Whole Cell Biocatalysis of 5-Hydroxymethylfurfural for Sustainable Biorefineries
Abstract
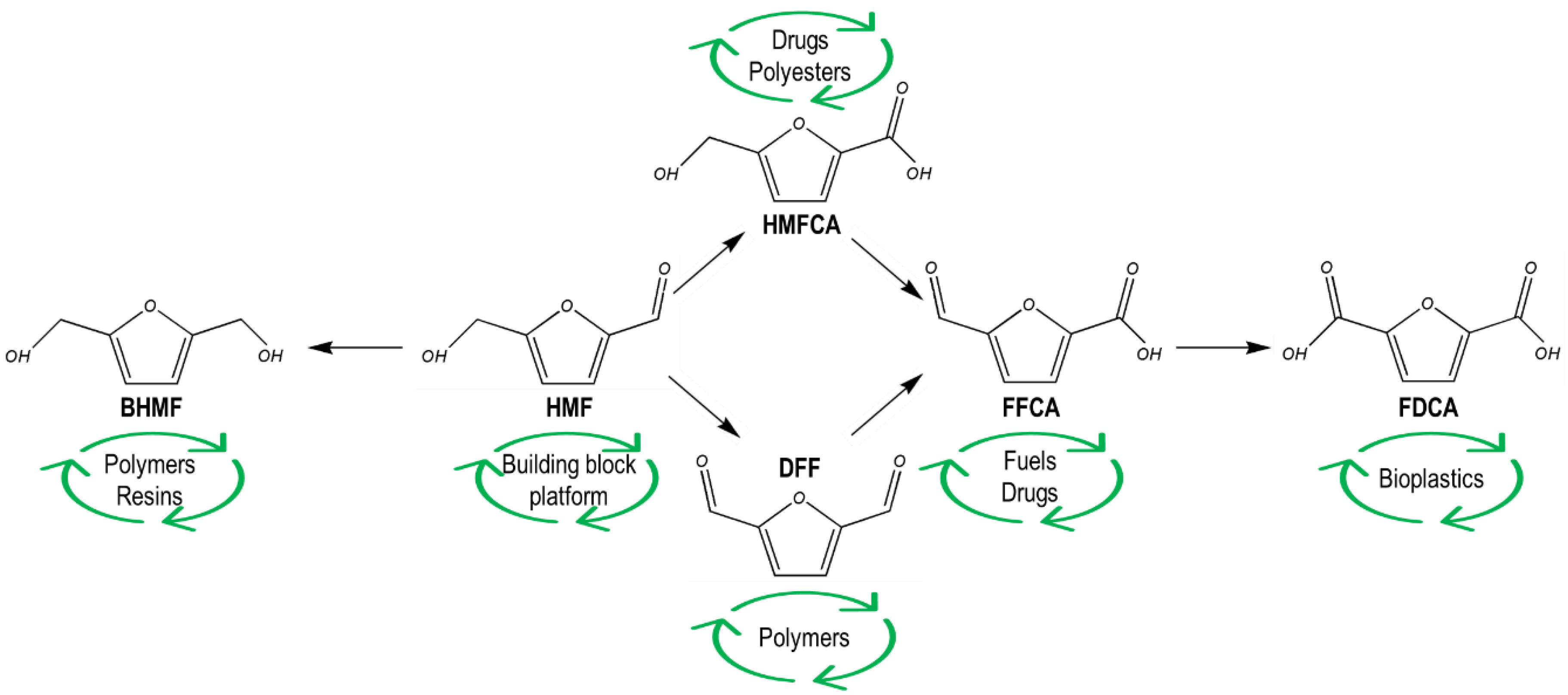
:1. Introduction
2. 5-Hydroxymethylfurfural Derivatives
2.1. 2,5-Bis(Hydroxymethyl)Furan (BHMF)
2.2. 2,5-Furandicarboxylic Acid (FDCA)
2.3. 5-Hydroxymethyl-Furan-2-Carboxylic Acid (HMFCA)
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- de Jong, E.; Higson, A.; Walsh, P.; Wellisch, M. Bio-based chemicals value added products from biorefineries. IEA Bioenergy 2012, 34, 1–36. [Google Scholar]
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “Top 10” revisited. Green Chem. 2010, 12, 539–554. [Google Scholar] [CrossRef]
- Werpy, T.; Petersen, G. Top Value Added Chemicals from Biomass: Volume I—Results of Screening for Potential Candidates from Sugars and Synthesis Gas; US Department of Energy: Washington, DC, USA, 2004.
- Market Study Report. 2019. Global 5-hydroxymethylfurfural (5-HMF) (CAS 67-47-0) Market 2019 by Manufacturers, Regions, Type and Application, Forecast to 2024. Available online: https://www.marketstudyreport.com/reports/global-5-hydroxymethylfurfural-5-hmf-cas-67-47-0-market-2019-by-manufacturers-regions-type-and-application-forecast-to-2024 (accessed on 13 December 2021).
- Cunha, J.T.; Romaní, A.; Costa, C.E.; Sá-Correia, I.; Domingues, L. Molecular and physiological basis of Saccharomyces cerevisiae tolerance to adverse lignocellulose-based process conditions. Appl. Microbiol. Biotechnol. 2019, 103, 159–175. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-C.; Lin, Y.-C.; Chu, I.M.; Wang, L.-F.; Tsai, S.-L.; Wei, Y.-H. Feasibility of enhancing production of 5-hydroxymethylfurfural using deep eutectic solvents as reaction media in a high-pressure reactor. Biochem. Eng. J. 2020, 154, 107440. [Google Scholar] [CrossRef]
- Ji, Q.; Yu, X.; Yagoub, A.E.A.; Chen, L.; Zhou, C. Efficient and environmental-friendly dehydration of fructose to 5-HMF in ultrasound assisted Ionic Liquids/Deep Eutectic Solvents. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Steinbach, D.; Kruse, A.; Sauer, J. Pretreatment technologies of lignocellulosic biomass in water in view of furfural and 5-hydroxymethylfurfural production—A review. Biomass Convers. Biorefin. 2017, 7, 247–274. [Google Scholar] [CrossRef]
- Sweygers, N.; Harrer, J.; Dewil, R.; Appels, L. A microwave-assisted process for the in-situ production of 5-hydroxymethylfurfural and furfural from lignocellulosic polysaccharides in a biphasic reaction system. J. Clean. Prod. 2018, 187, 1014–1024. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Z.K. Microwave-assisted conversion of lignocellulosic biomass into furans in ionic liquid. Bioresour. Technol. 2010, 101, 1111–1114. [Google Scholar] [CrossRef]
- Baptista, S.L.; Costa, C.E.; Cunha, J.T.; Soares, P.O.; Domingues, L. Metabolic engineering of Saccharomyces cerevisiae for the production of top value chemicals from biorefinery carbohydrates. Biotechnol. Adv. 2021, 47, 107697. [Google Scholar] [CrossRef]
- Kong, X.; Zhu, Y.; Fang, Z.; Kozinski, J.A.; Butler, I.S.; Xu, L.; Songa, H.; Weia, X. Catalytic conversion of 5-hydroxymethylfurfural to some value-added derivatives. Green Chem. 2018, 20, 3657–3682. [Google Scholar] [CrossRef]
- Acumen Research and Consulting. 2,5-Furandicarboxylic Acid (FDCA) Market Size Worth Around $850 Million By 2025: Acumen Research and Consulting. 2019. Available online: https://www.acumenresearchandconsulting.com/press-releases/2-5-furandicarboxylic-acid-fdca-market (accessed on 13 December 2021).
- Godan, T.K.; Rajesh, R.O.; Loreni, P.C.; Kumar Rai, A.; Sahoo, D.; Pandey, A.; Binod, P. Biotransformation of 5-hydroxymethylfurfural by Acinetobacter oleivorans S27 for the synthesis of furan derivatives. Bioresour. Technol. 2019, 282, 88–93. [Google Scholar] [CrossRef]
- Rajesh, R.O.; Godan, T.K.; Rai, A.K.; Sahoo, D.; Pandey, A.; Binod, P. Biosynthesis of 2,5-furan dicarboxylic acid by Aspergillus flavus APLS-1: Process optimization and intermediate product analysis. Bioresour. Technol. 2019, 284, 155–160. [Google Scholar] [CrossRef]
- Yang, C.-F.; Huang, C.-R. Biotransformation of 5-hydroxy-methylfurfural into 2,5-furan-dicarboxylic acid by bacterial isolate using thermal acid algal hydrolysate. Bioresour. Technol. 2016, 214, 311–318. [Google Scholar] [CrossRef]
- Sajid, M.; Zhao, X.; Liu, D. Production of 2,5-furandicarboxylic acid (FDCA) from 5-hydroxymethylfurfural (HMF): Recent progress focusing on the chemical-catalytic routes. Green Chem. 2018, 20, 5427–5453. [Google Scholar] [CrossRef]
- Saikia, K.; Rathankumar, A.K.; Kumar, P.S.; Varjani, S.; Nizar, M.; Lenin, R.; George, J.; Vaidyanathan, V.K. Recent advances in biotransformation of 5-Hydroxymethylfurfural: Challenges and future aspects. J. Chem. Technol. Biotechnol. 2021, 97, 409–419. [Google Scholar] [CrossRef]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Factories 2017, 16, 106. [Google Scholar] [CrossRef] [Green Version]
- Gomes, D.; Cunha, J.; Zanuso, E.; Teixeira, J.; Domingues, L. Strategies towards reduction of cellulases consumption: Debottlenecking the economics of lignocellulosics valorization processes. Polysaccharides 2021, 2, 20. [Google Scholar] [CrossRef]
- Liu, Z.L.; Slininger, P.J.; Dien, B.S.; Berhow, M.A.; Kurtzman, C.P.; Gorsich, S.W. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 2004, 31, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; He, A.; Wu, B.; Hu, L.; Liu, X.; Wu, Z.; Xia, J.; Xu, J.; Zhou, S. Redox-switchable biocatalyst for controllable oxidation or reduction of 5-hydroxymethylfurfural into high-value derivatives. ACS Omega 2020, 5, 19625–19632. [Google Scholar] [CrossRef]
- Li, Y.-M.; Zhang, X.-Y.; Li, N.; Xu, P.; Lou, W.-Y.; Zong, M.-H. Biocatalytic reduction of HMF to 2,5-bis(hydroxymethyl)furan by HMF-tolerant whole cells. ChemSusChem 2017, 10, 372–378. [Google Scholar] [CrossRef]
- Xu, Z.-H.; Cheng, A.-D.; Xing, X.-P.; Zong, M.-H.; Bai, Y.-P.; Li, N. Improved synthesis of 2,5-bis(hydroxymethyl)furan from 5-hydroxymethylfurfural using acclimatized whole cells entrapped in calcium alginate. Bioresour. Technol. 2018, 262, 177–183. [Google Scholar] [CrossRef] [PubMed]
- He, Y.C.; Jiang, C.X.; Chong, G.G.; Di, J.H.; Ma, C.L. Biological synthesis of 2,5-bis(hydroxymethyl)furan from biomass-derived 5-hydroxymethylfurfural by E. coli CCZU-K14 whole cells. Bioresour. Technol. 2018, 247, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.-H.; Zong, M.-H.; Li, N. Catalytic synthesis of 2,5-bis(hydroxymethyl)furan from 5-hydroxymethylfurfual by recombinant Saccharomyces cerevisiae. Enzym. Microb. Technol. 2020, 134, 109491. [Google Scholar] [CrossRef]
- Chen, D.; Cang, R.; Zhang, Z.-D.; Huang, H.; Zhang, Z.-G.; Ji, X.-J. Efficient reduction of 5-hydroxymethylfurfural to 2, 5-bis (hydroxymethyl) furan by a fungal whole-cell biocatalyst. Mol. Catal. 2021, 500, 111341. [Google Scholar] [CrossRef]
- Millán, A.; Sala, N.; Torres, M.; Canela-Garayoa, R. Biocatalytic transformation of 5-hydroxymethylfurfural into 2,5-di(hydroxymethyl)furan by a newly isolated Fusarium striatum strain. Catalysts 2021, 11, 216. [Google Scholar] [CrossRef]
- Chang, S.; He, X.; Li, B.; Pan, X. Improved bio-synthesis of 2,5-bis(hydroxymethyl)furan by Burkholderia contaminans NJPI-15 with co-substrate. Front. Chem. 2021, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Baptista, M.; Cunha, J.T.; Domingues, L. Establishment of Kluyveromyces marxianus as a microbial cell factory for lignocellulosic processes: Production of high value furan derivatives. J. Fungi 2021, 7, 1047. [Google Scholar] [CrossRef]
- Motagamwala, A.H.; Won, W.; Sener, C.; Alonso, D.M.; Maravelias, C.T.; Dumesic, J.A. Toward biomass-derived renewable plastics: Production of 2,5-furandicarboxylic acid from fructose. Sci. Adv. 2018, 4, eaap9722. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; He, A.; Liu, X.; Xia, J.; Xu, J.; Zhou, S.; Xu, J. Biocatalytic transformation of 5-hydroxymethylfurfural into high-value derivatives: Recent advances and future aspects. ACS Sustain. Chem. Eng. 2018, 6, 15915–15935. [Google Scholar] [CrossRef]
- Koopman, F.; Wierckx, N.; de Winde, J.H.; Ruijssenaars, H.J. Efficient whole-cell biotransformation of 5-(hydroxymethyl)furfural into FDCA, 2,5-furandicarboxylic acid. Bioresour. Technol. 2010, 101, 6291–6296. [Google Scholar] [CrossRef]
- Hsu, C.-T.; Kuo, Y.-C.; Liu, Y.-C.; Tsai, S.-L. Green conversion of 5-hydroxymethylfurfural to furan-2,5-dicarboxylic acid by heterogeneous expression of 5-hydroxymethylfurfural oxidase in Pseudomonas putida S12. Microb. Biotechnol. 2020, 13, 1094–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, N.N.; Chen, C.-Y.; Li, H.; Nguyen, M.T.T.; Nguyen, P.K.P.; Tsai, S.-L.; Chou, J.-Y.; Ramli, T.C.; Hu, Y.-C. Engineering stable Pseudomonas putida S12 by CRISPR for 2,5-furandicarboxylic acid (FDCA) production. ACS Synth. Biol. 2020, 9, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Wen, R.C.; Shen, C.R.; Tsai, S.-L. Biotransformation of 5-hydroxymethylfurfural to 2,5-furandicarboxylic acid by a syntrophic consortium of engineered Synechococcus elongatus and Pseudomonas putida. Biotechnol. J. 2020, 15, 1900357. [Google Scholar] [CrossRef]
- Tan, H.; Zhou, F.; Liao, D.; Ouyang, J.; Zheng, Z. Improved biosynthesis of 2,5-Furandicarboxylic acid through coupling of heterologous pathways in Escherichia coli and native pathways in Pseudomonas putida. Biochem. Eng. J. 2020, 161, 107657. [Google Scholar] [CrossRef]
- Hossain, G.S.; Yuan, H.; Li, J.; Shin, H.-D.; Wang, M.; Du, G.; Chen, J.; Liu, L. Metabolic engineering of Raoultella ornithinolytica BF60 for production of 2,5-furandicarboxylic acid from 5-hydroxymethylfurfural. Appl. Environ. Microbiol. 2016, 83, e02312-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.; Li, J.; Shin, H.D.; Du, G.; Chen, J.; Shi, Z.; Liu, L. Improved production of 2,5-furandicarboxylic acid by overexpression of 5-hydroxymethylfurfural oxidase and 5-hydroxymethylfurfural/furfural oxidoreductase in Raoultella ornithinolytica BF60. Bioresour. Technol. 2018, 247, 1184–1188. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Y.; Li, J.; Shin, H.D.; Du, G.; Shi, Z.; Chen, J.; Liu, L. Combinatorial synthetic pathway fine-tuning and comparative transcriptomics for metabolic engineering of Raoultella ornithinolytica BF60 to efficiently synthesize 2,5-furandicarboxylic acid. Biotechnol. Bioeng. 2018, 115, 2148–2155. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Y.; Lv, X.; Li, J.; Du, G.; Shi, Z.; Liu, L. Enhanced 2,5-Furandicarboxylic Acid (FDCA) Production in Raoultella ornithinolytica BF60 by Manipulation of the Key Genes in FDCA Biosynthesis Pathway. J. Microbiol. Biotechnol. 2018, 28, 1999–2008. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.-Y.; Zong, M.-H.; Li, N. Sacrificial substrate-free whole-cell biocatalysis for the synthesis of 2,5-furandicarboxylic acid by engineered Escherichia coli. ACS Sustain. Chem. Eng. 2020, 8, 4341–4345. [Google Scholar] [CrossRef]
- Yang, C.-F.; Huang, C.-R. Isolation of 5-hydroxymethylfurfural biotransforming bacteria to produce 2,5-furan dicarboxylic acid in algal acid hydrolysate. J. Biosci. Bioeng. 2018, 125, 407–412. [Google Scholar] [CrossRef]
- Rajesh, R.O.; Godan, T.K.; Ashok, P.; Binod, P. Whole cell based biocatalytic production of 2,5-furan dicarboxylic acid. Indian J. Exp. Biol. 2018, 56, 493–497. [Google Scholar]
- Sheng, Y.; Tan, X.; Zhou, X.; Xu, Y. Bioconversion of 5-hydroxymethylfurfural (hmf) to 2,5-furandicarboxylic acid (fdca) by a native obligate aerobic bacterium, Acinetobacter calcoaceticus NL14. Appl. Biochem. Biotechnol. 2020, 192, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Becerra, M.L.; Lizarazo, L.M.; Rojas, H.A.; Prieto, G.A.; Martinez, J.J. Biotransformation of 5-hydroxymethylfurfural and furfural with bacteria of bacillus genus. Biocatal. Agric. Biotechnol. 2022, 39, 102281. [Google Scholar] [CrossRef]
- Troiano, D.; Orsat, V.; Dumont, M.-J. Use of filamentous fungi as biocatalysts in the oxidation of 5-(hydroxymethyl)furfural (HMF). Bioresour. Technol. 2022, 344, 126169. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-Y.; Lu, P.-Y.; Yang, C.-F. Lignocellulosic acid hydrolysis inhibitor impact on 5-hydroxymethylfurfural biotransformation into 2, 5-furandicarboxylic acid using immobilised Burkholderia cells. Biocatal. Biotransformation 2021, 1–11. [Google Scholar] [CrossRef]
- Munekata, M.; Tamura, G. Antitumor activity of 5-hydroxymethyl-2-furoic acid. Agric. Biol. Chem. 1981, 45, 2149–2150. [Google Scholar]
- Braisted, A.C.; Oslob, J.D.; Delano, W.L.; Hyde, J.; McDowell, R.S.; Waal, N.; Yu, C.; Arkin, M.R.; Raimundo, B.C. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J. Am. Chem. Soc. 2003, 125, 3714–3715. [Google Scholar] [CrossRef]
- Hirai, H. Oligomers from hydroxymethylfurancarboxylic acid. J. Macromol. Sci. Part A—Chem. 1984, 21, 1165–1179. [Google Scholar] [CrossRef]
- Ran, H.; Zhang, J.; Gao, Q.; Lin, Z.; Bao, J. Analysis of biodegradation performance of furfural and 5-hydroxymethylfurfural by Amorphotheca resinae ZN1. Biotechnol. Biofuels 2014, 7, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taherzadeh, M.J.; Gustafsson, L.; Niklasson, C.; Lidén, G. Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2000, 53, 701–708. [Google Scholar] [CrossRef]
- Mitsukura, K.; Sato, Y.; Yoshida, T.; Nagasawa, T. Oxidation of heterocyclic and aromatic aldehydes to the corresponding carboxylic acids by Acetobacter and Serratia strains. Biotechnol. Lett. 2004, 26, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- De Bont, J.A.M.; Ruijssenaars, H.J.; Werij, J. Fungal Production of FDCA. U.S. Patent WO2017050815, 30 March 2017. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2017050815 (accessed on 13 December 2021).
- Wen, M.; Zhang, X.-Y.; Zong, M.-H.; Li, N. Significantly improved oxidation of bio-based furans into furan carboxylic acids using substrate-adapted whole cells. J. Energy Chem. 2020, 41, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.-S.; Zhang, X.-Y.; Zong, M.-H.; Wang, C.-F.; Li, N. Selective synthesis of 2-furoic acid and 5-hydroxymethyl-2-furancarboxylic acid from bio-based furans by recombinant Escherichia coli cells. Mol. Catal. 2019, 469, 68–74. [Google Scholar] [CrossRef]
- Cheng, A.-D.; Shi, S.-S.; Li, Y.; Zong, M.-H.; Li, N. Biocatalytic oxidation of biobased furan aldehydes: Comparison of toxicity and inhibition of furans toward a whole-cell biocatalyst. ACS Sustain. Chem. Eng. 2020, 8, 1437–1444. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Ou, X.-Y.; Fu, Y.-J.; Zong, M.-H.; Li, N. Efficient synthesis of 5-hydroxymethyl-2-furancarboxylic acid by Escherichia coli overexpressing aldehyde dehydrogenases. J. Biotechnol. 2020, 307, 125–130. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Wang, X.; Li, N.-W.; Guo, Z.-W.; Zong, M.-H.; Li, N. Furan carboxylic acids production with high productivity by cofactor-engineered whole-cell biocatalysts. ChemCatChem 2020, 12, 3257–3264. [Google Scholar] [CrossRef]
- Wang, Z.-W.; Gong, C.-J.; He, Y.-C. Improved biosynthesis of 5-hydroxymethyl-2-furancarboxylic acid and furoic acid from biomass-derived furans with high substrate tolerance of recombinant Escherichia coli HMFOMUT whole-cells. Bioresour. Technol. 2020, 303, 122930. [Google Scholar] [CrossRef]
- Muñoz, T.; Rache, L.Y.; Rojas, H.A.; Romanelli, G.P.; Martinez, J.J.; Luque, R. Production of 5-hydroxymethyl-2-furan carboxylic acid by Serratia marcescens from crude 5-hydroxymethylfurfural. Biochem. Eng. J. 2020, 154, 107421. [Google Scholar] [CrossRef]
- Liu, P.; Xie, J.; Tan, H.; Zhou, F.; Zou, L.; Ouyang, J. Valorization of Gelidium amansii for dual production of D-galactonic acid and 5-hydroxymethyl-2-furancarboxylic acid by chemo-biological approach. Microb. Cell Factories 2020, 19, 104. [Google Scholar] [CrossRef]
- Chang, S.; He, X.; Wang, X.; Li, B.; Liu, L.; Qin, J.; Yao, Z.; Pan, X. Exploring the optimized strategy for 5-hydroxymethyl-2-furancarboxylic acid production from agriculture wastes using Pseudomonas aeruginosa PC-1. Process Biochem. 2021, 102, 417–422. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zong, M.-H.; Li, N. Whole-cell biocatalytic selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid. Green Chem. 2017, 19, 4544–4551. [Google Scholar] [CrossRef]
- Cang, R.; Shen, L.-Q.; Yang, G.; Zhang, Z.-D.; Huang, H.; Zhang, Z.-G. highly selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid by a robust whole-cell biocatalyst. Catalysts 2019, 9, 526. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Zheng, Z.; Zou, L.; Zhang, C.; Yang, F.; Zhou, K.; Ouyang, J. A versatile Pseudomonas putida KT2440 with new ability: Selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid. Bioprocess Biosyst. Eng. 2020, 43, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Sayed, M.; Pyo, S.-H.; Rehnberg, N.; Hatti-Kaul, R. Selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid using Gluconobacter oxydans. ACS Sustain. Chem. Eng. 2019, 7, 4406–4413. [Google Scholar] [CrossRef]
- Pan, X.; Wu, S.; Yao, D.; Liu, L.; Zhang, L.; Yao, Z.; Pan, Y.; Chang, S.; Li, B. Efficient biotransformation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid by a new whole-cell biocatalyst Pseudomonas aeruginosa PC-1. React. Chem. Eng. 2020, 5, 1397–1404. [Google Scholar] [CrossRef]
| Strain | Conditions | Cell Inoculum | Medium | Initial [HMF] (mM) | Process | Final [BHMF] (mM) | Yield (%) | Productivity (mM/h) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Paraburkholderia azotifigens F18 | 30 °C, pH 7, 150 rpm | 67 g/L wet cells | 100 mM phosphate buffer with 40 mM glucose | 40 mM | Batch, anaerobic | 36.9 | 92 | - | [22] |
| Meyerozyma guilliermondii SC1103 | 35 °C, pH 7.2, 200 rpm | 20 g/L wet cells | 100 mM phosphate buffer with glucose | 100 | Batch | 86 | 86 | 7.17 | [23] |
| 50 | Fed-batch: ~217 mM total HMF | 191 | ~88 | 7.80 | |||||
| Meyerozyma guilliermondii SC1103 | 35 °C, pH 8, 200 rpm | 20 g/L wet cells | 100 mM Tris-HCl buffer with glucose | 213 | Batch. Cell acclimatization and immobilization calcium alginate beads | 181 | 85 | 25.8 | [24] |
| Escherichia coli CCZU-K14 harboring NADH-dependent reductase from Candida magnoliae | 30 °C, pH 6.5, 160 rpm | 100 g/L wet cells | Glucose, xylose, l-glutamic acid, Mg2+, β-cyclodextrin, and CTAB | 200 | Batch. | 181 | 91 | 2.51 | [25] |
| Saccharomyces cerevisiae harboring an aryl alcohol dehydrogenase from M. guilliermondii | 30 °C, pH 8, 200 rpm | 60 g/L wet cells | 100 mM Tris-HCl buffer and glucose | 250 | Batch | 235 | 94 | 9.79 | [26] |
| 150 | Fed-batch: ~450 mM total HMF | 345 | ~77 | 15.0 | |||||
| Aureobasidium subglaciale F134 | 30 °C, pH 7, 850 rpm | 200 g/L wet cells | 100 mM phosphate buffer with Zn2+ ion | 180 | Batch | 148 | 82 | 16.4 | [27] |
| 100 | Fed-batch: ~500 mM total HMF | 430 | 86 | 28.7 | |||||
| Fusarium striatum | 28 °C, pH 7, 160 rpm | 4.0 × 106 spores/mL | Malt extract media with glucose | 75 | Fed-batch: 150 mM total HMF | 145 | 97 | 2.42 | [28] |
| Burkholderia contaminans NJPI-15 | 35 °C, pH 7, 180 rpm | 20 g/L wet cells | 50 mM PBS with glutamine, sucrose and Mn2+ | 100 | Batch | 95 | 95 | nd | [29] |
| 125 | Fed-batch: 700 mM total HMF | 656 | 94 | 13.7 | |||||
| Kluyveromyces marxianus | 37 °C, 150 rpm | 100 g/L wet cells | YPD medium (111 mM glucose) | 55.5 | Batch | 55.3 | 100 | 4.61 | [30] |
| Strain | Modification | Conditions | Cell Inoculum | Medium | Initial [HMF] (mM) | Process | Final [FDCA] (mM) | Yield (%) | Productivity (mM/h) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Burkholderia cepacia H-2 | None | 28°, pH 7, 120 rpm | OD600 = 0.1 | Mineral salt media | 12.8 | Batch | 8.17 | 64 | 0.340 | [16] |
| OD600 = 0.25 | Undiluted algal acid hydrolysate (supplemented with HMF) | 6.34 | 50 | 0.264 | ||||||
| Methylobacterium radiotolerans G-2 | None | 26°, pH 7, 120 rpm | OD600 = 0.1 | Mineral salt media | 6.41 | 3.29 | 51 | 0.137 | [43] | |
| OD600 = 0.25 | Two-fold diluted algal acid hydrolysate (supplemented with HMF) | nd | 2.94 | nd | 0.0817 | |||||
| Pseudomonas putida S12 | hmfH from Cupriavidus basilensis HMF14, encoding HMF/furfural oxidoreductase | 30 °C, pH 7, 150 rpm | 0.2 g/L CDW | Mineral medium with glycerol | 0 | Fed-batch: 188 mmol total HMF | 193 | 97 | 1.26 | [33] |
| P. putida S12 | hmfo from Methylovorus sp. strain MP688 encoding HMF oxidase | 30 °C, pH 8 | Near 0 g/L CDW | Mineral media with glycerol and MgCl2 | 0 | Fed-batch: total HMF not defined | 545 | nd | 7.57 | [34] |
| P. putida S12 | hmfH (encoding HMF/furfural oxidoreductase) and HMFT1 (encoding a HMF transporter) from C. basilensis HMF14 | 30 °C, pH 7, 180 rpm | OD600 = 20 | Mineral media with MnO2 and CaCO3 | 250 | Batch | 196 | 78 | 8.17 | [35] |
| Synechococcus elongatus PCC7942 and P. putida S12 | Sucrose symporter (CscB) of Escherichia coli in S. elongatus. Sucrose-6-phosphate hydrolase (CscA) from E. coli W and hmfH from C. basilensis HMF14 in P. putida | 30 °C, pH 7.3 | S. elongatus (OD750 ~0.8) and P. putida (OD600 of 1) | 30 mm HEPES-NaOH | ~1 | Fed-batch: total HMF > 6 mM | ~4.6 mM | nd | Nd | [36] |
| Escherichia coli M and P. putida KT2440 | Galactose oxidase (GOase M3−5) in E. coli | 37 °C, pH 7, 200 rpm | E. coli M (8.5 g/L DCW) and P. putida (6 g/L DCW) | 100 mM sodium phosphate buffer, HRP, catalase and CaCO3 | 100 | Batch. Stepwise addition of E. coli followed by P. putida | >99 | >99 | >15.6 | [37] |
| Raoultella ornithinolytica BF60 | None | 30 °C, pH 8, 150 rpm | 45 g/L wet cells | 50 mM phosphate buffer | 100 | Batch | 50.6 | 51 | 0.294 | [38] |
| Mutation of dcaD, encoding dicarboxylic acid decarboxylase. Mutation of aldR, encoding aldehyde reductase. Overexpression of the gene encoding aldehyde dehydrogenase 1. | 89.0 | 89 | 0.517 | |||||||
| R. ornithinolytica BF60 | hmfo from Methylovorus sp. strain MP688 encoding HMF oxidase and hmfH from C. basilensis HMF14 encoding for HMF/Furfural oxidoreductase | 30 °C, pH 8, 220 rpm | 45 g/L wet cells | 50 mM phosphate buffer | 100 | Batch | 93.6 | 94 | 0.979 | [39] |
| R. ornithinolytica BF60 | Combinatorial synthetic pathway fine-tuning of hmfo and hmfH. Deletion of aldR, dkgA, akR, AdhP1 and AdhP2, involved in the reduction of HMF to BHMF. | 30 °C, pH 8, 220 rpm | OD600 = 100 | 50 mM phosphate buffer with CaCO3 | 100 | Pulse addition: 250 mM total HMF | 222 | 89 | 1.66 | [40] |
| R. ornithinolytica BF60 | Deletion of adhP3 and alkR, involved in the reduction of HMF to BHMF. Overexpression of aldH, responsible for the oxidation of FFCA into FDCA | 30 °C, pH 8 | OD600 = 100 | 50 mM phosphate buffer with CaCO3 | 50 | Pulse addition: 275 mM total HMF | 265 | 96 | 1.84 | [41] |
| E. coli | hmfH from C. basilensis HMF14 and vanillin dehydrogenase (VDH1) from Comamonas testosteroni SC1588 | 30 °C, pH 7, 150 rpm | 150 g/L wet cells | 200 mM phosphate buffer | 150 | Batch | 144 | 96 | 2 | [42] |
| Strain | Conditions | Cell Inoculum | Medium | Initial [HMF] (mM) | Process | Final [HMFCA] (mM) | Yield (%) | Productivity (mM/h) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Serratia liquefaciens LF14 | 30 °C, pH 7, 150 rpm | ~12 g/L CDW | 100 mM potassium buffer | nd | Fed-batch: 173 mM total HMF | 168 | 97 | nd | [54] |
| Saccharomyces cerevisiae CEN.PK113-1A | pH 7, 150 rpm | Nd | Mineral medium with glucose | 3.05 | Batch | 2.84 | 93 | nd | [55] |
| C. testosteroni SC1588 | 30 °C, pH 7, 160 rpm | 70 g/L wet cells | 200 mM phosphate buffer and histidine | 200 | Batch with substrate adaptation strategy | ~200 | ~100 | ~8.3 | [56] |
| E. coli modified with 3-succinoylsemialdehyde-pyridine dehydrogenase from C. testosteroni SC1588 | 30 °C, pH 7, 160 rpm | 50 g/L wet cells | 200 mM phosphate buffer | 50 | Batch | 47.5 | 95 | 9.5 | [57] |
| 175 | nd | >90 | nd | [58] | |||||
| 200 | nd | <80 | nd | ||||||
| E. coli modified with vanillin dehydrogenase from C. testosteroni SC1588 | 30 °C, pH 7, 150 rpm | 50 g/L wet cells | 200 mM phosphate buffer | 200 | Batch | ~184 | ~92 | ~15.3 | [59] |
| 200 mM phosphate buffer and NaHCO3 | ~50 | Fed-batch 1st run: 333 mM total HMF | 292 | 88 | 14.2 | ||||
| ~50 | Fed-batch 2nd run: >160 mM total HMF | 149 | nd | nd | |||||
| E. coli modified with vanillin dehydrogenase from C. testosteroni SC1588 and NADH oxidase from Lactobacillus brevis | 30 °C, pH 7, 150 rpm | 50 g/L wet cells | 200 mM phosphate buffer | 250 | Batch | 238 | 95 | 26.0 | [60] |
| E. coli modified with mutated hmfo from Methylovorus sp. strain MP688 | 30 °C, pH 7, 200 rpm | 50 g/L wet cells | 100 mM phosphate buffer | 150 | Batch | 145 | 97 | 1.5 | [61] |
| Paraburkholderia azotifigens F18 with deletion of genes encoding HMF oxidoreductase/oxidase | 30 °C, pH 7, 200 rpm | 133 g/L wet cells | Reaction buffer with glucose | 150 | Batch | 147.5 | 98 | 3.07 | [22] |
| Serratia marcescens | 30 °C, pH 8, 200 rpm | OD = 0.05 | Minimal salt medium | 3 (crude HMF) | Fed-batch: total HMF not defined | 5.56 | 28 | 0.278 | [62] |
| Pseudomonas putida ATCC 47054 | 35 °C, pH 6, 200 rpm | 8 g/L DCW | 200 mM phosphate buffer | 75.3 | Batch | 73.9 | 98 | 73.9 | [63] |
| G. amansii hydrolysates | 79.1 (biomass-derived) | Batch | 78.0 | 99 | 44.6 | ||||
| Pseudomonas aeruginosa PC-1 | 35 °C, pH 7, 180 rpm | 5 g/L wet cells | Soybean dreg hydrolysate, NaCl, corn cob residue with cellulase, and rhamnolipid; pH tuning | 100 | Fed-batch: 900 mM total HMF | 809 | 90 | 13.0 | [64] |
| Comamonas testosteroni SC1588 | 30 °C, pH 7, 150 rpm | 30 g/L wet cells | 200 mM phosphate buffer with histidine and pH tuning | 160 | Batch with substrate adaptation strategy | 157 | 98 | 4.36 | [65] |
| Deinococcus wulumuqiensis R12 | 35 °C, pH 7 | 200 g/L wet cells | 100 mM phosphate buffer | 300 | Batch | ~270 | ~90 | 7.5 | [66] |
| 150 | Fed-batch: 601 mM total HMF | 511 | 85 | 25.6 | |||||
| Pseudomonas putida KT2440 | 35 °C, pH 6.5, 200 rpm | OD600 = 25 | 200 mM phosphate buffer and CaCO3 | 160 | Batch | 155 | 97 | 12.9 | [67] |
| Gluconobacter oxydans DSM 50049 | 24 g/L DCW | sodium phosphate buffer and controlled pH | 250 | Batch | 250 | 100 | 41.7 | [68] | |
| 250 | Fed-batch: 345 mM total HMF | 314 | 91 | 13.6 | |||||
| Pseudomonas aeruginosa PC-1 | 30 °C, pH 7, 180 rpm | 5 g/L wet cells | Sucrose, peptone and NaCl; pH tuning | 100 | Fed-batch: 800 mM total HMF | 721 | 90 | 12.4 | [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cunha, J.T.; Romaní, A.; Domingues, L. Whole Cell Biocatalysis of 5-Hydroxymethylfurfural for Sustainable Biorefineries. Catalysts 2022, 12, 202. https://doi.org/10.3390/catal12020202
Cunha JT, Romaní A, Domingues L. Whole Cell Biocatalysis of 5-Hydroxymethylfurfural for Sustainable Biorefineries. Catalysts. 2022; 12(2):202. https://doi.org/10.3390/catal12020202
Chicago/Turabian StyleCunha, Joana T., Aloia Romaní, and Lucília Domingues. 2022. "Whole Cell Biocatalysis of 5-Hydroxymethylfurfural for Sustainable Biorefineries" Catalysts 12, no. 2: 202. https://doi.org/10.3390/catal12020202
APA StyleCunha, J. T., Romaní, A., & Domingues, L. (2022). Whole Cell Biocatalysis of 5-Hydroxymethylfurfural for Sustainable Biorefineries. Catalysts, 12(2), 202. https://doi.org/10.3390/catal12020202






