Abstract
The implementation of cost-effective and sustainable biorefineries to substitute the petroleum-based economy is dependent on coupling the production of bioenergy with high-value chemicals. For this purpose, the US Department of Energy identified a group of key target compounds to be produced from renewable biomass. Among them, 5-hydroxymethylfurfural (HMF) can be obtained by dehydration of the hexoses present in biomass and is an extremely versatile molecule that can be further converted into a wide range of higher value compounds. HMF derivatives include 2,5-bis(hydroxymethyl)furan (BHMF), 5-hydroxymethyl-furan-2-carboxylic acid (HMFCA), 2,5-diformylfuran (DFF), 5-formyl-2-furancarboxylic acid (FFCA) and 2,5-furandicarboxylic acid (FDCA), all presenting valuable applications, in polymers, bioplastics and pharmaceuticals. Biocatalysis conversion of HMF into its derivatives emerges as a green alternative, taking into account the high selectivity of enzymes and the mild reaction conditions used. Considering these factors, this work reviews the use of microorganisms as whole-cell biocatalysts for the production of HMF derivatives. In the last years, a large number of whole-cell biocatalysts have been discovered and developed for HMF conversion into BHMF, FDCA and HMFCA, however there are no reports on microbial production of DFF and FFCA. While the production of BHMF and HMFCA mainly relies on wild type microorganisms, FDCA production, which requires multiple bioconversion steps from HMF, is strongly dependent on genetic engineering strategies. Together, the information gathered supports the possibility for the development of cell factories to produce high-value compounds, envisioning economical viable biorefineries.
1. Introduction
The world faces the progressive depletion of its fossil fuels reserves, with excessive consumption leading to an increase in the emission of greenhouse gases and in climate changes promoted by global warming. These factors, coupled with the growing demand for energy for transportation, heating, industrial processes, among others, highlight the necessity to transition from a petroleum-based towards a bio-based economy. This bioeconomy is based on the biorefinery concept, defined as “the sustainable processing of biomass into a spectrum of marketable products (food, feed, materials, chemicals) and energy (fuels, power, heat)” [1].
Biorefineries have been mostly conceptualized around energy and biofuels. More recently, process integration and conversion of all fractions of the biomasses into value-added products has been recognized as key elements for a profitable biorefinery operation. This encompasses the two main goals of a biorefinery: (1) the energetic goal, focusing on the replacement of petroleum by renewable carbons for fuel production, and (2) the economic goal, directing efforts towards the efficient production of biobased chemicals providing the financial incentive to maintain and expand the biorefining industry.
Aiming at the economic viability of biorefineries, Bozell and Petersen suggested that the manufacture of low-value biofuels should be supplemented with the production of high-value biobased chemicals [2]. They also presented an updated list of biobased product opportunities from renewable carbohydrates, based on the one published by the US Department of Energy in 2004 [3], which consists of ethanol, furans (5-hydroxymethylfurfural, furfural and FDCA), glycerol and derivatives, biohydrocarbons (isoprene), organic acids (lactic, succinic, levulinic and 3-hydroxypropanoic acids) and sugar alcohols (xylitol and sorbitol). This selection was made considering the type of raw material, production costs, sales prices and availability of processes and technologies. Considering that the conversion of renewable carbon into chemicals is the more challenging and least developed step of all biorefinery operations, there is an urgent necessity to identify and develop microbial catalysts for an efficient production of biofuels (e.g., bioethanol), as well as other top value compounds from renewable biomass.
2. 5-Hydroxymethylfurfural Derivatives
The versatile composition of 5-hydroxymethylfurfural (HMF)—consisting of an aromatic furan ring and reactive functional groups (aldehyde and alcohol groups)—makes it an attractive building block platform, as it can be transformed into higher value derivative compounds, having applications in several areas such as plastic, pharmaceutical, fragrance and textile industries (Figure 1). Accordingly, the global market of HMF is expected to reach $61 million by 2024 [4]. HMF is commonly generated during pretreatment of lignocellulosic biomass and is usually regarded as a microbial inhibitor [5]. It derives from the dehydration of hexoses (such as the cellulose-derived glucose), and its accumulation depends on the biomass used and the type and severity of pretreatment and hydrolysis applied. As its value has been receiving growing attention, several studies focused on optimizing pretreatment methods to increase HMF accumulation from different biomasses by using different reaction media and/or catalysts (e.g., solid acid catalysts, Lewis acids, Brönsted acids, ionic liquids, salts, deep eutectic solvents and biphasic systems) as well as alternative heating methods (e.g., ultrasound and microwave) [6,7,8,9,10].
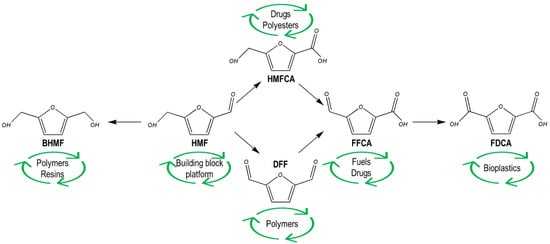
Figure 1.
Reduction and oxidation derivatives of HMF and corresponding application. BHMF: 2,5-bis(hydroxymethyl)furan. HMF: 5-hydroxymethylfurfural. HMFCA: 5-hydroxymethyl-furan-2-carboxylic acid. DFF: 2,5-diformylfuran. FFCA: 5-formyl-2-furancarboxylic acid. FDCA: 2,5-furandicarboxylic acid.
Accordingly, increased importance has been given to the conversion of HMF into higher value derivatives [11,12]. Among these, 2,5-furandicarboxylic acid (FDCA) was also identified as one of the top products to be obtained from biomass [2], and its worldwide market is expected to reach $850 million by 2025 [13]. Its main application is the polymerization with ethylene glycol to produce polyethylene furanoate (PEF), a substitute to petroleum-derived polyethylene terephthalate (PET) plastic. In fact, the European Union’s Horizon 2020 funded a consortium of eleven companies named to establish an innovative biobased production of FDCA and PEF (https://peference.eu/ (accessed on 9 December 2021)). FDCA is obtained by oxidation of HMF, and some microorganisms such as Acinetobacter oleivorans, Aspergillus flavus and Burkholderia cepacian [14,15,16] can produce it natively. Nevertheless, chemical processes for its production are applied in most industries, with Corbion being a pioneer in using microbial biocatalyst to produce FDCA from HMF. In fact, while chemocatalytic processes are still the main transformation strategy used, their application requires the use of metals (mostly noble) as catalysts and harsh operating conditions, generating hazardous by-products [17]. Considering these, and aiming at sustainable development, biocatalysis has been receiving increased attention due to its milder conditions, higher selectivity and environmental friendliness [18]. Moreover, considering these factors, the use of whole-cell biocatalysts presents advantages over enzyme biocatalysis, such as regeneration of co-factors and ease of catalyst recycling [19,20].
The chemistry of HMF, containing aldehyde and alcohol functional groups, makes it highly reactive towards oxidation–reduction and other reactions. Besides the already mentioned FDCA, other derivatives with valuable applications can be obtained from HMF, such as 2,5-bis(hydroxymethyl)furan (BHMF), 5-hydroxymethyl-furan-2-carboxylic acid (HMFCA), 2,5-diformylfuran (DFF) and 5-formyl-2-furancarboxylic acid (FFCA) (Figure 1). Considering these, and to contribute to the replacement of chemical approaches and development of sustainable biorefineries, this review focuses on the reported microorganisms used as whole-cell biocatalysts for the production of these HMF derivatives. For that, each HMF derivative will be approached, including the most relevant results obtained.
2.1. 2,5-Bis(Hydroxymethyl)Furan (BHMF)
BHMF has a potential application in the synthesis of polymers, resins and ethers [18]. BHMF was first reported by Liu et al. [21] as the reduction product of HMF by S. cerevisiae. More recently, fungi, yeast and bacteria strains have been described as biocatalysts for its production from HMF (Table 1). The bacteria Paraburkholderia azotifigens F18 was found to produce 36.9 mM of BHMF from HMF, with a 92% yield, under anaerobic conditions [22]. The yeast Meyerozyma guilliermondii SC1103 was identified as a BHMF producer [23], and its conversion abilities were further improved by cell acclimatization and immobilization in calcium alginate beads, reaching BHMF titers of 181 mM, with 85% and a productivity of 25.8 mM/h [24]. Furthermore, in a process with cell recycling the yeast maintained good catalytic activities during four runs with yields between 81% and 88% and a cell viability of up to 70% after the 4th run [24]. An Escherichia coli strain was modified to harbor a NADH-dependent reductase from Candida magnoliae and produced 181 mM of BHMF with 91% yield [25]. In another genetic engineering strategy, a S. cerevisiae strain harboring an aryl alcohol dehydrogenase from M. guilliermondii reached a BHMF titer of 345 mM with a productivity of 15 mM/h, presenting however a low yield of ~77% [26]. Moreover, in this work, an inexpensive corncob hydrolysate was described as a promising alternative to glucose as co-substrate for the biocatalysis. The fungus Aureobasidium subglaciale F134 was identified for the production of BHMF, reaching the highest reported productivity of 28.7 mM/h with a fed-batch process [27]. Another fungus, Fusarium striatum, was reported to produce 145 mM of BHMF with high yield of 97% [28]. The bacterium Burkholderia contaminans NJPI-15 reached the highest BHMF titer of 656 mM in a fed-batch with 94% yield [29]. More recently, the highest BHMF yield of 99.65% was reported using a Kluyveromyces marxianus yeast strain isolated from cocoa fermentation, which reached an BHMF titer of 55.3 mM in a batch process [30].

Table 1.
Whole-cell biocatalysts reported for the conversion of 5-hydroxymethylfurfural (HMF) into 2,5-bis(hydroxymethyl)furan (BHMF). Yield is presented as percentage of the theoretical molar yield. OD: optical density. CDW: cell dry weight. CTAB: cetyltrimethyl ammonium bromide. YPD: Yeast extract peptone dextrose.
2.2. 2,5-Furandicarboxylic Acid (FDCA)
As already mentioned, the main application of FDCA is as a substitute of terephthalic acid, which can be used to synthesize several polyesters, with PEF being the most studied [31]. In addition, it can also be used as a building block in the production of medicines and polyamides [32].
The first microbial biocatalyst reported for FDCA production from HMF was a Pseudomonas putida S12 strain modified to express the hmfH gene from Cupriavidus basilensis (encoding an HMF/furfural oxidoreductase), which resulted in the production of 193 mM with 97% yield in a fed-batch strategy [33]. More recently, the hmfo from Methylovorus sp. strain MP688, encoding a HMF oxidase, was expressed in P. putida S12, which resulted in the production of 545 mM of FDCA in a fed-batch [34]. P. putida S12 was also modified by CRISPR for the development of more stable strains [35]. With this technique, this bacterium was modified to express hmfH and HMFT1 (encoding a HMF transporter) genes from C. basilensis, which allowed the production of 196 mM of FDCA with 78% yield. In a consortium approach, a Synechococcus elongatus strain engineered to export sucrose via CO2 fixation was used as carbon source for a P. putida S12 strain modified for sucrose consumption and improved FDCA production [36]. Despite being innovative and having promise for reducing feedstocks costs within a biorefinery, this strategy only resulted in the production of ~4.6 mM of FDCA. Tan and collaborators [37] reported a whole-cell cascade, where an engineered E. coli was coupled with a wild type P. putida in a stepwise reaction, where HMF would be oxidized into DFF by E. coli, which would be further oxidized into FDCA by addition of a P. putida strain. This allowed the conversion of 100 mM of HMF with a FDCA yield higher than 99%, however the reaction required the addition of the commercial enzymes horseradish peroxidase (as an enzyme activator) and catalase (to decompose H2O2) [37].
Hossain et al. [38] isolated a Raoultella ornithinolytica BF60 strain capable of converting HMF into FDCA with 51.0% yield. This bacterium was modified to prevent FDCA degradation to furoic acid and HMF reduction into BHMF, and to improve HMF oxidation into FDCA by overexpression of the gene encoding aldehyde dehydrogenase 1, which increased FDCA yield to 89%, corresponding to a titer of 89.1 mM [38]. The same research group also modified the R. ornithinolytica BF60 strain to express the genes hmfo from Methylovorus sp. strain MP688, encoding HMF oxidase, and hmfH from C. basilensis HMF14, encoding HMF/furfural oxidoreductase, which improved the FDCA titer to 93.6 mM and yield to 94% [39]. In a different study using Raoultella ornithinolytica BF60 as host, combinatorial synthetic pathway fine-tuning of hmfo and hmfH, as well as deletion of five genes associated with HMF reduction, resulted in increased FDCA titers of 221.5 mM with an 89% yield in a fed-batch strategy [40]. In another fed-batch approach, FDCA production was further improved to titers of 264.7 mM with 96% yield by using a modified R. ornithinolytica BF60 strain with deletion of two genes responsible for the reduction of HMF into BHMF and overexpression of AldH, encoding an enzyme responsible for the oxidation of FFCA into FDCA [41].
Using the bacteria E. coli as cell host, FDCA production was attained by modification with hmfH from C. basilensis HMF14 and vanillin dehydrogenase (VDH1) from Comamonas testosteroni SC1588, resulting in the production of 144 mM of FDCA with a yield of 96% [42]. These values are in line with the ones obtained with modified P. putida S12 and R. ornithinolytica BF60 (Table 2).

Table 2.
Whole-cell biocatalysts reported for the conversion of 5-hydroxymethylfurfural (HMF) into 2,5-furandicarboxylic acid (FDCA). Yield is presented as percentage of the theoretical molar yield. OD: optical density. CDW: cell dry weight.
Several other microorganisms have been described to convert HMF into FDCA, such as the bacteria Burkholderia cepacia H-2 [16], Methylobacterium radiotolerans G-2 [43], Enterobacter sp. [44], Acinetobacter oleivorans S27 [14], Acinetobacter calcoaceticus NL14 [45] and Bacillus toyonensis [46] and the filamentous fungi Aspergillus flavus APLS-1 [15], Aspergillus niger [47] and Trichoderma reesei [47]. Nonetheless, FDCA titers obtained with these biocatalysts were low (<8.2 mM), highlighting the importance of genetic engineering approaches for the production of this HMF derivative. Despite the low values obtained, these reports broaden the pool of possible hosts for further modifications for improved FDCA production. Furthermore, note that two of these species, Burkholderia cepacia H-2 and Methylobacterium radiotolerans G-2, were used as biocatalysts in undiluted and 2-fold diluted macroalgae acid hydrolysate supplemented with HMF, showing their capacity to maintain HMF conversion even in the presence of other microbial inhibitors such as furfural and weak acids [43]. Furthermore, the same group immobilized B. cepacia H-2 in calcium-alginate beads and evaluated its capacity to produce FDCA from HMF in the presence of lignocellulosic-derived inhibitors [48]. The immobilized strain was able to produce FDCA in the presence of up to 5 g/L formic acid, 4.39 g/L acetic acid, 5 g/L levulinic acid, 0.5 g/L furfural and 0.5 g/L phenol. This is relevant because the severity of the conditions needed to generate high HMF quantities from biomass would also result in high accumulation of other inhibitory compounds. While the Corbion Company is known to produce FDCA from HMF with microbial catalysts, there is not enough information regarding the HMF source, and to the extent of our knowledge, there is still no reports of FDCA production by whole-cell biocatalysts from biomass-derived HMF.
2.3. 5-Hydroxymethyl-Furan-2-Carboxylic Acid (HMFCA)
HMFCA is an antitumor agent [49] and interleukin inhibitor [50], and it can be also used as an intermediate in the preparation of polyesters [51]. Early reports on HMFCA microbial production refer to it as a by-product or intermediate and are exploratory regarding HMF oxidation/degradation products [33,52,53]. As an exception, in 2004 Mitsukura and collaborators [54] focused on obtaining carboxylic acids from aromatic aldehydes and reported a Serratia liquefaciens LF14 strain capable of attaining 168 mM of HMFCA from HMF with 97% yield [54]. More recently, great effort has been put into HMFCA and finding whole-cell biocatalysts capable of efficiently produce it from HMF (Table 3). Mostly, bacteria species have been described for this purpose, e.g., Serratia spp., Comamonas testosteroni, Deinococcus wulumuqiensis, Pseudomonas spp. and Gluconobacter oxydans. In a patent that focuses on fungal production of FDCA, the S. cerevisiae CEN.PK113-1A strain has been reported to produce 2.84 mM of HMFCA with a 93% yield [55]. Fed-batch strategies have been recurrently used to diminish the toxic effect of HMF on the cells, allowing for higher HMFCA titers and productivity, up to 809 mM and 25.6 mM/h, respectively (Table 3). In the case of Comamonas testosteroni SC1588, its catalytic performance (e.g., substrate tolerance and catalytic efficiency) was improved by a substrate adaptation strategy, reaching a HMFCA yield near 100% in 24 h from 200 mM of HMF [56].

Table 3.
Whole-cell biocatalysts reported for the conversion of 5-hydroxymethylfurfural (HMF) into 5-hydroxymethyl-furan-2-carboxylic acid (HMFCA). Yield is presented as a percentage of the theoretical molar yield. OD: optical density. CDW: cell dry weight.
Differently from the microorganisms reported for FDCA production (Table 2), most of the whole-cell biocatalysts for HMFCA production require no genetic modification, but rather optimization of reaction conditions (e.g., temperature, pH and inoculum) to achieve high titers and yields. Despite the satisfactory values obtained with wild type strains, efforts have been made to genetically engineer bacteria to produce HMFCA. An E. coli strain modified with 3-succinoylsemialdehyde-pyridine dehydrogenase (SAPDH) from C. testosteroni SC1588 was able to produce HMFCA from 50 mM HMF with 95% yield [57]. Later, this strain was found to maintain high HMFCA yields even from higher HMF concentrations: ~90% from 175 mM HMF and almost 80% from 200 mM HMF [58]. A vanillin dehydrogenase from C. testosteroni SC1588 was also introduced in an E. coli strain by the same research group, resulting in the conversion of 200 mM HMF with 92% yield, which was further improved by a fed-batch strategy resulting in the synthesis of 292 mM of HMFCA [59]. Furthermore, these authors attempted a cell recycling strategy after the fed-batch, and despite the decrease in cell viability, 149 mM of HMFCA was produced in 18 h in the second run, maintaining the yields of the first run [59]. In a subsequent work of the same group, a cofactor-engineered E. coli with co-expression of vanillin dehydrogenase and NADH oxidase resulted in improved HMFCA titers and productivities [60]. The significant improvement in HMFCA productivity and HMF tolerance of these E. coli strains harboring C. testosteroni enzymes in comparison with the use of wild type C. testosteroni as a biocatalyst (Table 3) should be highlighted. Wang et al. [61] constructed a recombinant E. coli harboring a hmfo from Methylovorus sp. strain MP688 mutated for specificity for HMFCA rather than FDCA production, achieving HMFCA titers of 145 mM with 97% yield. Despite presenting higher HMFCA yield than the other recombinant E. coli strains, its productivity is significantly (up to 10 times) lower (Table 3). Surprisingly, in that work the authors did not explore the potential of the E. coli modified with the original hmfo for FDCA production, which presented yields of ~80%. The Paraburkholderia azotifigens F18 strain, capable of producing BHMF under anaerobic conditions, was found to accumulate HMFCA in more aerobic conditions, and with deletion of genes encoding HMF oxidoreductase/oxidase (to prevent oxidation of HMFCA to FDCA) it produced 147.5 mM of HMFCA with 98% yield [22].
The majority of the reports on whole-cell production of HMFCA use commercial HMF as substrate. Muñoz and collaborators [62] reported the bioconversion of crude HMF, obtained by dehydration of fructose, into HMFCA by the bacteria Serratia marcescens. The use of crude HMF, which more closely mimics real conditions and contains other by-products (such as furfural), severely hinders HMFCA yields and microbial tolerance [62], resulting in low HMFCA titers even with fed-batch strategies (Table 3). Liu and collaborators [63] produced HMF from the macroalgae Gelidium amansii with a thermal pretreatment with 2% (w/w) HCl at 120 °C for 40 min and identified P. putida ATCC 47054 as an efficient biocatalyst, which was capable of producing 78.0 mM of HMFCA from G. amansii hydrolysate with an 99% yield and the remarkable productivity of 44.6 mM/h. Differently from the results with crude HMF obtained from fructose, the HMFCA yield obtained from G. amansii hydrolysates was equal to the results with commercial HMF, with only HMFCA productivity being affected (Table 3). These results prove that the use of whole-cell biocatalysts for the conversion of biomass-derived HMF into higher value compounds is a promising strategy and should be further explored. Furthermore, the material cost of HMFCA production can be decreased to 11.47% of the traditional method by using corncob residue and soybean dreg as nutrient sources, which resulted in a fed-batch production of 809 mM HMFCA, from commercial HMF, with 90% yield and using a Pseudomonas aeruginosa PC-1 strain [64].
3. Conclusions
HMF is a building block platform with potential to be explored by microbial conversion. This review discusses the array of microorganisms used as whole-cell biocatalysts for the production of HMF derivatives. While there are no reports on microbial production of DFF and FFCA, several whole-cell biocatalysts have been discovered and developed for HMF conversion into BHMF, FDCA and HMFCA. With those, the use of fed-batch and pulse addition approaches resulted in the production of high compound titers, circumventing the toxic effect of HMF. The toxicity of the HMF derivatives should also be considered, and this can be lowered by using two-phase systems for an in situ extraction of the product, which also facilitates the downstream process. Regarding the optimization of process conditions, much of the focus is given to the optimal temperature and pH of biocatalysis. However, other parameters, such as aeration and presence of co-substrates, have been poorly explored despite being reported to play an important role on HMF conversion (e.g., favoring either the reduction or the oxidation of HMF). This review also shows that while the production of BHMF and HMFCA mainly relies on wild type microorganisms, FDCA production, which requires more bioconversion steps from HMF, is strongly dependent on genetic engineering strategies. Genetic engineering strategies were also found to play an important role in preventing the accumulation of by-products (e.g., preventing further oxidation of HMFCA), and should be further explored to increase the specificity of the process (e.g., to avoid BHMF accumulation when aiming at producing HMFCA and FDCA, and vice versa). Furthermore, genetic modifications should also be used to develop more efficient whole-cell biocatalysts (e.g., following approaches already used in different hosts or expressing enzymes with reported activity for conversion of HMF into the target derivative). Furthermore, note that the vast majority of works discussed in this review report production of HMF derivatives using commercial HMF, where no other inhibitors are present. It is inevitable that the production of HMF from renewables biomasses would also result in the co-production of other inhibitory compounds, such as weak acids and furfural, which are known to affect the microbial conversion of HMF. In this sense, the success of using whole-cell biocatalysts for the production of HMF derivatives is also dependent on the evolution of the techniques used to produce HMF from renewable biomasses: envisioning HMF-enriched media with or without low concentrations of inhibitors and containing appropriate amounts of co-substrates (e.g., glucose for BHMF production). Alternatively, HMF purification techniques can be applied, but these will represent an increase in the cost of the process. Together, this information shows that the establishment of economically viable microbial biorefineries based on HMF derivatives is dependent not only on more efficient whole-cell biocatalysts, but also on the development of all the steps of the process, with important focus on the production of HMF from raw materials and also downstream processing.
Author Contributions
Conceptualization, J.T.C., A.R. and L.D.; writing—original draft preparation, J.T.C.; writing—review and editing, A.R. and L.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UIDB/04469/2020, and by the Ministry of Science and Innovation (MICIN) through the grant RYC2020-030690-I (to AR).
Conflicts of Interest
The authors declare no conflict of interest.
References
- de Jong, E.; Higson, A.; Walsh, P.; Wellisch, M. Bio-based chemicals value added products from biorefineries. IEA Bioenergy 2012, 34, 1–36. [Google Scholar]
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “Top 10” revisited. Green Chem. 2010, 12, 539–554. [Google Scholar] [CrossRef]
- Werpy, T.; Petersen, G. Top Value Added Chemicals from Biomass: Volume I—Results of Screening for Potential Candidates from Sugars and Synthesis Gas; US Department of Energy: Washington, DC, USA, 2004.
- Market Study Report. 2019. Global 5-hydroxymethylfurfural (5-HMF) (CAS 67-47-0) Market 2019 by Manufacturers, Regions, Type and Application, Forecast to 2024. Available online: https://www.marketstudyreport.com/reports/global-5-hydroxymethylfurfural-5-hmf-cas-67-47-0-market-2019-by-manufacturers-regions-type-and-application-forecast-to-2024 (accessed on 13 December 2021).
- Cunha, J.T.; Romaní, A.; Costa, C.E.; Sá-Correia, I.; Domingues, L. Molecular and physiological basis of Saccharomyces cerevisiae tolerance to adverse lignocellulose-based process conditions. Appl. Microbiol. Biotechnol. 2019, 103, 159–175. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-C.; Lin, Y.-C.; Chu, I.M.; Wang, L.-F.; Tsai, S.-L.; Wei, Y.-H. Feasibility of enhancing production of 5-hydroxymethylfurfural using deep eutectic solvents as reaction media in a high-pressure reactor. Biochem. Eng. J. 2020, 154, 107440. [Google Scholar] [CrossRef]
- Ji, Q.; Yu, X.; Yagoub, A.E.A.; Chen, L.; Zhou, C. Efficient and environmental-friendly dehydration of fructose to 5-HMF in ultrasound assisted Ionic Liquids/Deep Eutectic Solvents. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Steinbach, D.; Kruse, A.; Sauer, J. Pretreatment technologies of lignocellulosic biomass in water in view of furfural and 5-hydroxymethylfurfural production—A review. Biomass Convers. Biorefin. 2017, 7, 247–274. [Google Scholar] [CrossRef]
- Sweygers, N.; Harrer, J.; Dewil, R.; Appels, L. A microwave-assisted process for the in-situ production of 5-hydroxymethylfurfural and furfural from lignocellulosic polysaccharides in a biphasic reaction system. J. Clean. Prod. 2018, 187, 1014–1024. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Z.K. Microwave-assisted conversion of lignocellulosic biomass into furans in ionic liquid. Bioresour. Technol. 2010, 101, 1111–1114. [Google Scholar] [CrossRef]
- Baptista, S.L.; Costa, C.E.; Cunha, J.T.; Soares, P.O.; Domingues, L. Metabolic engineering of Saccharomyces cerevisiae for the production of top value chemicals from biorefinery carbohydrates. Biotechnol. Adv. 2021, 47, 107697. [Google Scholar] [CrossRef]
- Kong, X.; Zhu, Y.; Fang, Z.; Kozinski, J.A.; Butler, I.S.; Xu, L.; Songa, H.; Weia, X. Catalytic conversion of 5-hydroxymethylfurfural to some value-added derivatives. Green Chem. 2018, 20, 3657–3682. [Google Scholar] [CrossRef]
- Acumen Research and Consulting. 2,5-Furandicarboxylic Acid (FDCA) Market Size Worth Around $850 Million By 2025: Acumen Research and Consulting. 2019. Available online: https://www.acumenresearchandconsulting.com/press-releases/2-5-furandicarboxylic-acid-fdca-market (accessed on 13 December 2021).
- Godan, T.K.; Rajesh, R.O.; Loreni, P.C.; Kumar Rai, A.; Sahoo, D.; Pandey, A.; Binod, P. Biotransformation of 5-hydroxymethylfurfural by Acinetobacter oleivorans S27 for the synthesis of furan derivatives. Bioresour. Technol. 2019, 282, 88–93. [Google Scholar] [CrossRef]
- Rajesh, R.O.; Godan, T.K.; Rai, A.K.; Sahoo, D.; Pandey, A.; Binod, P. Biosynthesis of 2,5-furan dicarboxylic acid by Aspergillus flavus APLS-1: Process optimization and intermediate product analysis. Bioresour. Technol. 2019, 284, 155–160. [Google Scholar] [CrossRef]
- Yang, C.-F.; Huang, C.-R. Biotransformation of 5-hydroxy-methylfurfural into 2,5-furan-dicarboxylic acid by bacterial isolate using thermal acid algal hydrolysate. Bioresour. Technol. 2016, 214, 311–318. [Google Scholar] [CrossRef]
- Sajid, M.; Zhao, X.; Liu, D. Production of 2,5-furandicarboxylic acid (FDCA) from 5-hydroxymethylfurfural (HMF): Recent progress focusing on the chemical-catalytic routes. Green Chem. 2018, 20, 5427–5453. [Google Scholar] [CrossRef]
- Saikia, K.; Rathankumar, A.K.; Kumar, P.S.; Varjani, S.; Nizar, M.; Lenin, R.; George, J.; Vaidyanathan, V.K. Recent advances in biotransformation of 5-Hydroxymethylfurfural: Challenges and future aspects. J. Chem. Technol. Biotechnol. 2021, 97, 409–419. [Google Scholar] [CrossRef]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Factories 2017, 16, 106. [Google Scholar] [CrossRef] [Green Version]
- Gomes, D.; Cunha, J.; Zanuso, E.; Teixeira, J.; Domingues, L. Strategies towards reduction of cellulases consumption: Debottlenecking the economics of lignocellulosics valorization processes. Polysaccharides 2021, 2, 20. [Google Scholar] [CrossRef]
- Liu, Z.L.; Slininger, P.J.; Dien, B.S.; Berhow, M.A.; Kurtzman, C.P.; Gorsich, S.W. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 2004, 31, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; He, A.; Wu, B.; Hu, L.; Liu, X.; Wu, Z.; Xia, J.; Xu, J.; Zhou, S. Redox-switchable biocatalyst for controllable oxidation or reduction of 5-hydroxymethylfurfural into high-value derivatives. ACS Omega 2020, 5, 19625–19632. [Google Scholar] [CrossRef]
- Li, Y.-M.; Zhang, X.-Y.; Li, N.; Xu, P.; Lou, W.-Y.; Zong, M.-H. Biocatalytic reduction of HMF to 2,5-bis(hydroxymethyl)furan by HMF-tolerant whole cells. ChemSusChem 2017, 10, 372–378. [Google Scholar] [CrossRef]
- Xu, Z.-H.; Cheng, A.-D.; Xing, X.-P.; Zong, M.-H.; Bai, Y.-P.; Li, N. Improved synthesis of 2,5-bis(hydroxymethyl)furan from 5-hydroxymethylfurfural using acclimatized whole cells entrapped in calcium alginate. Bioresour. Technol. 2018, 262, 177–183. [Google Scholar] [CrossRef] [PubMed]
- He, Y.C.; Jiang, C.X.; Chong, G.G.; Di, J.H.; Ma, C.L. Biological synthesis of 2,5-bis(hydroxymethyl)furan from biomass-derived 5-hydroxymethylfurfural by E. coli CCZU-K14 whole cells. Bioresour. Technol. 2018, 247, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.-H.; Zong, M.-H.; Li, N. Catalytic synthesis of 2,5-bis(hydroxymethyl)furan from 5-hydroxymethylfurfual by recombinant Saccharomyces cerevisiae. Enzym. Microb. Technol. 2020, 134, 109491. [Google Scholar] [CrossRef]
- Chen, D.; Cang, R.; Zhang, Z.-D.; Huang, H.; Zhang, Z.-G.; Ji, X.-J. Efficient reduction of 5-hydroxymethylfurfural to 2, 5-bis (hydroxymethyl) furan by a fungal whole-cell biocatalyst. Mol. Catal. 2021, 500, 111341. [Google Scholar] [CrossRef]
- Millán, A.; Sala, N.; Torres, M.; Canela-Garayoa, R. Biocatalytic transformation of 5-hydroxymethylfurfural into 2,5-di(hydroxymethyl)furan by a newly isolated Fusarium striatum strain. Catalysts 2021, 11, 216. [Google Scholar] [CrossRef]
- Chang, S.; He, X.; Li, B.; Pan, X. Improved bio-synthesis of 2,5-bis(hydroxymethyl)furan by Burkholderia contaminans NJPI-15 with co-substrate. Front. Chem. 2021, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Baptista, M.; Cunha, J.T.; Domingues, L. Establishment of Kluyveromyces marxianus as a microbial cell factory for lignocellulosic processes: Production of high value furan derivatives. J. Fungi 2021, 7, 1047. [Google Scholar] [CrossRef]
- Motagamwala, A.H.; Won, W.; Sener, C.; Alonso, D.M.; Maravelias, C.T.; Dumesic, J.A. Toward biomass-derived renewable plastics: Production of 2,5-furandicarboxylic acid from fructose. Sci. Adv. 2018, 4, eaap9722. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; He, A.; Liu, X.; Xia, J.; Xu, J.; Zhou, S.; Xu, J. Biocatalytic transformation of 5-hydroxymethylfurfural into high-value derivatives: Recent advances and future aspects. ACS Sustain. Chem. Eng. 2018, 6, 15915–15935. [Google Scholar] [CrossRef]
- Koopman, F.; Wierckx, N.; de Winde, J.H.; Ruijssenaars, H.J. Efficient whole-cell biotransformation of 5-(hydroxymethyl)furfural into FDCA, 2,5-furandicarboxylic acid. Bioresour. Technol. 2010, 101, 6291–6296. [Google Scholar] [CrossRef]
- Hsu, C.-T.; Kuo, Y.-C.; Liu, Y.-C.; Tsai, S.-L. Green conversion of 5-hydroxymethylfurfural to furan-2,5-dicarboxylic acid by heterogeneous expression of 5-hydroxymethylfurfural oxidase in Pseudomonas putida S12. Microb. Biotechnol. 2020, 13, 1094–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, N.N.; Chen, C.-Y.; Li, H.; Nguyen, M.T.T.; Nguyen, P.K.P.; Tsai, S.-L.; Chou, J.-Y.; Ramli, T.C.; Hu, Y.-C. Engineering stable Pseudomonas putida S12 by CRISPR for 2,5-furandicarboxylic acid (FDCA) production. ACS Synth. Biol. 2020, 9, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Wen, R.C.; Shen, C.R.; Tsai, S.-L. Biotransformation of 5-hydroxymethylfurfural to 2,5-furandicarboxylic acid by a syntrophic consortium of engineered Synechococcus elongatus and Pseudomonas putida. Biotechnol. J. 2020, 15, 1900357. [Google Scholar] [CrossRef]
- Tan, H.; Zhou, F.; Liao, D.; Ouyang, J.; Zheng, Z. Improved biosynthesis of 2,5-Furandicarboxylic acid through coupling of heterologous pathways in Escherichia coli and native pathways in Pseudomonas putida. Biochem. Eng. J. 2020, 161, 107657. [Google Scholar] [CrossRef]
- Hossain, G.S.; Yuan, H.; Li, J.; Shin, H.-D.; Wang, M.; Du, G.; Chen, J.; Liu, L. Metabolic engineering of Raoultella ornithinolytica BF60 for production of 2,5-furandicarboxylic acid from 5-hydroxymethylfurfural. Appl. Environ. Microbiol. 2016, 83, e02312-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.; Li, J.; Shin, H.D.; Du, G.; Chen, J.; Shi, Z.; Liu, L. Improved production of 2,5-furandicarboxylic acid by overexpression of 5-hydroxymethylfurfural oxidase and 5-hydroxymethylfurfural/furfural oxidoreductase in Raoultella ornithinolytica BF60. Bioresour. Technol. 2018, 247, 1184–1188. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Y.; Li, J.; Shin, H.D.; Du, G.; Shi, Z.; Chen, J.; Liu, L. Combinatorial synthetic pathway fine-tuning and comparative transcriptomics for metabolic engineering of Raoultella ornithinolytica BF60 to efficiently synthesize 2,5-furandicarboxylic acid. Biotechnol. Bioeng. 2018, 115, 2148–2155. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, Y.; Lv, X.; Li, J.; Du, G.; Shi, Z.; Liu, L. Enhanced 2,5-Furandicarboxylic Acid (FDCA) Production in Raoultella ornithinolytica BF60 by Manipulation of the Key Genes in FDCA Biosynthesis Pathway. J. Microbiol. Biotechnol. 2018, 28, 1999–2008. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.-Y.; Zong, M.-H.; Li, N. Sacrificial substrate-free whole-cell biocatalysis for the synthesis of 2,5-furandicarboxylic acid by engineered Escherichia coli. ACS Sustain. Chem. Eng. 2020, 8, 4341–4345. [Google Scholar] [CrossRef]
- Yang, C.-F.; Huang, C.-R. Isolation of 5-hydroxymethylfurfural biotransforming bacteria to produce 2,5-furan dicarboxylic acid in algal acid hydrolysate. J. Biosci. Bioeng. 2018, 125, 407–412. [Google Scholar] [CrossRef]
- Rajesh, R.O.; Godan, T.K.; Ashok, P.; Binod, P. Whole cell based biocatalytic production of 2,5-furan dicarboxylic acid. Indian J. Exp. Biol. 2018, 56, 493–497. [Google Scholar]
- Sheng, Y.; Tan, X.; Zhou, X.; Xu, Y. Bioconversion of 5-hydroxymethylfurfural (hmf) to 2,5-furandicarboxylic acid (fdca) by a native obligate aerobic bacterium, Acinetobacter calcoaceticus NL14. Appl. Biochem. Biotechnol. 2020, 192, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Becerra, M.L.; Lizarazo, L.M.; Rojas, H.A.; Prieto, G.A.; Martinez, J.J. Biotransformation of 5-hydroxymethylfurfural and furfural with bacteria of bacillus genus. Biocatal. Agric. Biotechnol. 2022, 39, 102281. [Google Scholar] [CrossRef]
- Troiano, D.; Orsat, V.; Dumont, M.-J. Use of filamentous fungi as biocatalysts in the oxidation of 5-(hydroxymethyl)furfural (HMF). Bioresour. Technol. 2022, 344, 126169. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-Y.; Lu, P.-Y.; Yang, C.-F. Lignocellulosic acid hydrolysis inhibitor impact on 5-hydroxymethylfurfural biotransformation into 2, 5-furandicarboxylic acid using immobilised Burkholderia cells. Biocatal. Biotransformation 2021, 1–11. [Google Scholar] [CrossRef]
- Munekata, M.; Tamura, G. Antitumor activity of 5-hydroxymethyl-2-furoic acid. Agric. Biol. Chem. 1981, 45, 2149–2150. [Google Scholar]
- Braisted, A.C.; Oslob, J.D.; Delano, W.L.; Hyde, J.; McDowell, R.S.; Waal, N.; Yu, C.; Arkin, M.R.; Raimundo, B.C. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J. Am. Chem. Soc. 2003, 125, 3714–3715. [Google Scholar] [CrossRef]
- Hirai, H. Oligomers from hydroxymethylfurancarboxylic acid. J. Macromol. Sci. Part A—Chem. 1984, 21, 1165–1179. [Google Scholar] [CrossRef]
- Ran, H.; Zhang, J.; Gao, Q.; Lin, Z.; Bao, J. Analysis of biodegradation performance of furfural and 5-hydroxymethylfurfural by Amorphotheca resinae ZN1. Biotechnol. Biofuels 2014, 7, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taherzadeh, M.J.; Gustafsson, L.; Niklasson, C.; Lidén, G. Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2000, 53, 701–708. [Google Scholar] [CrossRef]
- Mitsukura, K.; Sato, Y.; Yoshida, T.; Nagasawa, T. Oxidation of heterocyclic and aromatic aldehydes to the corresponding carboxylic acids by Acetobacter and Serratia strains. Biotechnol. Lett. 2004, 26, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- De Bont, J.A.M.; Ruijssenaars, H.J.; Werij, J. Fungal Production of FDCA. U.S. Patent WO2017050815, 30 March 2017. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2017050815 (accessed on 13 December 2021).
- Wen, M.; Zhang, X.-Y.; Zong, M.-H.; Li, N. Significantly improved oxidation of bio-based furans into furan carboxylic acids using substrate-adapted whole cells. J. Energy Chem. 2020, 41, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.-S.; Zhang, X.-Y.; Zong, M.-H.; Wang, C.-F.; Li, N. Selective synthesis of 2-furoic acid and 5-hydroxymethyl-2-furancarboxylic acid from bio-based furans by recombinant Escherichia coli cells. Mol. Catal. 2019, 469, 68–74. [Google Scholar] [CrossRef]
- Cheng, A.-D.; Shi, S.-S.; Li, Y.; Zong, M.-H.; Li, N. Biocatalytic oxidation of biobased furan aldehydes: Comparison of toxicity and inhibition of furans toward a whole-cell biocatalyst. ACS Sustain. Chem. Eng. 2020, 8, 1437–1444. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Ou, X.-Y.; Fu, Y.-J.; Zong, M.-H.; Li, N. Efficient synthesis of 5-hydroxymethyl-2-furancarboxylic acid by Escherichia coli overexpressing aldehyde dehydrogenases. J. Biotechnol. 2020, 307, 125–130. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Wang, X.; Li, N.-W.; Guo, Z.-W.; Zong, M.-H.; Li, N. Furan carboxylic acids production with high productivity by cofactor-engineered whole-cell biocatalysts. ChemCatChem 2020, 12, 3257–3264. [Google Scholar] [CrossRef]
- Wang, Z.-W.; Gong, C.-J.; He, Y.-C. Improved biosynthesis of 5-hydroxymethyl-2-furancarboxylic acid and furoic acid from biomass-derived furans with high substrate tolerance of recombinant Escherichia coli HMFOMUT whole-cells. Bioresour. Technol. 2020, 303, 122930. [Google Scholar] [CrossRef]
- Muñoz, T.; Rache, L.Y.; Rojas, H.A.; Romanelli, G.P.; Martinez, J.J.; Luque, R. Production of 5-hydroxymethyl-2-furan carboxylic acid by Serratia marcescens from crude 5-hydroxymethylfurfural. Biochem. Eng. J. 2020, 154, 107421. [Google Scholar] [CrossRef]
- Liu, P.; Xie, J.; Tan, H.; Zhou, F.; Zou, L.; Ouyang, J. Valorization of Gelidium amansii for dual production of D-galactonic acid and 5-hydroxymethyl-2-furancarboxylic acid by chemo-biological approach. Microb. Cell Factories 2020, 19, 104. [Google Scholar] [CrossRef]
- Chang, S.; He, X.; Wang, X.; Li, B.; Liu, L.; Qin, J.; Yao, Z.; Pan, X. Exploring the optimized strategy for 5-hydroxymethyl-2-furancarboxylic acid production from agriculture wastes using Pseudomonas aeruginosa PC-1. Process Biochem. 2021, 102, 417–422. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zong, M.-H.; Li, N. Whole-cell biocatalytic selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid. Green Chem. 2017, 19, 4544–4551. [Google Scholar] [CrossRef]
- Cang, R.; Shen, L.-Q.; Yang, G.; Zhang, Z.-D.; Huang, H.; Zhang, Z.-G. highly selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid by a robust whole-cell biocatalyst. Catalysts 2019, 9, 526. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Zheng, Z.; Zou, L.; Zhang, C.; Yang, F.; Zhou, K.; Ouyang, J. A versatile Pseudomonas putida KT2440 with new ability: Selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid. Bioprocess Biosyst. Eng. 2020, 43, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Sayed, M.; Pyo, S.-H.; Rehnberg, N.; Hatti-Kaul, R. Selective oxidation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid using Gluconobacter oxydans. ACS Sustain. Chem. Eng. 2019, 7, 4406–4413. [Google Scholar] [CrossRef]
- Pan, X.; Wu, S.; Yao, D.; Liu, L.; Zhang, L.; Yao, Z.; Pan, Y.; Chang, S.; Li, B. Efficient biotransformation of 5-hydroxymethylfurfural to 5-hydroxymethyl-2-furancarboxylic acid by a new whole-cell biocatalyst Pseudomonas aeruginosa PC-1. React. Chem. Eng. 2020, 5, 1397–1404. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).