Identification of Five Robust Novel Ene-Reductases from Thermophilic Fungi
Abstract
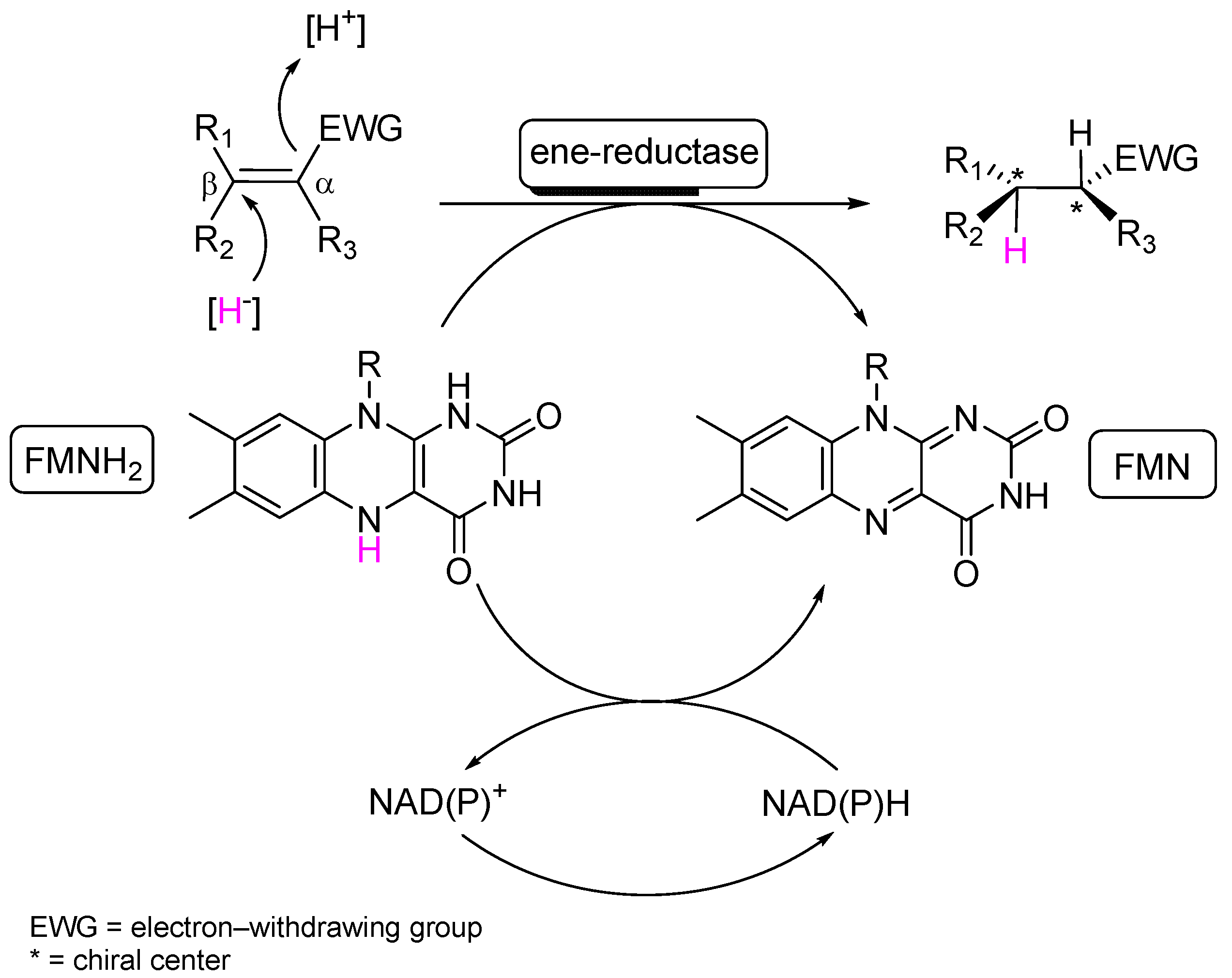
1. Introduction
2. Results and Discussion
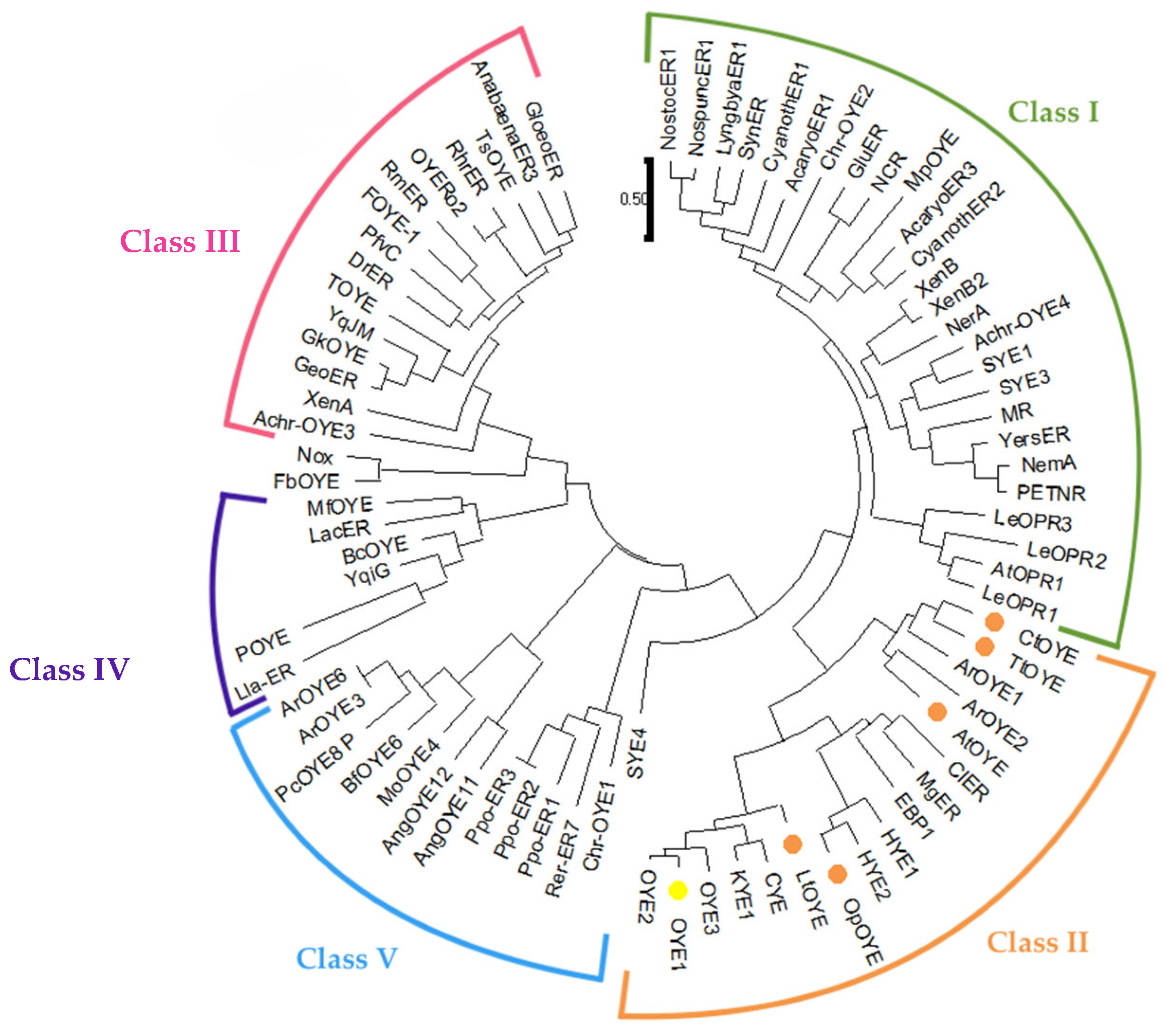
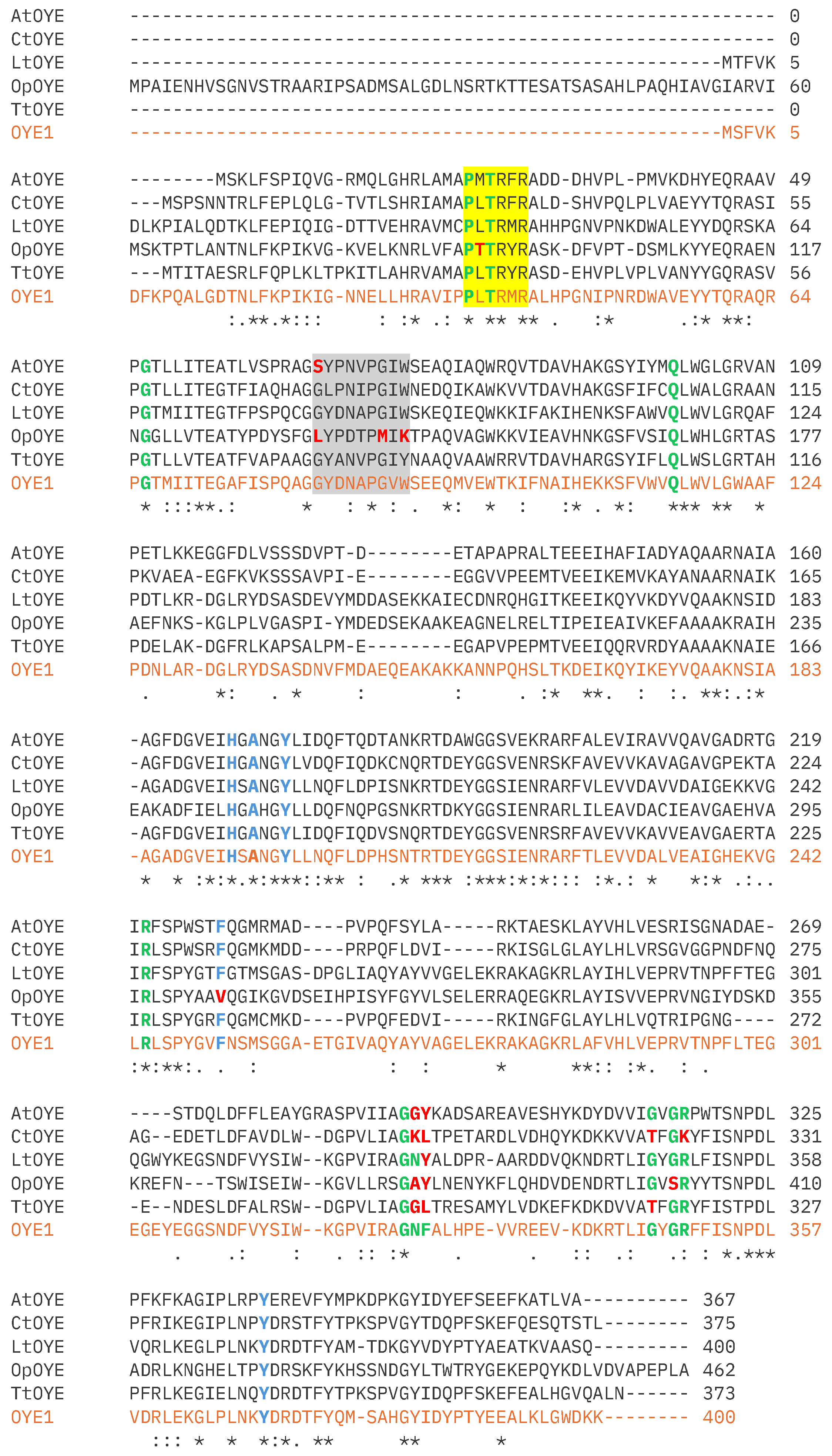
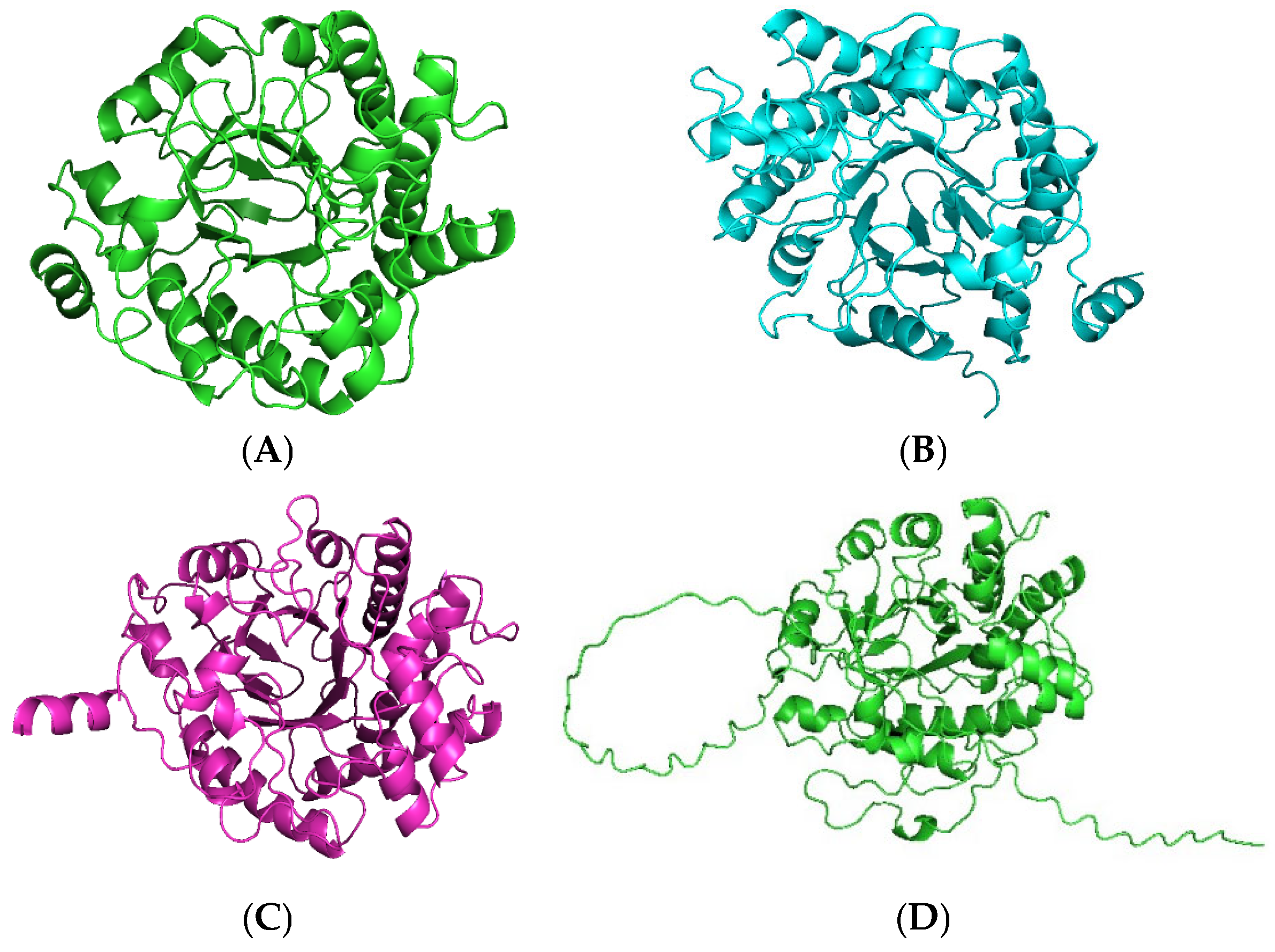
2.1. Identification and Sequence Analyses of New Putative ERs
2.2. Production and Purification of Fungal ERs
2.3. Substrate Acceptance
2.4. Steady-State Kinetic Parameters
2.5. Bioconversion
2.6. Thermostability
3. Materials and Methods
3.1. Chemicals, Strains, and Enzymes
3.2. Bioinformatic Analysis
3.3. Plasmid Construction
3.4. Expression
3.5. Purification
3.6. Spectral Analysis
3.7. Enzyme Analysis
3.8. Bioconversions
3.9. Analysis of Thermostability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gatti, F.G.; Parmeggiani, F.; Sacchetti, A. Synthetic strategies based on C=C bioreductions for the preparation of biologically active molecules. In Synthetic Methods for Biologically Active Molecules, 1st ed.; Brenna, E., Ed.; Wiley: Hoboken, NJ, USA, 2013; pp. 49–84. [Google Scholar]
- Toogood, H.S.; Scrutton, N.S. Discovery, characterization, engineering, and applications of ene-reductases for industrial biocatalysis. ACS Catal. 2018, 8, 3532–3549. [Google Scholar] [PubMed]
- Hecht, K.; Buller, R. Ene-reductases in pharmaceutical chemistry. In Pharmaceutical Biocatalysis: Chemoenzymatic Synthesis of Active Pharmaceutical Ingredients; Grunwald, P., Ed.; Jenny Stanford: Singapore, 2019. [Google Scholar]
- Mathew, S.; Trajkovic, M.; Kumar, H.; Nguyen, Q.T.; Fraaije, M.W. Enantio- and regioselective ene -reductions using F420H2-dependent enzymes. Chem. Commun. 2018, 54, 11208–11211. [Google Scholar]
- Warburg, O.; Christian, W. Yellow enzyme and its effects. Biochemistry 1933, 266, 377–411. [Google Scholar]
- Knaus, T.; Toogood, H.S.; Scrutton, N.S. Ene-reductases and their applications. In Green Biocatalysis; Patel, R.N., Ed.; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2016; pp. 473–488. [Google Scholar]
- Scholtissek, A.; Tischler, D.; Westphal, A.; Van Berkel, W.; Paul, C. Old yellow enzyme-catalysed asymmetric hydrogenation: Linking family roots with improved catalysis. Catalysts 2017, 7, 130. [Google Scholar] [CrossRef]
- Kitzing, K.; Fitzpatrick, T.B.; Wilken, C.; Sawa, J.; Bourenkov, G.P.; Macheroux, P.; Clausen, T. The 1.3 Å crystal structure of the flavoprotein YqJM reveals a novel class of old yellow enzymes. J. Biol. Chem. 2005, 280, 27904–27913. [Google Scholar]
- Böhmer, S.; Marx, C.; Gómez-Baraibar, Á.; Nowaczyk, M.M.; Tischler, D.; Hemschemeier, A.; Happe, T. Evolutionary diverse Chlamydomonas reinhardtii old yellow enzymes reveal distinctive catalytic properties and potential for whole cell biotransformation. Algal Res. 2020, 50, 101970. [Google Scholar]
- Stuermer, R.; Hauer, B.; Hall, M.; Faber, K. Asymmetric bioreduction of activated C=C bonds using enoate reductases from the old yellow enzyme family. Curr. Opin. Chem. Biol. 2007, 11, 203–213. [Google Scholar]
- Williams, R.E.; Bruce, N.C. ‘New uses for an old enzyme’—The old yellow enzyme family of flavoenzymes. Microbiology 2002, 148, 1607–1614. [Google Scholar]
- Knowles, W.S.; Noyori, R. Pioneering perspectives on asymmetric hydrogenation. Acc. Chem. Res. 2007, 40, 1238–1239. [Google Scholar]
- Kumar Roy, T.; Sreedharan, R.; Ghosh, P.; Gandhi, T.; Maiti, D. Ene-reductase: A multifaceted biocatalyst in organic synthesis. Chem.—A Eur. J. 2022, 28, e202103949. [Google Scholar]
- Kohli, R.M.; Massey, V. The oxidative half-reaction of old yellow enzyme. J. Biol. Chem. 1998, 273, 32763–32770. [Google Scholar] [CrossRef]
- Parmeggiani, F.; Brenna, E.; Colombo, D.; Gatti, F.G.; Tentori, F.; Tessaro, D. “A study in yellow”: Investigations in the stereoselectivity of ene-reductases. ChemBioChem 2022, 23, e202100445. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.K.; Tasnádi, G.; Clay, D.; Hall, M.; Faber, K. Asymmetric bioreduction of activated alkenes to industrially relevant optically active compounds. J. Biotechnol. 2012, 162, 381–389. [Google Scholar] [CrossRef]
- Atalah, J.; Cáceres-Moreno, P.; Espina, G.; Blamey, J.M. Thermophiles and the applications of their enzymes as new biocatalysts. Bioresour. Technol. 2019, 280, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Littlechild, J.A. Improving the ‘toolbox’ for robust industrial enzymes. J. Ind. Microbiol. Biotechnol. 2017, 44, 711–720. [Google Scholar] [CrossRef]
- Robescu, M.S.; Loprete, G.; Gasparotto, M.; Vascon, F.; Filippini, F.; Cendron, L.; Bergantino, E. The family keeps on growing: Four novel fungal OYEs characterized. Int. J. Mol. Sci. 2022, 23, 3050. [Google Scholar] [CrossRef]
- Robescu, M.S.; Niero, M.; Loprete, G.; Cendron, L.; Bergantino, E. A new thermophilic ene-reductase from the filamentous anoxygenic phototrophic bacterium Chloroflexus aggregans. Microorganisms 2021, 9, 953. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bharadwaj, G.; Bhat, M.K. Thermophilic fungi: Their physiology and enzymes. Microbiol. Mol. Biol. Rev. 2000, 64, 461–488. [Google Scholar] [CrossRef]
- Ejmal, M.A.; Holland, D.J.; MacDiarmid, R.M.; Pearson, M.N. The effect of Aspergillus thermomutatus chrysovirus 1 on the biology of three Aspergillus species. Viruses 2018, 10, 539. [Google Scholar] [CrossRef]
- Benassi, V.M.; Lucas, R.C.D.; Jorge, J.A.; Polizeli, M.d.L.T.d.M. Screening of thermotolerant and thermophilic fungi aiming β-xylosidase and arabinanase production. Braz. J. Microbiol. 2014, 45, 1459–1467. [Google Scholar] [CrossRef]
- Tansey, M.R. Effect of temperature on growth rate and development of the thermophilic fungus Chaetomium Thermophile. Mycologia 1972, 64, 1290–1299. [Google Scholar] [CrossRef]
- Velasco, J.; Oliva, B.; Gonçalves, A.L.; Lima, A.S.; Ferreira, G.; França, B.A.; Mulinari, E.J.; Gonçalves, T.A.; Squina, F.M.; Kadowaki MA, S.; et al. Functional characterization of a novel thermophilic exo-arabinanase from Thermothielavioides terrestris. Appl. Microbiol. Biotechnol. 2020, 104, 8309–8326. [Google Scholar] [CrossRef] [PubMed]
- Morata, A.; Loira, I.; Tesfaye, W.; Bañuelos, M.; González, C.; Suárez Lepe, J. Lachancea thermotolerans applications in wine technology. Fermentation 2018, 4, 53. [Google Scholar] [CrossRef]
- Liebal, U.W.; Fabry, B.A.; Ravikrishnan, A.; Schedel, C.V.; Schmitz, S.; Blank, L.M.; Ebert, B.E. Genome-scale model reconstruction of the methylotrophic yeast Ogataea polymorpha. BMC Biotechnol. 2021, 21, 23. [Google Scholar] [CrossRef]
- Oberdorfer, G.; Steinkellner, G.; Stueckler, C.; Faber, K.; Gruber, K. Stereopreferences of old yellow enzymes: Structure correlations and sequence patterns in enoate reductases. ChemCatChem 2011, 3, 1562–1566. [Google Scholar] [CrossRef]
- Nizam, S.; Verma, S.; Borah, N.N.; Gazara, R.K.; Verma, P.K. Comprehensive genome-wide analysis reveals different classes of enigmatic old yellow enzyme in fungi. Sci. Rep. 2014, 4, 4013. [Google Scholar] [CrossRef]
- Brown, B.J.; Deng, Z.; Karplus, P.A.; Massey, V. On the active site of old yellow enzyme. J. Biol. Chem. 1998, 273, 32753–32762. [Google Scholar] [CrossRef]
- Nizam, S.; Gazara, R.K.; Verma, S.; Singh, K.; Verma, P.K. Comparative structural modeling of six old yellow enzymes (OYEs) from the necrotrophic fungus Ascochyta rabiei: Insight into novel OYE classes with differences in cofactor binding, organization of active site residues and stereopreferences. PLoS ONE 2014, 9, e95989. [Google Scholar] [CrossRef]
- Riedel, A.; Mehnert, M.; Paul, C.E.; Westphal, A.H.; Van Berkel, W.J.H.; Tischler, D. Functional characterization and stability improvement of a ‘thermophilic-like’ ene-reductase from Rhodococcus opacus 1CP. Front. Microbiol. 2015, 6, 1073. [Google Scholar] [CrossRef]
- Aregger, D.; Peters, C.; Buller, R.M. Characterization of the novel ene reductase Ppo-ER1 from Paenibacillus polymyxa. Catalysts 2020, 10, 254. [Google Scholar] [CrossRef]
- Fu, Y.; Castiglione, K.; Weuster-Botz, D. Comparative characterization of novel ene-reductases from cyanobacteria. Biotechnol. Bioeng. 2013, 110, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Yu, H.L.; Lin, G.Q.; Xu, J.H. An ene reductase from Clavispora lusitaniae for asymmetric reduction of activated alkenes. Enzym. Microb. Technol. 2014, 56, 40–45. [Google Scholar] [CrossRef]
- Tischler, D.; Gädke, E.; Eggerichs, D.; Gomez Baraibar, A.; Mügge, C.; Scholtissek, A.; Paul, C.E. Asymmetric reduction of (R)-carvone through a thermostable and organic-solvent-tolerant ene-reductase. ChemBioChem 2020, 21, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Torres Pazmiño, D.E.; Riebel, A.; De Lange, J.; Rudroff, F.; Mihovilovic, M.D.; Fraaije, M.W. Efficient biooxidations catalyzed by a new generation of self-sufficient Baeyer-Villiger monooxygenases. ChemBioChem 2009, 10, 2595–2598. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef]
- Forneris, F.; Orru, R.; Bonivento, D.; Chiarelli, L.R.; Mattevi, A. ThermoFAD, a ThermoFluor®-adapted flavin ad hoc detection system for protein folding and ligand binding. FEBS J. 2009, 276, 2833–2840. [Google Scholar] [CrossRef]
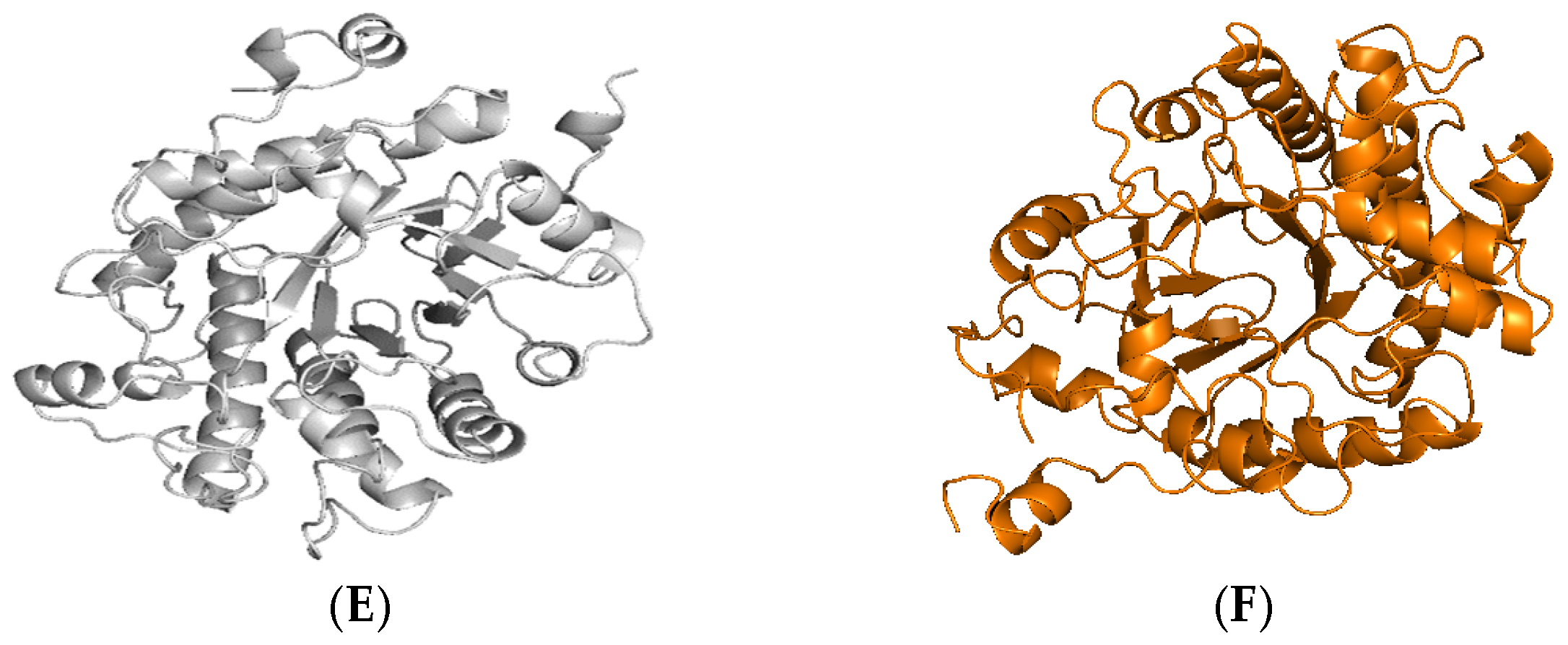

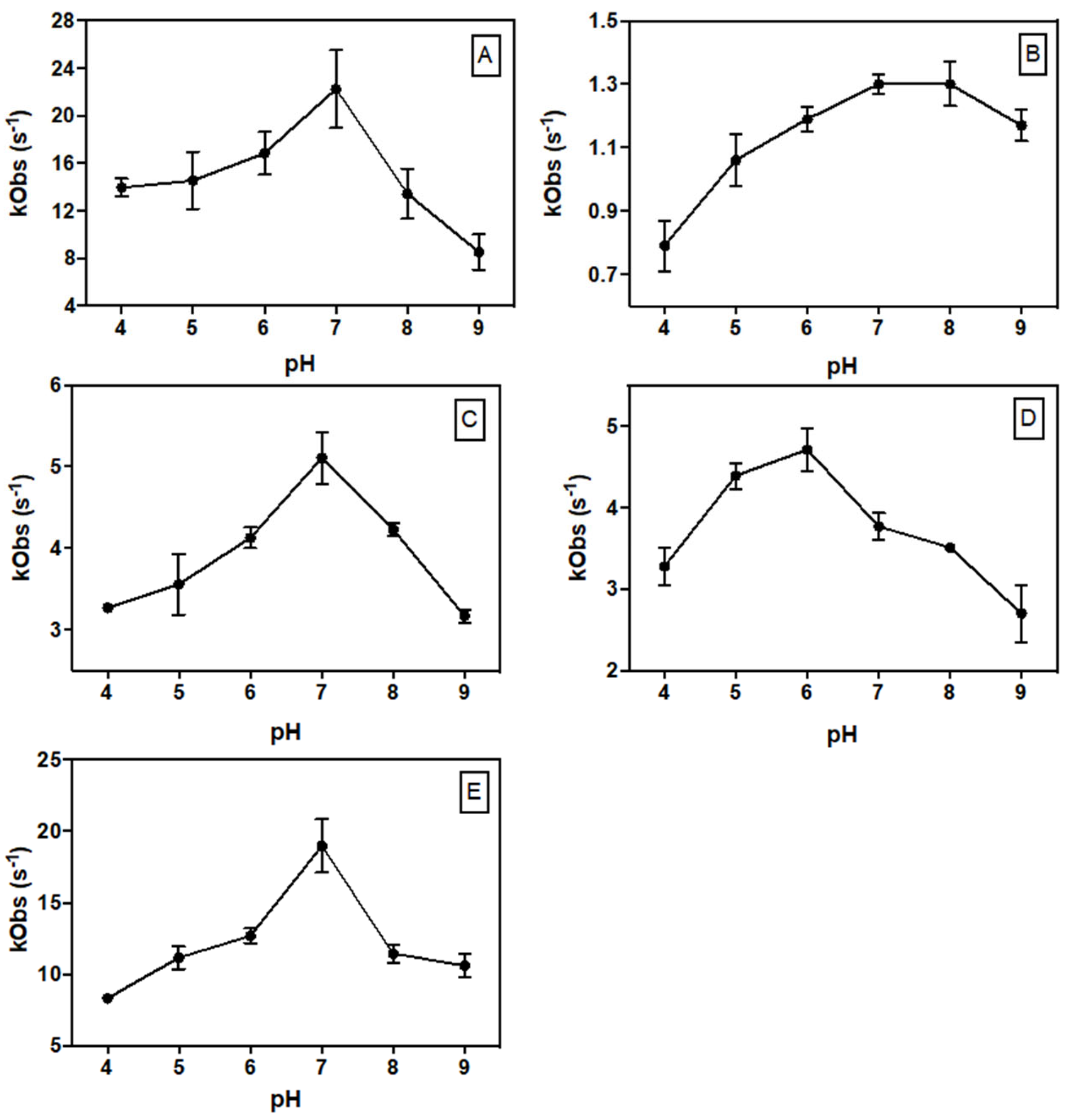
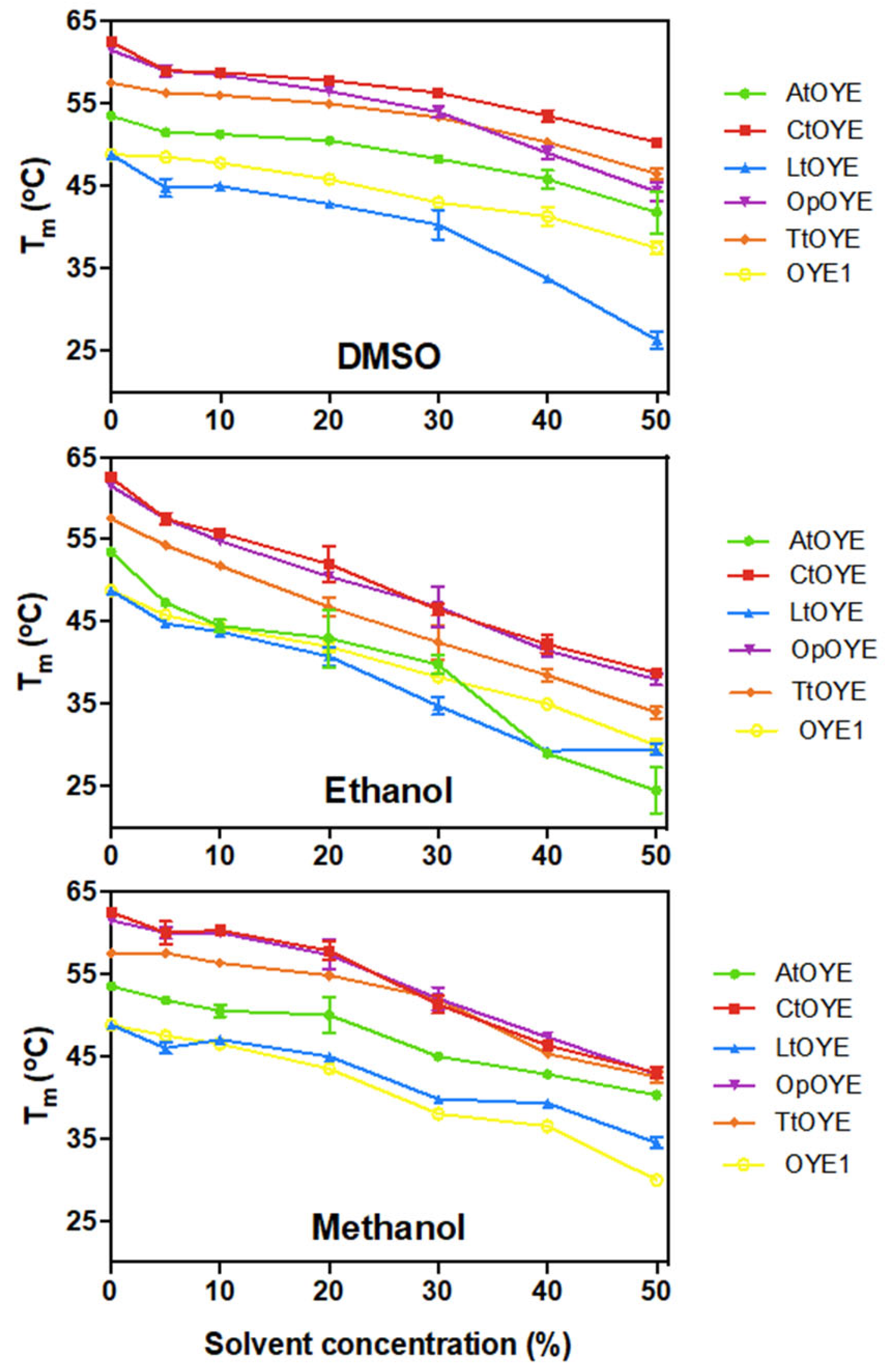
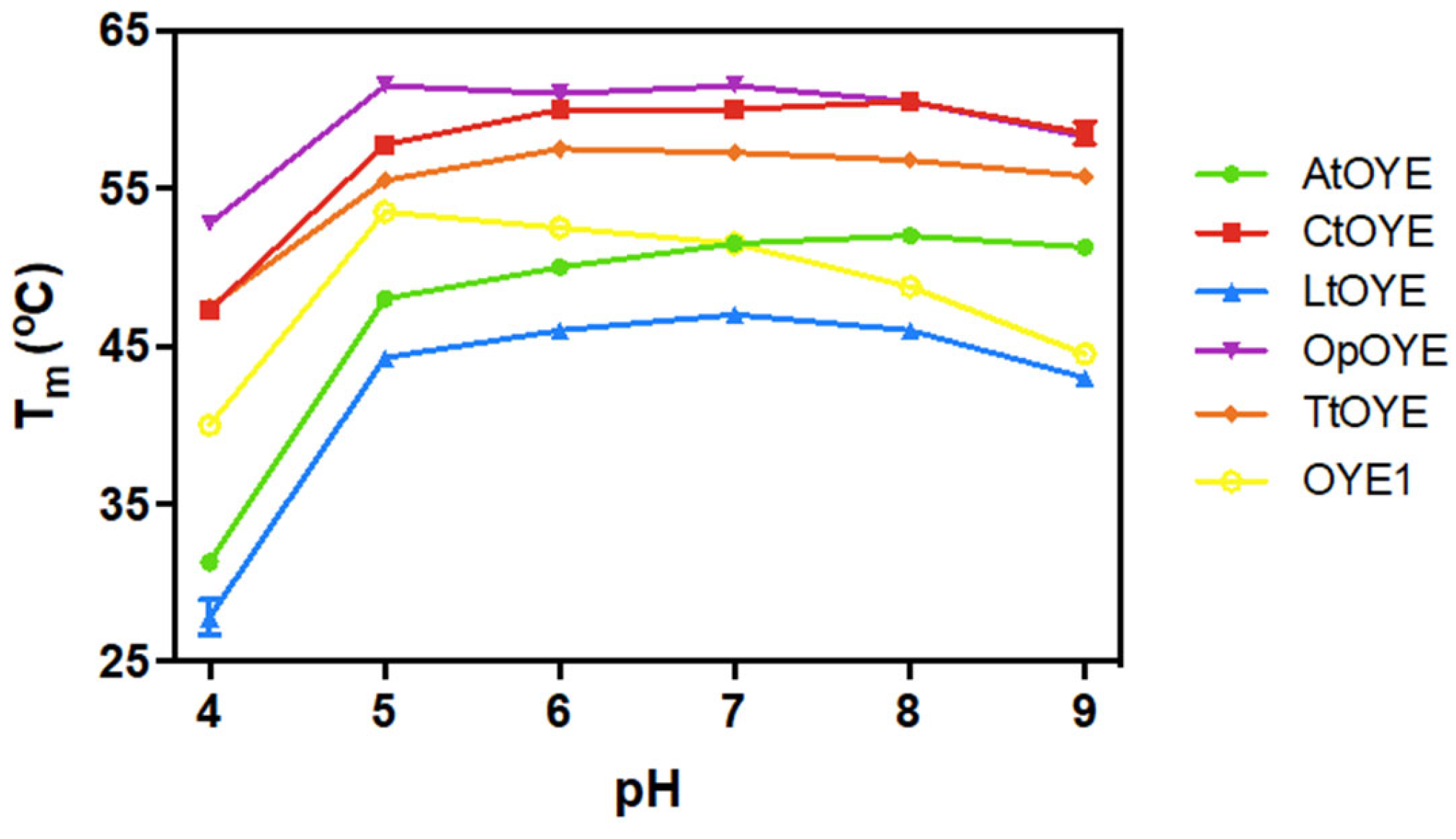
| Enzyme | Absorbance Maxima (nm) | Extinction Coefficient (mM−1 cm−1) | Yield (mg L−1) |
|---|---|---|---|
| AtOYE | 370, 463 | 13.4 | 147 |
| CtOYE | 376, 466 | 11.9 | 166 |
| LtOYE | 378, 459 | 13.0 | 136 |
| OpOYE | 381, 462 | 11.6 | 136 |
| TtOYE | 379, 465 | 12.9 | 117 |
| Enzyme | AtOYE (U mg−1) | CtOYE (U mg−1) | LtOYE (U mg−1) | OpOYE (U mg−1) | TtOYE (U mg−1) |
|---|---|---|---|---|---|
| Cofactor | NADPH/ NADH | NADPH/ NADH | NADPH/ NADH | NADPH/ NADH | NADPH/ NADH |
| O2 | 0.2 ± 0.1/ 2.1 ± 0.8 | 0.2 ± 0.0/ 1.1 ± 0.2 | 0.3 ± 0.0/ 0.5 ± 0.0 | 0.3 ± 0.0/ 0.3 ± 0.0 | 1.0 ± 0.1/ 1.1 ± 0.9 |
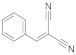 1 | 12.1 ± 1.0 | 1.0 ± 0.8 | 1.7 ± 0.4 | 1.9 ± 0.3 | 1.1 ± 0.1 |
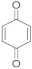 2 | 14.0 ± 2.0/ 6.7 ± 3.0 | 6.3 ± 0.1/ 2.0 ± 1.0 | 7.1 ± 0.6/ 2.1 ± 1.0 | 7.2 ± 1.0/ 3.1 ± 0.3 | 25.0 ± 2.0/ 11.7 ± 1.0 |
 3 | 8.0 ± 0.2 | n.d. | 3.0 ± 0.0 | 0.9 ± 0.0 | n.d. |
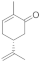 4 | 0.5 ± 0.0 | 0.1 ± 0.0 | 0.3 ± 0.0 | 0.1 ± 0.0 | 7.3 ± 0.5 |
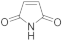 5 | 12.0 ± 1.0 | 0.8 ± 0.0 | 3.1 ± 0.2 | 2.3 ± 0.1 | 12.0 ± 1.0 |
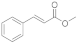 6 | 0.4 ± 0.1 | n.d. | 0.1 ± 0.0 | 0.3 ± 0.1 | 1.7 ± 0.3 |
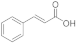 7 | 0.3 ± 0.0 | n.d. | n.d. | 0.1 ± 0.0 | 0.3 ± 0.3 |
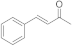 8 | 1.5 ± 0.2 | n.d. | 0.2 ± 0.0 | 0.4 ± 0.1 | 2.9 ± 0.5 |
| Enzyme | Substrate | KM (mM) | kcat (s−1) | kcat/KM (mM−1s−1) |
|---|---|---|---|---|
| AtOYE | maleimide (5) | 0.011 ± 0.003 | 25.0 ± 2.0 | 2340 |
| p-benzoquinone (2) | 0.9 ± 0.2 | 33.0 ± 1.0 | 36 | |
| NADPH | 0.043 ± 0.005 | 23.2 ± 0.8 | 540 | |
| CtOYE | maleimide (5) | 0.004 ± 0.001 | 1.5 ± 0.1 | 380 |
| p-benzoquinone (2) | 1.4 ± 0.2 | 18.9 ± 0.9 | 13 | |
| NADPH | 0.046 ± 0.002 | 2.7 ± 0.1 | 59 | |
| LtOYE | maleimide (5) | 0.004 ± 0.001 | 5.3 ± 0.1 | 1300 |
| p-benzoquinone (2) | 2.0 ± 0.3 | 25.0 ± 2.0 | 13 | |
| NADPH | 0.012 ± 0.002 | 5.2 ± 0.3 | 430 | |
| OpOYE | maleimide (5) | 0.007 ± 0.001 | 4.1 ± 0.1 | 590 |
| p-benzoquinone (2) | 1.0 ± 0.1 | 23.0 ± 0.8 | 23 | |
| NADPH | 0.011 ± 0.001 | 4.5 ± 0.1 | 400 | |
| TtOYE | maleimide (5) | 0.005 ± 0.001 | 16.3 ± 0.8 | 3300 |
| p-benzoquinone (2) | 1.6 ± 0.2 | 96.7 ± 4.0 | 60 | |
| NADPH | 0.012 ± 0.002 | 25.6 ± 1.0 | 2100 | |
| OYE1 | maleimide (5) | 0.003 ± 0.001 | 4.4 ± 0.1 | 1500 |
| p-benzoquinone (2) | - | - | - | |
| NADPH | 0.013 ± 0.003 | 4.8 ± 0.3 | 370 |
| Enzyme | Conversion (%) | d.e. (%)/(config.) |
|---|---|---|
| AtOYE | 99 | >99/R,R |
| CtOYE | 73 | >99/R,R |
| LtOYE | 70 | >99/R,R |
| OpOYE | 43 | 92/R,R |
| TtOYE | 91 | >99/R,R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damada, P.H.; Fraaije, M.W. Identification of Five Robust Novel Ene-Reductases from Thermophilic Fungi. Catalysts 2024, 14, 764. https://doi.org/10.3390/catal14110764
Damada PH, Fraaije MW. Identification of Five Robust Novel Ene-Reductases from Thermophilic Fungi. Catalysts. 2024; 14(11):764. https://doi.org/10.3390/catal14110764
Chicago/Turabian StyleDamada, Pedro H., and Marco W. Fraaije. 2024. "Identification of Five Robust Novel Ene-Reductases from Thermophilic Fungi" Catalysts 14, no. 11: 764. https://doi.org/10.3390/catal14110764
APA StyleDamada, P. H., & Fraaije, M. W. (2024). Identification of Five Robust Novel Ene-Reductases from Thermophilic Fungi. Catalysts, 14(11), 764. https://doi.org/10.3390/catal14110764










