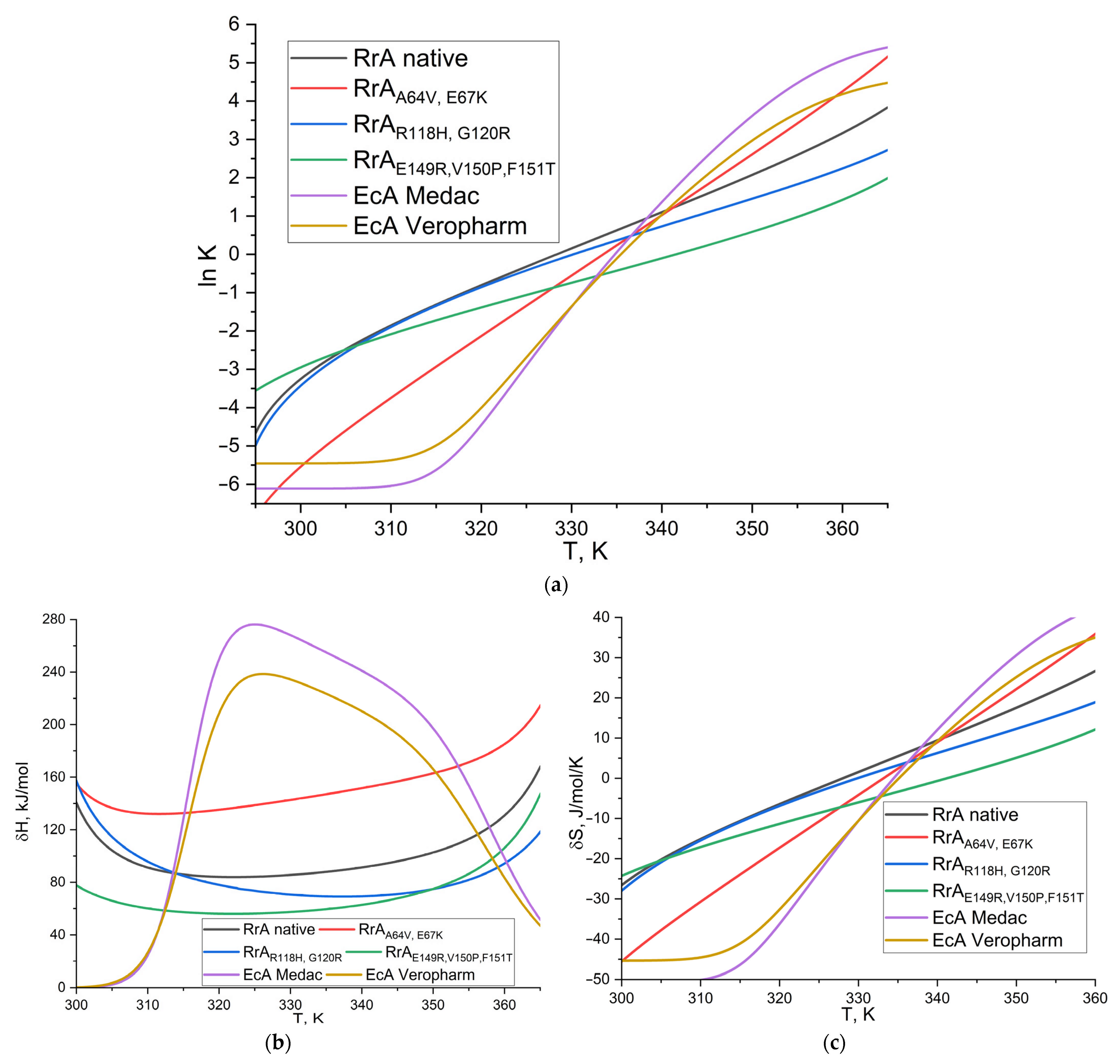
2.3. The Thermograms of L-Asparaginases and Parameters of Its Thermodenaturation
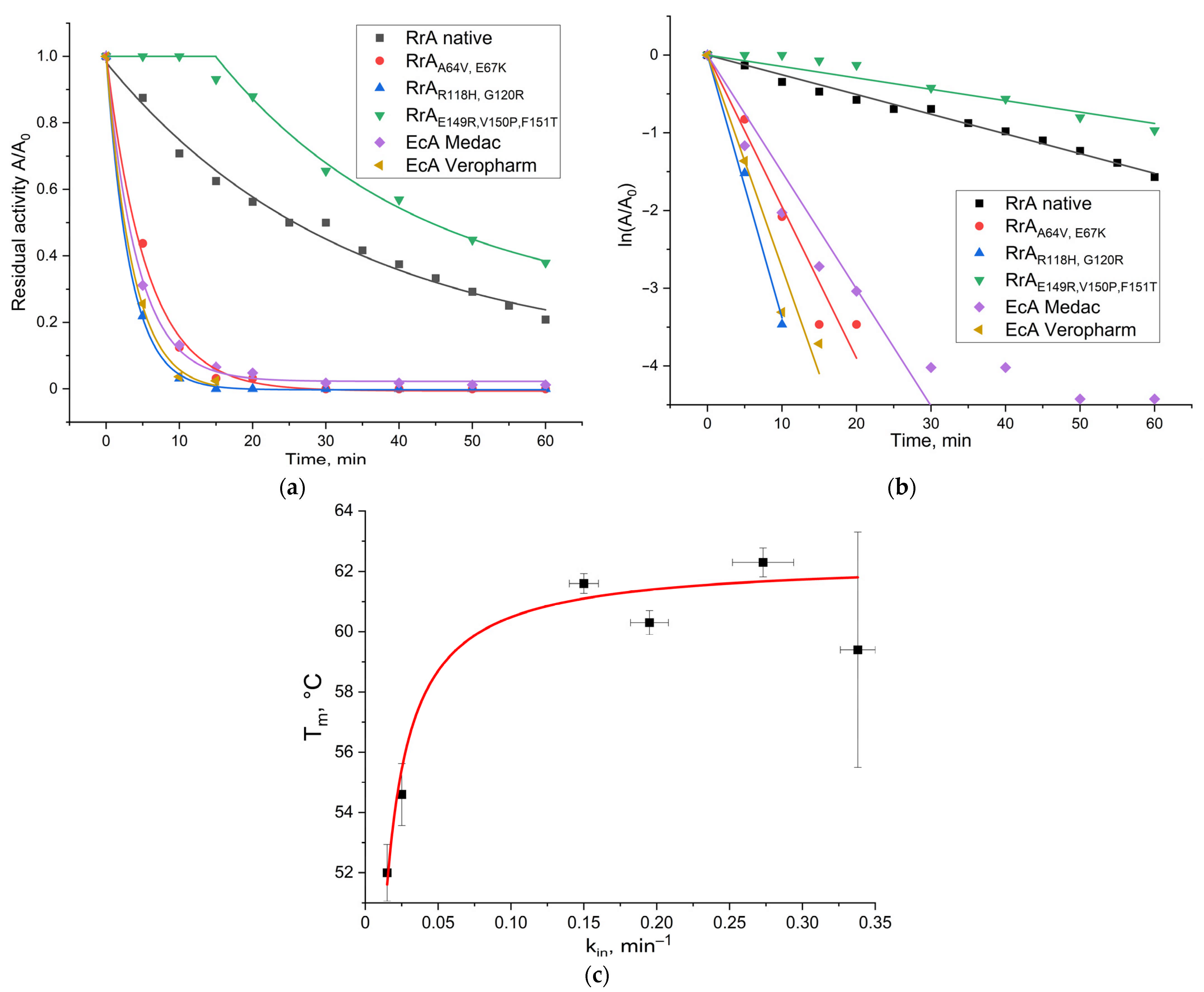
2.4. Resistance of L-Asparaginases to Trypsinolysis
2.5. Cytostatic Effect of L-Asparaginases on Cancer Cells
3. Materials and Methods
3.1. Reagents
3.2. Enzymes
3.3. FTIR Spectroscopy of Enzymes and Its Secondary Structure
3.4. Circular Dichroism (CD) Spectroscopy
3.5. Catalytic Activity Determination by CD Spectroscopy
3.6. L-Asparaginase Thermodenaturation Parameter Determination
3.7. Trypsinolysis Stability Assay
3.8. Cell Culture and Cytotoxicity Assay
3.9. FTIR Spectroscopy for Studying Enzyme-Cell Interactions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASP | L-asparaginase |
| CD | Circular dichroism |
| EcA | Escherichia coli L-asparaginase |
| FTIR | Fourier-transform infrared |
| RrA | Rhodospirillum rubrum L-asparaginase |
| Vmax | Maximum reaction rate |
References
- Burke, M.J.; Zalewska-Szewczyk, B. Hypersensitivity Reactions to Asparaginase Therapy in Acute Lymphoblastic Leukemia: Immunology and Clinical Consequences. Futur. Oncol. 2022, 18, 1285–1299. [Google Scholar] [CrossRef] [PubMed]
- Maese, L.; Rau, R.E. Current Use of Asparaginase in Acute Lymphoblastic Leukemia/Lymphoblastic Lymphoma. Front. Pediatr. 2022, 10, 902117. [Google Scholar] [CrossRef] [PubMed]
- Pokrovskii, V.S.; Komarova, M.V.; Aleksandrova, S.S.; Pokrovskaya, M.V.; Kalish’yan, S.S.K.; Treshchalina, E.M. The Role of Enzymatic Activity in the Realization of the Antiproliferative Effect of L-Asparaginases. Wedge. Oncogematol. 2015, 8, 120–128. [Google Scholar]
- Nikolaos, L. (Ed.) Therapeutic Enzymes: Function and Clinical Implications; Springer: Singapore, 2019; Volume 1148, ISBN 978-981-13-7708-2. [Google Scholar]
- Emadi, A.; Zokaee, H.; Sausville, E. a Asparaginase in the Treatment of Non-ALL Hematologic Malignancies. Cancer Chemother. Pharmacol. 2014, 73, 875–883. [Google Scholar] [CrossRef]
- Juluri, K.R.; Siu, C.; Cassaday, R.D. Asparaginase in the Treatment of Acute Lymphoblastic Leukemia in Adults: Current Evidence and Place in Therapy. Blood Lymphat. Cancer Targets Ther. 2022, 12, 55–79. [Google Scholar] [CrossRef]
- Appel, I.M.; Hop, W.C.J.; Pieters, R. Changes in Hypercoagulability by Asparaginase: A Randomized Study between Two Asparaginases. Blood Coagul. Fibrinolysis 2006, 17, 139–146. [Google Scholar] [CrossRef]
- Lorenzi, P.L.; Llamas, J.; Gunsior, M.; Ozbun, L.; Reinhold, W.C.; Varma, S.; Ji, H.; Kim, H.; Hutchinson, A.A.; Kohn, E.C.; et al. Asparagine Synthetase Is a Predictive Biomarker of L-Asparaginase Activity in Ovarian Cancer Cell Lines. Mol. Cancer Ther. 2008, 7, 3123–3128. [Google Scholar] [CrossRef]
- Lomelino, C.L.; Andring, J.T.; McKenna, R.; Kilberg, M.S. Asparagine Synthetase: Function, Structure, and Role in Disease. J. Biol. Chem. 2017, 292, 19952–19958. [Google Scholar] [CrossRef]
- Ren, Y.; Roy, S.; Ding, Y.; Iqbal, J.; Broome, J.D. Methylation of the Asparagine Synthetase Promoter in Human Leukemic Cell Lines Is Associated with a Specific Methyl Binding Protein. Oncogene 2004, 23, 3953–3961. [Google Scholar] [CrossRef]
- Richards, N.G.J.; Kilberg, M.S. Asparagine Synthetase Chemotherapy. Annu. Rev. Biochem. 2006, 76, 629–654. [Google Scholar] [CrossRef]
- Ikeuchi, H.; Ahn, Y.-M.; Otokawa, T.; Watanabe, B.; Hegazy, L.; Hiratake, J.; Richards, N.G.J. A Sulfoximine-Based Inhibitor of Human Asparagine Synthetase Kills L-Asparaginase-Resistant Leukemia Cells. Bioorg. Med. Chem. 2012, 20, 5915–5927. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Kilberg, M.S.; Bussolati, O. Asparagine Synthetase in Cancer: Beyond Acute Lymphoblastic Leukemia. Front. Oncol. 2020, 9, 1480. [Google Scholar] [CrossRef]
- Hutson, R.G.; Kitoh, T.; Moraga Amador, D.A.; Cosic, S.; Schuster, S.M.; Kilberg, M.S. Amino Acid Control of Asparagine Synthetase: Relation to Asparaginase Resistance in Human Leukemia Cells. Am. J. Physiol.-Cell Physiol. 1997, 272, 1691–1699. [Google Scholar] [CrossRef]
- Balasubramanian, M.N.; Butterworth, E.A.; Kilberg, M.S. Asparagine Synthetase: Regulation by Cell Stress and Involvement in Tumor Biology. Am. J. Physiol. Endocrinol. Metab. 2013, 304, 789–799. [Google Scholar] [CrossRef]
- Haskell, C.M.; Canellos, G.P. L-Asparaginase Resistance in Human Leukemia--Asparagine Synthetase. Biochem. Pharmacol. 1969, 18, 2578–2580. [Google Scholar] [CrossRef]
- Gutierrez, J.A.; Pan, Y.-X.; Koroniak, L.; Hiratake, J.; Kilberg, M.S.; Richards, N.G.J. An Inhibitor of Human Asparagine Synthetase Suppresses Proliferation of an L-Asparaginase-Resistant Leukemia Cell Line. Chem. Biol. 2006, 13, 1339–1347. [Google Scholar] [CrossRef]
- Hashimoto, K.E.N.; Suzuki, F.; Sakagami, H. Declined Asparagine Synthetase MRNA Expression and Enhanced Sensitivity to Asparaginase in HL-60 Cells Committed to Monocytic Differentiation. Anticancer Res. 2009, 29, 1303–1308. [Google Scholar]
- Muneer, F.; Siddique, M.H.; Azeem, F.; Rasul, I.; Muzammil, S.; Zubair, M.; Afzal, M.; Nadeem, H. Microbial L-Asparaginase: Purification, Characterization and Applications. Arch. Microbiol. 2020, 202, 967–981. [Google Scholar] [CrossRef]
- Dumina, M.; Zhgun, A.; Pokrovskaya, M.; Aleksandrova, S.; Zhdanov, D.; Sokolov, N.; El’darov, M. Highly Active Thermophilic L-Asparaginase from Melioribacter roseus Represents a Novel Large Group of Type II Bacterial L-Asparaginases from Chlorobi-Ignavibacteriae-Bacteroidetes Clade. Int. J. Mol. Sci. 2021, 22, 13632. [Google Scholar] [CrossRef]
- Lubkowski, J.; Wlodawer, A. Structural and Biochemical Properties of L-Asparaginase. FEBS J. 2021, 288, 4183–4209. [Google Scholar] [CrossRef]
- Zhang, D.; Czapinska, H.; Bochtler, M.; Wlodawer, A.; Lubkowski, J. RrA, an Enzyme from Rhodospirillum rubrum, is a Prototype of a New Family of Short-chain L-asparaginases. Protein Sci. 2024, 33, e4920. [Google Scholar] [CrossRef]
- Al-Hazmi, N.E.; Naguib, D.M. Plant Asparaginase versus Microbial Asparaginase as Anticancer Agent. Environ. Sci. Pollut. Res. 2022, 29, 27283–27293. [Google Scholar] [CrossRef]
- Dumina, M.; Zhgun, A.; Pokrovskaya, M.; Aleksandrova, S.; Zhdanov, D.; Sokolov, N.; El’darov, M. A Novel L-asparaginase from Hyperthermophilic Archaeon Thermococcus sibiricus: Heterologous Expression and Characterization for Biotechnology Application. Int. J. Mol. Sci. 2021, 22, 9894. [Google Scholar] [CrossRef]
- Batool, T.; Makky, E.A.; Jalal, M.; Yusoff, M.M. A Comprehensive Review on L-Asparaginase and Its Applications. Appl. Biochem. Biotechnol. 2016, 178, 900–923. [Google Scholar] [CrossRef]
- Chan, W.K.; Lorenzi, P.L.; Anishkin, A.; Purwaha, P.; Rogers, D.M.; Sukharev, S.; Rempe, S.B.; Weinstein, J.N. The Glutaminase Activity of L -Asparaginase Is Not Required for Anticancer Activity against ASNS-Negative Cells. Blood 2014, 123, 3596–3607. [Google Scholar] [CrossRef]
- Kudryashova, E.V.; Pokrovskaya, M.V.; Alexandrova, S.S.; Vinogradov, A.A.; Sokolov, N.N. FTIR-Based L-Asparaginase Activity Assay Enables Continuous Measurements in Optically Dense Media Including Blood Plasma. Anal. Biochem. 2020, 598, 113694. [Google Scholar] [CrossRef]
- Pokrovsky, V.S.; Kazanov, M.D.; Dyakov, I.N.; Pokrovskaya, M.V.; Aleksandrova, S.S. Comparative Immunogenicity and Structural Analysis of Epitopes of Different Bacterial L-Asparaginases. BMC Cancer 2016, 16, 89. [Google Scholar] [CrossRef]
- Dobryakova, N.V.; Zhdanov, D.D.; Sokolov, N.N.; Aleksandrova, S.S.; Pokrovskaya, M.V.; Kudryashova, E.V. Improvement of Biocatalytic Properties and Cytotoxic Activity of L-Asparaginase from Rhodospirillum rubrum by Conjugation with Chitosan-Based Cationic Polyelectrolytes. Pharmaceuticals 2022, 15, 406. [Google Scholar] [CrossRef]
- Pokrovskaya, M.V.; Pokrovsky, V.S.; Alexandrova, S.S.; Anisimova, N.Y.; Andrianov, R.M.; Treshkalina, E.M.; Ponomarev, G.V.; Sokolov, N.N. Recombinant Intracellular L-Asparaginase of Rhodospirillum rubrum with Low L-Glutaminase Activity and Antiproliferative effect. Biomed. Chem. 2013, 59, 192–198. [Google Scholar] [CrossRef][Green Version]
- Sokolov, N.N.; Eldarov, M.A.; Pokrovskaya, M.V.; Aleksandrova, S.S.; Abakumova, O.Y.; Podobed, O.V.; Melik-Nubarov, N.S.; Kudryashova, E.V.; Grishin, D.V.; Archakov, A.I. Bacterial Recombinant L-Asparaginases: Properties, Structure, and Anti-Proliferative Activity. Biochem. Suppl. Ser. B Biomed. Chem. 2015, 9, 325–338. [Google Scholar] [CrossRef]
- Pokrovskaya, M.V.; Zhdanov, D.D.; Eldarov, M.A.; Aleksandrova, S.S.; Veselovsky, A.V.; Pokrovskiy, V.S.; Grishin, D.V.; Gladilina, J.A.; Sokolov, N.N. Suppression of Telomerase Activity in Leukemic Cells by Mutant Forms of Rhodospirillum rubrum L-Asparaginase. Biochem. Suppl. Ser. B Biomed. Chem. 2017, 11, 219–233. [Google Scholar] [CrossRef]
- Plyasova, A.A.; Pokrovskaya, M.V.; Lisitsyna, O.M.; Pokrovsky, V.S.; Alexandrova, S.S.; Hilal, A.; Sokolov, N.N.; Zhdanov, D.D. Penetration into Cancer Cells via Clathrin-dependent Mechanism Allows L-asparaginase from Rhodospirillum rubrum to Inhibit Telomerase. Pharmaceuticals 2020, 13, 286. [Google Scholar] [CrossRef]
- Mezentsev, Y.V.; Molnar, A.A.; Sokolov, N.N.; Lisitsina, V.B.; Shatskaya, M.A.; Ivanov, A.S.; Archakov, A.I. Specificity of Molecular Recognition in Oligomerization of Bacterial L-Asparaginases. Biochem. Suppl. Ser. B Biomed. Chem. 2011, 5, 124–134. [Google Scholar] [CrossRef]
- Mezentsev, Y.V.; Molnar, A.A.; Gnedenko, O.V.; Krasotkina, Y.V.; Sokolov, N.N.; Ivanov, A.S. Oligomerization of L-Asparaginase from Erwinia Carotovora. Biomed. Khimiya 2006, 52, 258–271. [Google Scholar] [CrossRef]
- Ehrman, M.; Cedar, H.; Schwartz, J.H. L-Asparaginase II of Escherichia coli. J. Biol. Chem. 1971, 246, 88–94. [Google Scholar] [CrossRef]
- Dobryakova, N.V.; Zhdanov, D.D.; Sokolov, N.N.; Aleksandrova, S.S.; Pokrovskaya, M.V.; Kudryashova, E.V. Rhodospirillum rubrum L-Asparaginase Conjugates with Polyamines of Improved Biocatalytic Properties as a New Promising Drug for the Treatment of Leukemia. Appl. Sci. 2023, 13, 3373. [Google Scholar] [CrossRef]
- Pokrovskaya, M.V.; Aleksandrova, S.S.; Pokrovsky, V.S.; Veselovsky, A.V.; Grishin, D.V.; Abakumova, O.Y.; Podobed, O.V.; Mishin, A.A.; Zhdanov, D.D.; Sokolov, N.N. Identification of Functional Regions in the Rhodospirillum rubrum L-Asparaginase by Site-Directed Mutagenesis. Mol. Biotechnol. 2015, 57, 251–264. [Google Scholar] [CrossRef]
- Pokrovskaya, M.V.; Aleksandrova, S.S.; Veselovsky, A.V.; Zdanov, D.D.; Pokrovsky, V.S.; Eldarov, M.A.; Grishin, D.V.; Gladilina, Y.A.; Toropigin, I.Y.; Sokolov, N.N. Physical-Chemical Properties of L-Asparaginase Mutants From Rhodospirillum rubrum Which Showed Antitelomerase Activity. Biomed. Chem. Res. Methods 2019, 2, e00071. [Google Scholar] [CrossRef]
- Tatulian, S.A. Structural Characterization of Membrane Proteins and Peptides by FTIR and ATR-FTIR Spectroscopy. In Biotechnology and Bioengineering; Humana Press: Totowa, NJ, USA, 2013; Volume 84, pp. 177–218. [Google Scholar]
- Malakhova, M.A.; Pokrovskaya, M.V.; Alexandrova, S.S.; Sokolov, N.N.; Kudryashova, E.V. Regulation of Catalytic Activity of Recombinant L-Asparaginase from Rhodospirillum rubrum by Conjugation with a PEG-Chitosan Copolymer. Mosc. Univ. Chem. Bull. 2018, 73, 185–191. [Google Scholar] [CrossRef]
- de Moura, W.A.F.; Schultz, L.; Breyer, C.A.; de Oliveira, A.L.P.; Tairum, C.A.; Fernandes, G.C.; Toyama, M.H.; Pessoa-Jr, A.; Monteiro, G.; de Oliveira, M.A. Functional and Structural Evaluation of the Antileukaemic Enzyme L-Asparaginase II Expressed at Low Temperature by Different Escherichia Coli Strains. Biotechnol. Lett. 2020, 42, 2333–2344. [Google Scholar] [CrossRef]
- Kudryashova, E.V.; Sukhoverkov, K.V. “Reagent-Free” l-Asparaginase Activity Assay Based on CD Spectroscopy and Conductometry. Anal. Bioanal. Chem. 2016, 408, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Boos, J.; Werber, G.; Ahlke, E.; Schulze-Westhoff, P.; Nowak-Göttl, U.; Würthwein, G.; Verspohl, E.J.; Ritter, J.; Jürgens, H. Monitoring of Asparaginase Activity and Asparagine Levels in Children on Different Asparaginase Preparations. Eur. J. Cancer Part. A 1996, 32, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Fine, B.M.; Kaspers, G.J.L.; Ho, M.; Loonen, A.H.; Boxer, L.M. A Genome-Wide View of the In Vitro Response to L -Asparaginase in Acute Lymphoblastic Leukemia. Cancer Res. 2005, 65, 291–300. [Google Scholar] [CrossRef]
- Serravalle, S.; Bertuccio, S.N.; Astolfi, A.; Melchionda, F.; Pession, A. Synergistic Cytotoxic Effect of L-Asparaginase Combined with Decitabine as a Demethylating Agent in Pediatric T-ALL, with Specific Epigenetic Signature. BioMed. Res. Int. 2016, 2016, 1985750. [Google Scholar] [CrossRef]
- Van Trimpont, M.; Peeters, E.; De Visser, Y.; Schalk, A.M.; Mondelaers, V.; De Moerloose, B.; Lavie, A.; Lammens, T.; Goossens, S.; Van Vlierberghe, P. Novel Insights on the Use of L-Asparaginase as an Efficient and Safe Anti-Cancer Therapy. Cancers 2022, 14, 902. [Google Scholar] [CrossRef]
- Kusano-Arai, O.; Iwanari, H.; Mochizuki, Y.; Nakata, H.; Kodama, T.; Kitoh, T.; Hamakubo, T. Evaluation of the Asparagine Synthetase Level in Leukemia Cells by Monoclonal Antibodies. Hybridoma 2012, 31, 325–332. [Google Scholar] [CrossRef]
- Pan, Y.; Suzuki, T.; Sakai, K.; Hirano, Y.; Ikeda, H.; Hattori, A.; Dohmae, N.; Nishio, K.; Kakeya, H. Bisabosqual A: A Novel Asparagine Synthetase Inhibitor Suppressing the Proliferation and Migration of Human Non-Small Cell Lung Cancer A549 Cells. Eur. J. Pharmacol. 2023, 960, 176156. [Google Scholar] [CrossRef]
- Zhdanov, D.D.; Plyasova, A.A.; Gladilina, Y.A.; Pokrovsky, V.S.; Grishin, D.V.; Grachev, V.A.; Orlova, V.S.; Pokrovskaya, M.V.; Alexandrova, S.S.; Lobaeva, T.A.; et al. Inhibition of Telomerase Activity by Splice-Switching Oligonucleotides Targeting the MRNA of the Telomerase Catalytic Subunit Affects Proliferation of Human CD4+ T Lymphocytes. Biochem. Biophys. Res. Commun. 2019, 509, 790–796. [Google Scholar] [CrossRef]
- Byler, D.M.; Susi, H. Examination of the Secondary Structure of Proteins by sDeconvolved FTIR Spectra. Biopolymers 1986, 25, 469–487. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining Information about Protein Secondary Structures in Aqueous Solution Using Fourier Transform IR Spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Sukumaran, S. Protein Secondary Structure Elucidation Using FTIR Spectroscopy. 2018, 4. Available online: https://assets.thermofisher.com/TFS-Assets/MSD/Application-Notes/AN52985-protein-secondary-structure-elucidation-using-ftir-spectroscopy.pdf (accessed on 25 February 2025).
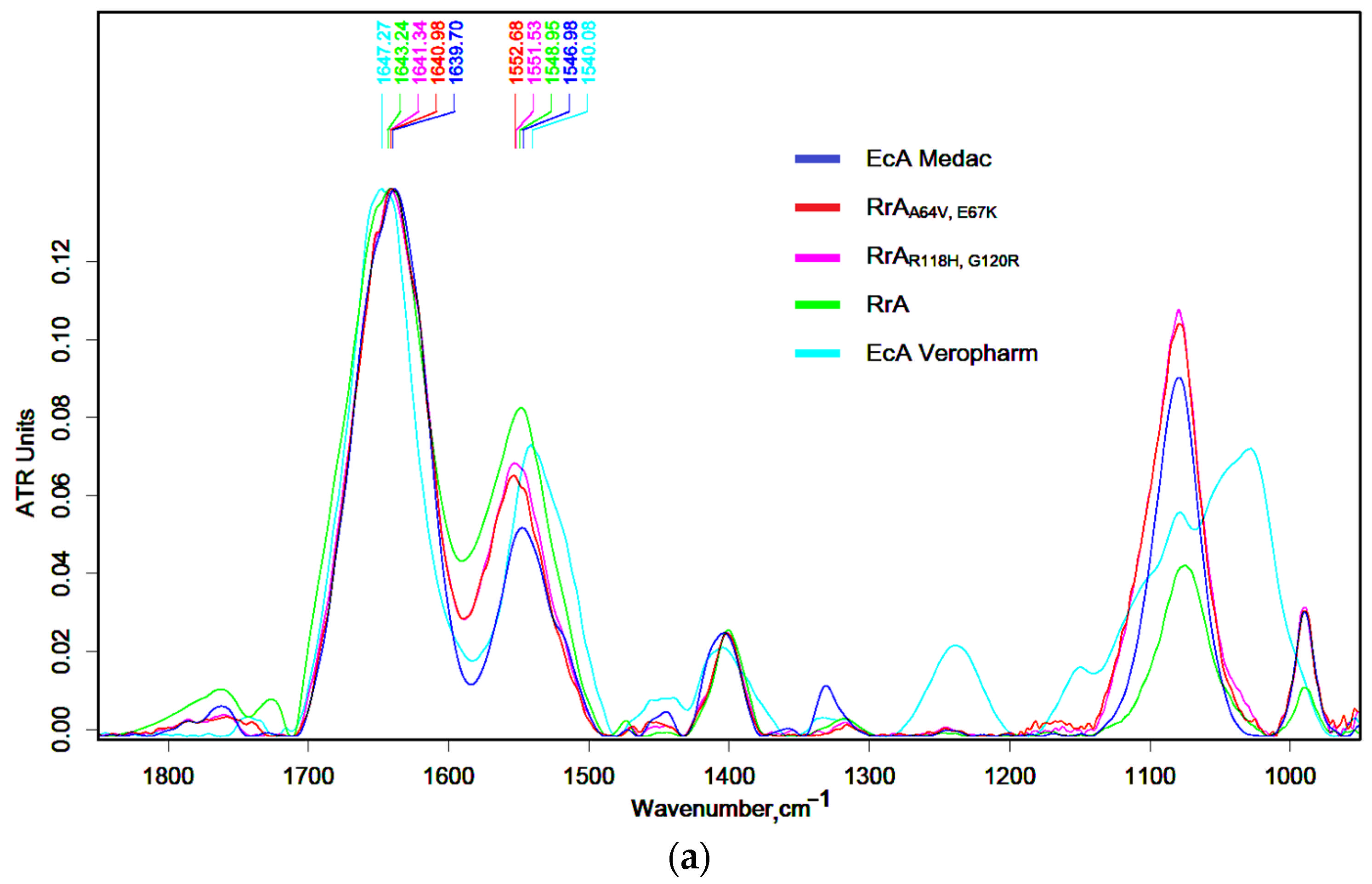

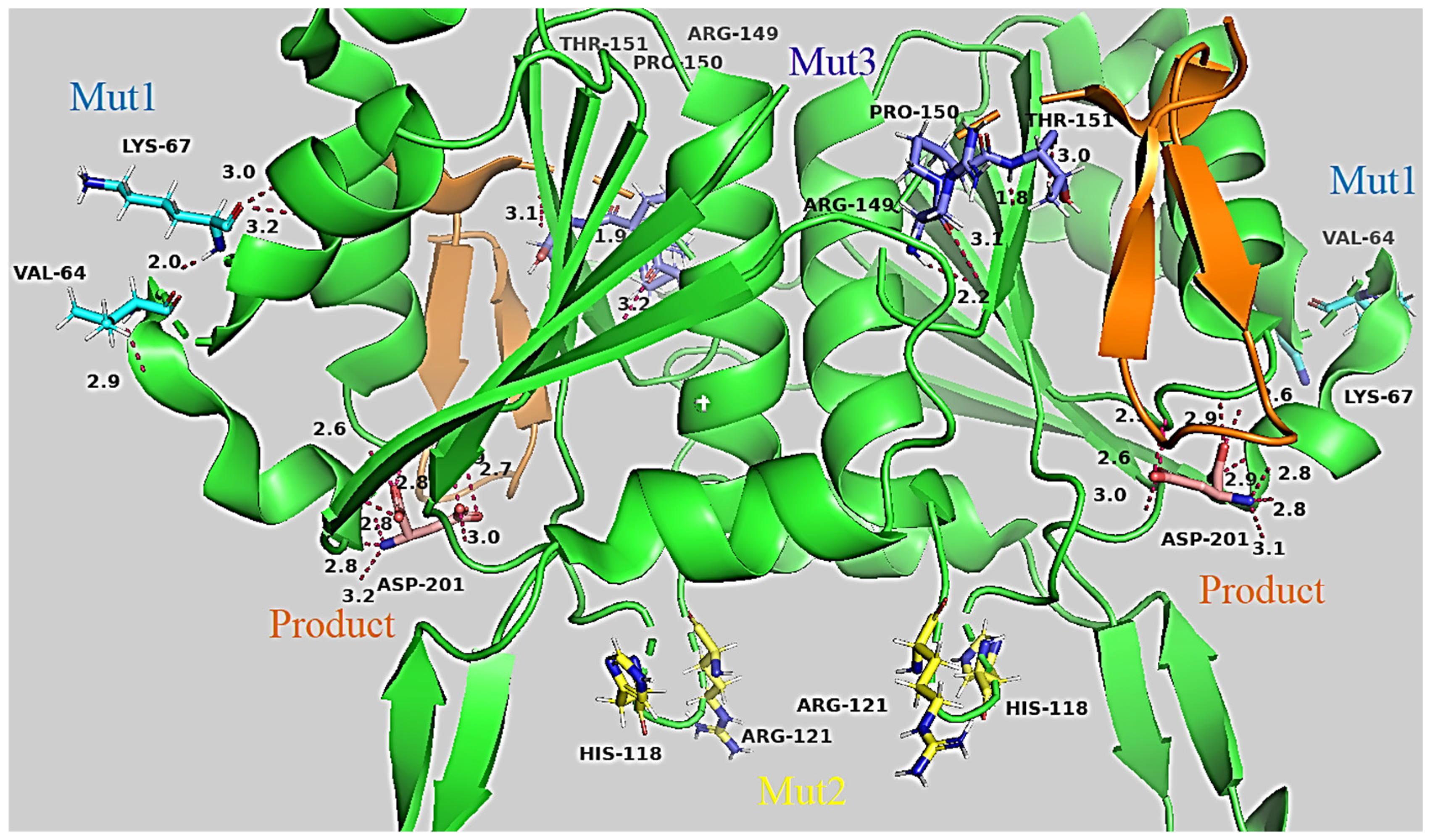


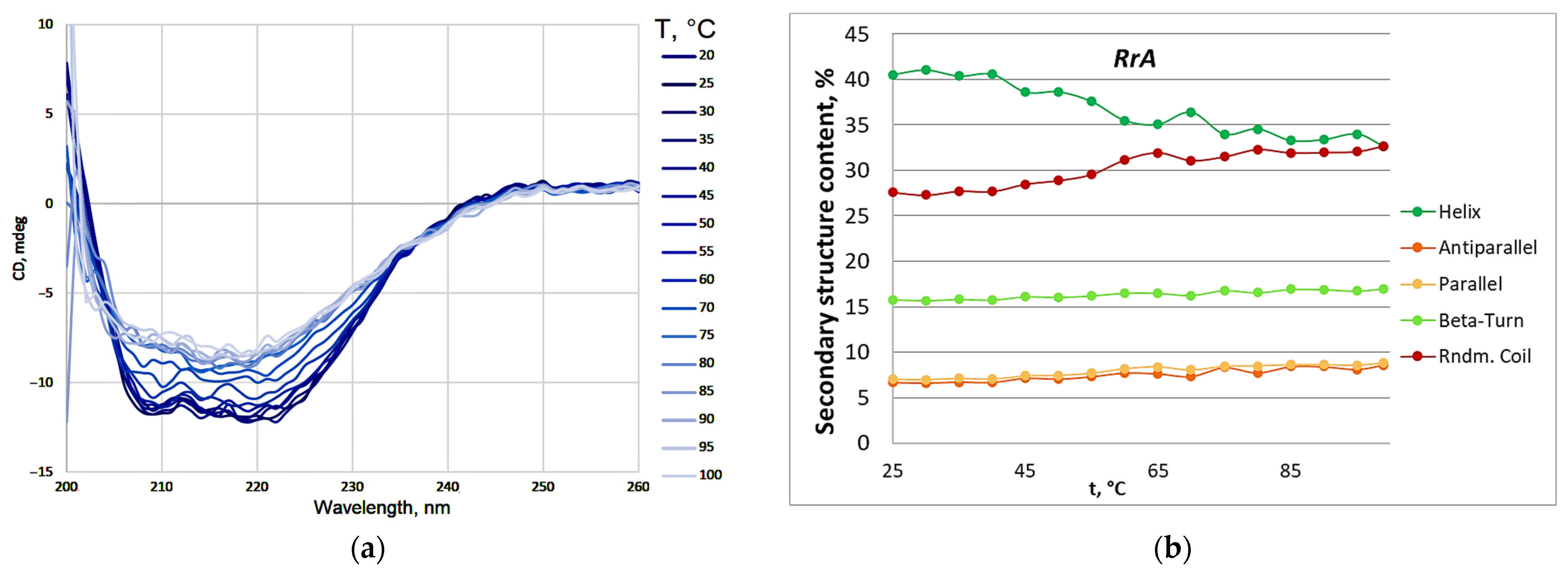
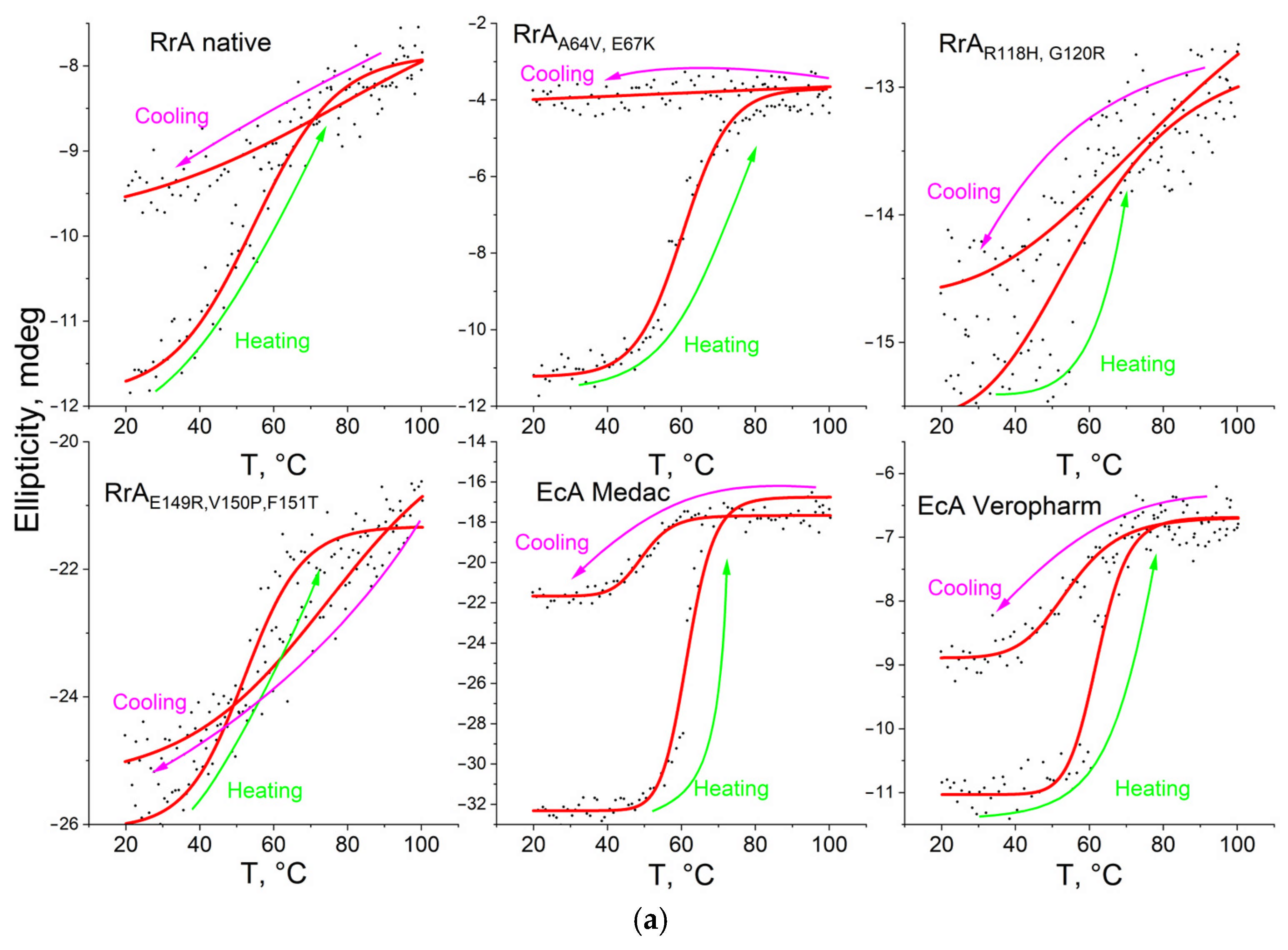

| Designation | Substitutions in DNA | Substitutions in Protein |
|---|---|---|
| Mut1 * | C191T, G199A | A64V, E67K |
| Mut2 * | G353A, G354C, G358A | R118H, G120R |
| Mut3 | G445A, A446G, G448C, T449C, T450C, T451A, T452C | E149R, V150P, F151T |
| Michaelis–Menten nonlinear regression analysis | ||||
| Enzyme | Native RrA | RrAA64V, E67K (RrA Mut1) | RrAR118H, G120R (RrA Mut2) | RrAE149R, V150P, F151T (RrA Mut3) |
| Vmax, U/mg | 29.6 ± 1.4 | 31.6 ± 0.7 | 34.9 ± 0.4 | 57.2 ± 2.4 |
| KM, mM | 4.5 ± 0.5 | 5.0 ± 0.3 | 6.7 ± 0.4 | 6.5 ± 0.7 |
| R-Square | 0.9940 | 0.9979 | 0.9998 | 0.9970 |
| Lineweaver–Burk plot | ||||
| Enzyme | Native RrA | RrAA64V, E67K (RrA Mut1) | RrAR118H, G120R (RrA Mut2) | RrAE149R, V150P, F151T (RrA Mut3) |
| Vmax, U/mg | 29.7 ± 1.8 | 31.7 ± 0.8 | 34.7 ± 0.4 | 56.4 ± 2.2 |
| KM, mM | 4.3 ± 0.7 | 5 ± 0.4 | 6.5 ± 0.4 | 6.1 ± 0.7 |
| R-Square | 0.9428 | 0.982 | 0.9832 | 0.9715 |
| Hanes–Woolf plot | ||||
| Enzyme | Native RrA | RrAA64V, E67K (RrA Mut1) | RrAR118H, G120R (RrA Mut2) | RrAE149R, V150P, F151T (RrA Mut3) |
| Vmax, U/mg | 29.7 ± 1.8 | 31.7 ± 0.8 | 34.7 ± 0.4 | 56.4 ± 2.2 |
| KM, mM | 4.3 ± 0.2 | 5 ± 0.1 | 6.5 ± 0.1 | 6.1 ± 0.1 |
| R-Square | 0.9793 | 0.9962 | 0.9991 | 0.9912 |
| Eadie–Hofstee diagram | ||||
| Enzyme | Native RrA | RrAA64V, E67K (RrA Mut1) | RrAR118H, G120R (RrA Mut2) | RrAE149R, V150P, F151T (RrA Mut3) |
| Vmax, U/mg | 30.9 ± 2.3 | 31.8 ± 0.8 | 33.9 ± 0.4 | 57.6 ± 5.6 |
| KM, mM | 5.1 ± 0.5 | 5.1 ± 0.2 | 5.7 ± 0.3 | 6.9 ± 1.2 |
| R-Square | 0.9454 | 0.9932 | 0.9843 | 0.8436 |
| Parameter | Native RrA | RrAA64V, E67K (RrA Mut1) * | RrAR118H, G120R (RrA Mut2) * | RrAE149R, V150P, F151T (RrA Mut3) | EcA Medac | EcA Veropharm |
|---|---|---|---|---|---|---|
| ln K | −1.85 | −3.73 | −1.89 | −2.08 | −6.04 | −5.37 |
| ΔH, kJ/mol | 91.1 | 132.2 | 97.8 | 60.2 | 25.1 | 26.6 |
| ΔS, J/mol/K | −15.1 | −30.6 | −15.4 | −17.1 | −50.1 | −44.6 |
| Enzyme | kin, min−1 |
|---|---|
| RrA native | 0.025 ± 0.001 |
| * RrAA64V, E67K | 0.195 ± 0.013 |
| * RrAR118H, G120R | 0.338 ± 0.012 |
| RrAE149R, V150P, F151T | 0.015 ± 0.001 |
| EcA-Medac | 0.150 ± 0.010 |
| EcA-Veropharm | 0.273 ± 0.021 |
| Enzyme | IC50 on K562 Cells, U/mL |
|---|---|
| RrA native | 15 ± 2 |
| RrAA64V, E67K * | 25 ± 3 |
| RrAR118H, G120R * | 11.5 ± 0.7 |
| RrAE149R, V150P, F151T | 10 ± 1 |
| EcA-Veropharm | 24 ± 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zlotnikov, I.D.; Shishparyonok, A.N.; Pokrovskaya, M.V.; Alexandrova, S.S.; Zhdanov, D.D.; Kudryashova, E.V. Structural Features Underlying the Mismatch Between Catalytic and Cytostatic Properties in L-Asparaginase from Rhodospirillum rubrum. Catalysts 2025, 15, 476. https://doi.org/10.3390/catal15050476
Zlotnikov ID, Shishparyonok AN, Pokrovskaya MV, Alexandrova SS, Zhdanov DD, Kudryashova EV. Structural Features Underlying the Mismatch Between Catalytic and Cytostatic Properties in L-Asparaginase from Rhodospirillum rubrum. Catalysts. 2025; 15(5):476. https://doi.org/10.3390/catal15050476
Chicago/Turabian StyleZlotnikov, Igor D., Anastasia N. Shishparyonok, Marina V. Pokrovskaya, Svetlana S. Alexandrova, Dmitry D. Zhdanov, and Elena V. Kudryashova. 2025. "Structural Features Underlying the Mismatch Between Catalytic and Cytostatic Properties in L-Asparaginase from Rhodospirillum rubrum" Catalysts 15, no. 5: 476. https://doi.org/10.3390/catal15050476
APA StyleZlotnikov, I. D., Shishparyonok, A. N., Pokrovskaya, M. V., Alexandrova, S. S., Zhdanov, D. D., & Kudryashova, E. V. (2025). Structural Features Underlying the Mismatch Between Catalytic and Cytostatic Properties in L-Asparaginase from Rhodospirillum rubrum. Catalysts, 15(5), 476. https://doi.org/10.3390/catal15050476







