Comparative Computational Assessment of Hydrocarbon Bioremediation Potential Using Catechol 2,3-Dioxygenases from Cytobacillus kochii and Marinobacter sp.
Abstract
1. Introduction
2. Results and Discussions
2.1. Enzyme Selection and Physiochemical Properties of Enzymes
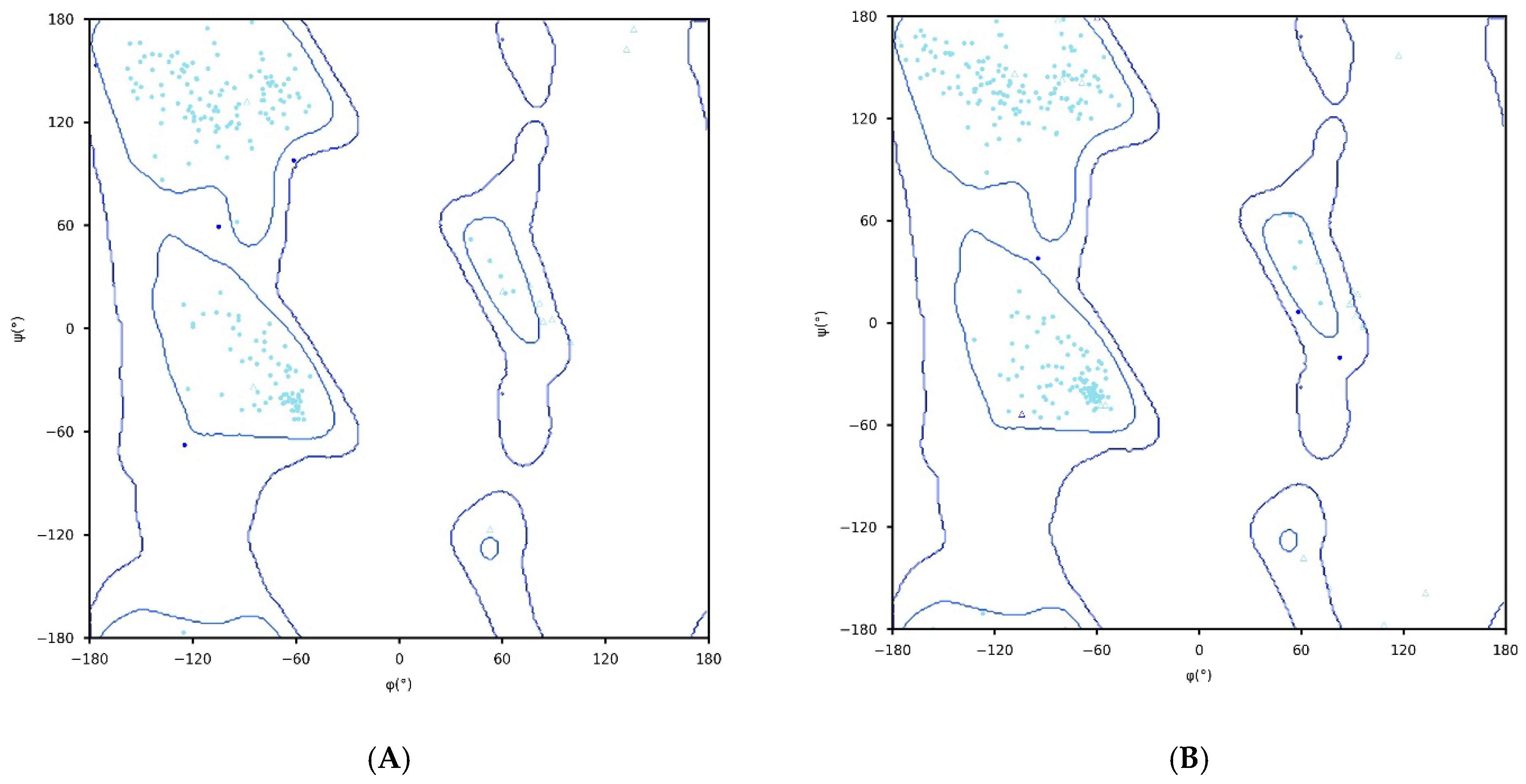
2.2. Three-Dimensional Modelling and the Validation
2.3. Virtual Screening of Hydrocarbon Pollutants Present in Environment
2.4. The Active Sites
2.5. Comparative Structural and Interaction Studies
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Structure Modelling and Physiochemical Properties
3.2.2. Structure Validation
3.2.3. Virtual Screening of Hydrocarbon Pollutants Present in Environment
3.2.4. Ligand Preparation
3.2.5. Comparative Structural and Interaction Studies
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef]
- Kalia, A.; Sharma, S.; Semor, N.; Babele, P.K.; Sagar, S.; Bhatia, R.K.; Walia, A. Recent advancements in hydrocarbon bioremediation and future challenges: A review. Biotech 2022, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Miles, S.M.; Gestler, R.; Dworatzek, S.M. Bioremediation of Petroleum Hydrocarbons in the Subsurface. In Advances in the Characterisation and Remediation of Sites Contaminated with Petroleum Hydrocarbons; García-Rincón, J., Gatsios, E., Lenhard, R.J., Atekwana, E.A., Naidu, R., Eds.; Springer: Cham, Switzerland, 2024; pp. 479–502. [Google Scholar]
- Harayama, S.; Kok, M.; Neidle, E.L. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 1992, 46, 565–601. [Google Scholar] [CrossRef] [PubMed]
- Duran, R. Marinobacter. In Handbook of Hydrocarbon and Lipid Microbiology; Eckey, C., Fabiani, S., Eds.; SPI-Publishing: Pondicherry, India, 2010; pp. 1725–1735. [Google Scholar]
- Chen, G.; Seukep, A.J.; Guo, M. Recent Advances in Molecular Docking for the Research and Discovery of Potential Marine Drugs. Mar. Drugs 2020, 18, 545. [Google Scholar] [CrossRef]
- Temelkovski, D.; Kiss, T.; Terstyanszky, G.; Greenwell, P. Building Science Gateways for Analysing Molecular Docking Results Using a Generic Framework and Methodology. J. Grid. Comput. 2020, 18, 529–546. [Google Scholar] [CrossRef]
- Raddadi, N.; Giacomucci, L.; Totaro, G.; Fava, F. Marinobacter sp. from marine sediments produce highly stable surface-active agents for combatting marine oil spills. Microb. Cell Fact. 2017, 16, 186. [Google Scholar] [CrossRef]
- Petra, P.H.; Bradshaw, R.A.; Walsh, K.A.; Neurath, H. Identification of the Amino Acid Replacements Characterizing the Allotypic Forms of Bovine Carboxypeptidase A. Biochemistry 1969, 8, 2762–2768. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, H.; Gao, C.; Zang, J.; Yang, D. The Distinct Properties of the Consecutive Disordered Regions Inside or Outside Protein Domains and Their Functional Significance. Int. J. Mol. Sci. 2021, 22, 10677. [Google Scholar] [CrossRef]
- Payá, G.; Bautista, V.; Camacho, M.; Esclapez, J.; Bonete, M.J. Comprehensive Bioinformatics Analysis of the Biodiversity of Lsm Proteins in the Archaea Domain. Microorganisms 2023, 11, 1196. [Google Scholar] [CrossRef]
- ATASDR. Toxicological Profile for Polycyclic Aromatic Hydrocarbons; Agency for Toxic Substances and Disease Registry: Atlanta, GA, USA, 1995; pp. 11–64. [Google Scholar]
- Chakrabarti, C.; Khimani, M.; Patel, V.; Parekh, P.; Pillai, S.; Mata, J.; Vekariya, R.L.; Bhadja, P.; Muddassir, M. Solubilization of polycyclic aromatic hydrocarbons (PAHs) in PEO-PPO-PEO type linear and star block copolymers. J. Mol. Liq. 2021, 325, 115177. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Zhang, J.; Li, S.; Liu, J.; Ghalib, H.B.; Zhang, X.; Ma, F. Biodegradation of n-hexadecane by Aspergillus sp. RFC-1 and its mechanism. Ecotoxicol. Environ. Saf. 2018, 164, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Hidayat, A.; Tachibana, S. Crude oil and n-octadecane degradation under saline conditions by Fusarium sp., F092. J. Environ. Sci. Technol. 2013, 6, 29–40. [Google Scholar] [CrossRef]
- National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554481/ (accessed on 6 May 2025).
- Zhang, Z.; Cai, Y.; Zheng, N.; Deng, Y.; Gao, L.; Wang, Q.; Xia, X. Diverse models of cavity engineering in enzyme modification: Creation, filling, and reshaping. Biotechnol. Adv. 2024, 72, 108346. [Google Scholar] [CrossRef]
- Weiß, R.G.; Chudoba, R.; Setny, P.; Dzubiella, J. Affinity, Kinetics, and Pathways of Anisotropic Ligands Binding to Hydrophobic Model Pockets. J. Chem. Phys. 2018, 149, 094902. [Google Scholar] [CrossRef]
- Vaillancourt, F.H.; Bolin, J.T.; Eltis, L.D. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 241–267. [Google Scholar] [CrossRef]
- Saragi, R.T.; Calabrese, C.; Juanes, M.; Pinacho, R.; Rubio, J.E.; Pérez, C.; Lesarri, A. π-Stacking Isomerism in Polycyclic Aromatic Hydrocarbons: The 2-Naphthalenethiol Dimer. J. Phys. Chem. Lett. 2023, 14, 207–213. [Google Scholar] [CrossRef]
- Vnučec, D.; Kutnar, A.; Goršek, A. Soy-based adhesives for wood-bonding—A review. J. Adhes. Sci. Technol. 2017, 31, 910–931. [Google Scholar] [CrossRef]
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein Binding Pocket Dynamics. Acc. Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Semana, P.; Powlowski, J. Four Aromatic Intradiol Ring Cleavage Dioxygenases from Aspergillus niger. Appl. Environ. Microbiol. 2019, 85, e01786-19. [Google Scholar] [CrossRef]
- Meckenstock, R.U.; Elsner, M.; Griebler, C.; Lueders, T.; Stumpp, C.; Aamand, J.; Agathos, S.N.; Albrechtsen, H.J.; Bastiaens, L.; Bjerg, P.L.; et al. Biodegradation: Updating the Concepts of Control for Microbial Cleanup in Contaminated Aquifers. Environ. Sci. Technol. 2015, 49, 7073–7081. [Google Scholar] [CrossRef]
- Jamal, M.T.; Pugazhendi, A. Isolation and characterization of halophilic bacterial consortium from seagrass, Jeddah coast, for the degradation of petroleum hydrocarbons and treatment of hydrocarbons—Contaminated boat fuel station wastewater. Clean Technol. Environ. Policy 2021, 23, 77–88. [Google Scholar] [CrossRef]
- Alim, M.B.; Oves, M.; Jamal, M.T.; Kunarso, K. Comparative Hydrocarbon Biodegradation by Bacillus haikouensis and Cytobacillus kochii: Pure Culture versus Consortium Performance. J. Pure Appl. Microbiol. 2025, 19, 2069–2086. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins Struct. Funct. Genet. 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Suzuki, N. Toxicities of Polycyclic Aromatic Hydrocarbons for Aquatic Animals. Int. J. Environ. Res. Public Health 2020, 17, 1363. [Google Scholar] [CrossRef] [PubMed]



| No. | Source | No. of Amino Acids | Theoretical pI | Molecular Formula | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|
| 1 | Marinobacter | 307 | 5.04 | C1569H2395N429O462S14 | 75.86 | −0.410 (non-polar) |
| 2 | Cytobacillus kochii | 214 | 5.90 | C1109H1723N293O328S3 | 90.70 | −0.347 (non-polar) |
| No. | Source | 3D Structure Obtained by AlphaFold | The Confidence Value in the AlphaFold (pLDDT) |
|---|---|---|---|
| 1 | Cytobacillus kochii |  | 96.5% |
| 2 | Marinobacter |  | 97.4% |
| No. | Compounds | 3D Structure | Molecular Weight (g/mol) | Molecular Formula |
|---|---|---|---|---|
| 1 | Benzene |  | 78.112 | C6H6 |
| 2 | Toluene | 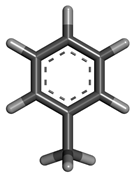 | 92.138 | C6H5CH3 |
| 3 | m-xylene | 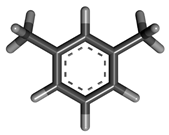 | 106.16 | C8H10 |
| 4 | Naphthalene |  | 128.171 | C10H8 |
| 5 | Anthracene | 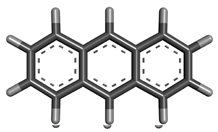 | 178.229 | C14H10 |
| 6 | Phenanthrene | 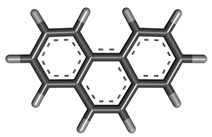 | 178.229 | C14H10 |
| 7 | Pyrene | 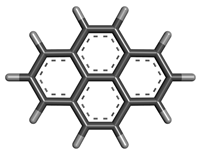 | 202.25 | C16H10 |
| 8 | Fluoranthene | 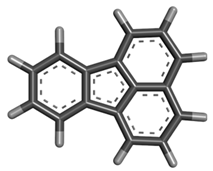 | 202.25 | C16H10 |
| 9 | Octadecane |  | 254.494 | C18H38 |
| 10 | Hexadecane |  | 226.441 | C16H34 |
| No. | Compound | Source | Binding (Kcal/mol) | Interaction (Number of Interactions) | Residues |
|---|---|---|---|---|---|
| 1 | Benzene | Cytobacillus kochii | −4 | pi-Alkyl (1) | ILE 183 |
| Van der Waals (7) | LEU 170, TRP 191, HIS 58, TYR 101, GLU 136, PHE 92, PHE 172 | ||||
| pi-Sigma (1) | ILE 192 | ||||
| Marinobacter | −4.1 | Van der Waals (2) | LEU 143, TRP 47 | ||
| pi-Cation (1) | LYS 147 | ||||
| 2 | Toluene | Cytobacillus kochii | −4.4 | pi-pi stacked (1) | PHE 92 |
| Pi-Alkyl (3) | PHE 172, ALA 95, PHE 92 | ||||
| Van der Waals (1) | ARG 115 | ||||
| Alkyl (1) | ALA 95 | ||||
| Marinobacter | −4.8 | pi-Alkyl (2) | PHE 191, LEU 155 | ||
| Van der Waals (7) | ILE 204, THR 249, HIS 246, GLU 265, HIS 199, HIS 153, HIS214 | ||||
| pi-pi T Shaped (1) | PHE 191 | ||||
| Alkyl | LEU 248, LEU 155, ILE 291 | ||||
| 3 | m-Xylene | Cytobacillus kochii | −5.2 | Pi-Alkyl (4) | TRP 191, PHE 172, ILE 183, PHE 92 |
| Van der Waals (3) | ARG 189, HIS 58, TYR 101 | ||||
| pi-Sigma (1) | ILE 192 | ||||
| Alkyl (2) | ILE 192, LEU 170 | ||||
| Marinobacter | −5.5 | pi-Alkyl (6) | HIS 214, HIS 199, LEU 155, PHE 191, PHE 191, ILE 291 | ||
| Van der Waals (2) | TYR 255, PHE 302 | ||||
| pi-Sigma (1) | LEU 248 | ||||
| pi-pi T Shaped (1) | HIS 246 | ||||
| Alkyl (5) | LEU 155, LEU 155, LEU 248, ILE 204, ILE 291 | ||||
| 4 | Naphthalene | Cytobacillus kochii | −6.3 | pi-pi stacked (2) | PHE 92, PHE 172 |
| pi-Alkyl (1) | ALA 95 | ||||
| pi-pi T-shaped | PHE 92, PHE 172 | ||||
| Van der Waals (7) | LEU 170, ILE 192, ILE 183, TYR 101, GLU 136, ILE 113, ARG 115 | ||||
| Marinobacter | −6.1 | pi-pi stacked (2) | TRP 139, TRP 139 | ||
| pi-Alkyl (1) | LYS 147 | ||||
| Van der Waals (5) | ARG 141, PRO 140, LEU 143, TRP 47, GLU 220 | ||||
| pi-Cation (1) | LYS 147 | ||||
| 5 | Anthracene | Cytobacillus kochii | −8 | pi-pi stacked (2) | PHE 92, PHE 92 |
| pi-Alkyl (3) | ALA 95, ILE 183, LEU 170 | ||||
| Van der Waals (6) | ILE 113, ARG 115, GLU 136, TYR 101, HIS 58, TRP 191 | ||||
| pi-sigma (1) | ILE 192 | ||||
| pi-pi T-Shaped (2) | PHE 172, PHE 172 | ||||
| Marinobacter | −7.1 | pi-pi stacked (3) | TRP 139, TRP 139, TRP 139 | ||
| pi-Alkyl (2) | LYS 147, ARG 141 | ||||
| Van der Waals (4) | GLU 220, TRP 47, LEU 143, PRO 140 | ||||
| pi-Cation (2) | LYS 147, LYS 147 | ||||
| 6 | Phenanthrene | Cytobacillus kochii | −8.2 | pi-pi stacked (3) | PHE 92, PHE 92, PHE 92 |
| pi-Alkyl (1) | ILE 183 | ||||
| Van der Waals (7) | LEU 170, TRP 191, TYR 101, GLU 111, HIS 58, GLU 136, ARG 115 | ||||
| pi-Sigma (1) | ILE 192 | ||||
| pi-pi T-Shaped (2) | PHE 172, PHE 172 | ||||
| Marinobacter | −7.2 | pi-pi stacked (3) | TRP 139, TRP 139, TRP 139 | ||
| pi-Alkyl (1) | ARG 141 | ||||
| Van der Waals (4) | PRO 140, LEU 143, TRP 47, GLU 220 | ||||
| pi-Cation (3) | LYS 147, LYS 147, LYS 147 | ||||
| 7 | Pyrene | Cytobacillus kochii | −8.5 | pi-pi Stacked (3) | PHE 92, PHE 92, PHE 92 |
| pi-Alkyl (3) | ILE 192, LEU 170, ALA 95 | ||||
| Van der Waals (6) | ARG 189, TYR 101, ILE 183, GLU 136, ILE 113, ARG 115 | ||||
| pi-pi T-shaped (2) | PHE 172, PHE 172 | ||||
| Marinobacter | −7.9 | pi-pi stacked (4) | TRP 139, TRP 139, TRP 139, TRP 139 | ||
| pi-Alkyl (1) | LYS 147 | ||||
| Van der Waals (5) | GLU 220, TRP 47, LEU 143, PRO 140, ARG 141 | ||||
| pi-Cation (2) | LYS 147, LYS 147 | ||||
| 8 | Fluoranthene | Cytobacillus kochii | −8.7 | pi-pi Stacked (4) | PHE 92, PHE 92, PHE 92, PHE 92 |
| pi-Alkyl (3) | ALA 95, ILE 183, ILE 192 | ||||
| Van der Waals (5) | ARG 115, GLU 136, TYR 101, HIS 58, ARG 189 | ||||
| pi-Sigma (2) | ILE 192, LEU 170 | ||||
| pi-pi T-shaped (2) | PHE 172, PHE 172 | ||||
| Marinobacter | −8 | pi-pi stacked (4) | TRP 139, TRP 139, TRP 139, TRP 139 | ||
| pi-Alkyl (1) | LYS 147 | ||||
| Van der Waals (5) | ARG 141, PRO 140, LEU 143, TRP 47, GLU 220 | ||||
| pi-Cation (2) | LYS 147, LYS 147 | ||||
| 9 | Octadecane | Cytobacillus kochii | −4.9 | pi-Alkyl (6) | TYR 169, PHE 172, TRP 191, PHE 92, PHE 92, PHE 172 |
| Van der Waals (6) | LYS 165, GLU 94, ARG 159, ILE 183, TYR 101, ARG 189 | ||||
| Alkyl (4) | LEU 170, LEU 170, ALA 95, ILE 192 | ||||
| Marinobacter | −4.2 | pi-Alkyl (3) | TRP 47, TRP 139, TRP 139 | ||
| Van der Waals (3) | LEU 143, GLU 220, ASP 224 | ||||
| Alkyl (4) | LYS 147, LYS 147, LYS 147, LYS 227 | ||||
| 10 | Hexadecane | Cytobacillus kochii | −4.8 | pi-Alkyl (6) | TYR 169, PHE 172, PHE 172, PHE 92, PHE 92, HIS 58 |
| Van der Waals (8) | GLU 162, ASN 171, SER 163, ARG 159, ARG 189, TRP 191, GLU 136, TYR 101 | ||||
| Alkyl (8) | LYS 165, LYS 165, LEU 170, LEU 170, LEU 170, ILE 192, ILE 192, ILE 183 | ||||
| Marinobacter | −4.3 | pi-Alkyl (3) | TRP 139, TRP 139, TRP 47 | ||
| Van der Waals (3) | MET 146, ASP 224, GLU 220 | ||||
| Alkyl (5) | LYS 227, LEU 143, LYS 147, LYS 147, LYS 147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alim, M.B.; Oves, M.; Jamal, M.T. Comparative Computational Assessment of Hydrocarbon Bioremediation Potential Using Catechol 2,3-Dioxygenases from Cytobacillus kochii and Marinobacter sp. Catalysts 2025, 15, 1100. https://doi.org/10.3390/catal15121100
Alim MB, Oves M, Jamal MT. Comparative Computational Assessment of Hydrocarbon Bioremediation Potential Using Catechol 2,3-Dioxygenases from Cytobacillus kochii and Marinobacter sp. Catalysts. 2025; 15(12):1100. https://doi.org/10.3390/catal15121100
Chicago/Turabian StyleAlim, Muhammad B., Mohamad Oves, and Mamdoh T. Jamal. 2025. "Comparative Computational Assessment of Hydrocarbon Bioremediation Potential Using Catechol 2,3-Dioxygenases from Cytobacillus kochii and Marinobacter sp." Catalysts 15, no. 12: 1100. https://doi.org/10.3390/catal15121100
APA StyleAlim, M. B., Oves, M., & Jamal, M. T. (2025). Comparative Computational Assessment of Hydrocarbon Bioremediation Potential Using Catechol 2,3-Dioxygenases from Cytobacillus kochii and Marinobacter sp. Catalysts, 15(12), 1100. https://doi.org/10.3390/catal15121100








