Resolution of Racemic Aryloxy-Propan-2-yl Acetates via Lipase-Catalyzed Hydrolysis: Preparation of Enantiomerically Pure/Enantioenriched Mexiletine Intermediates and Analogs
Abstract
:1. Introduction
2. Results and Discussion
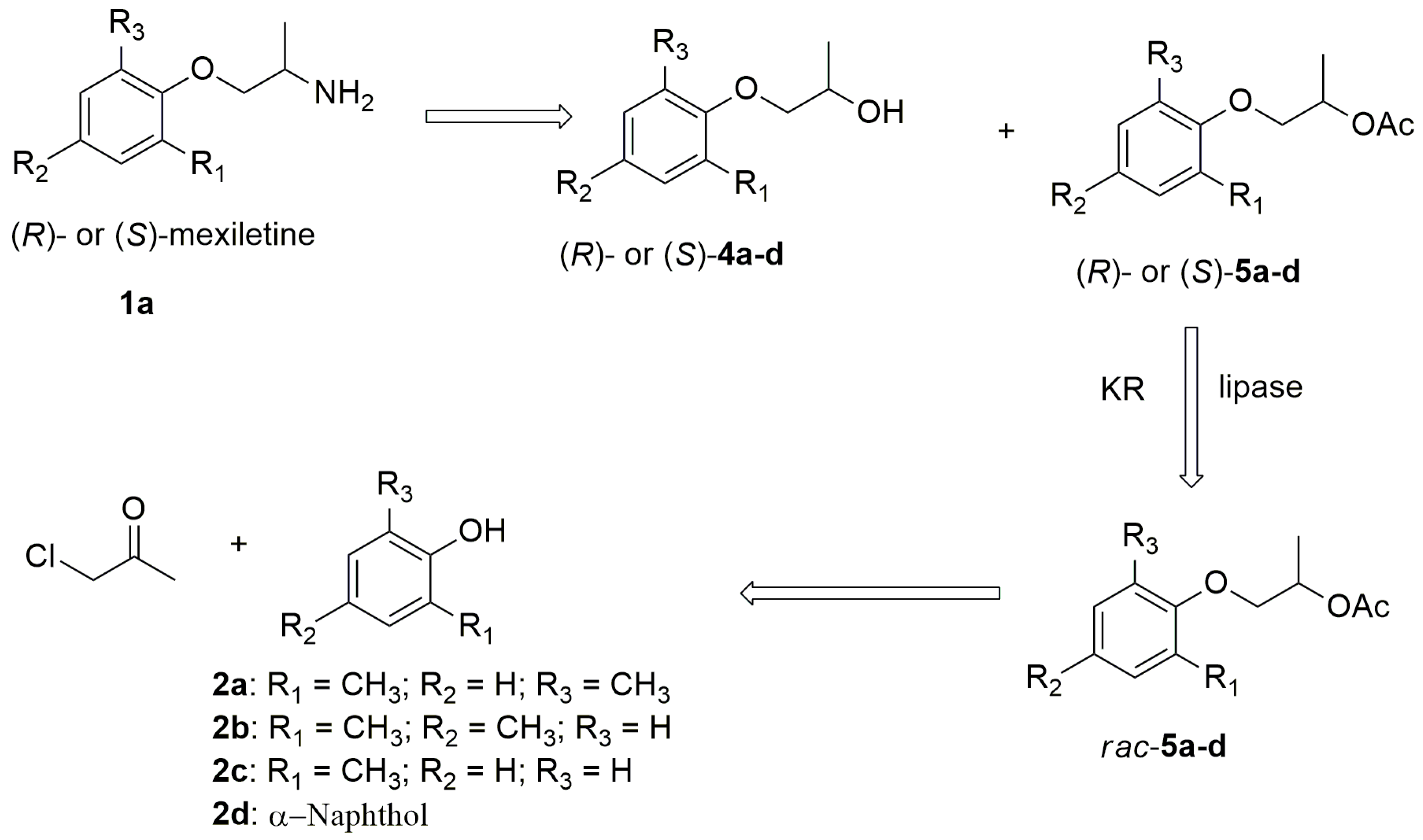
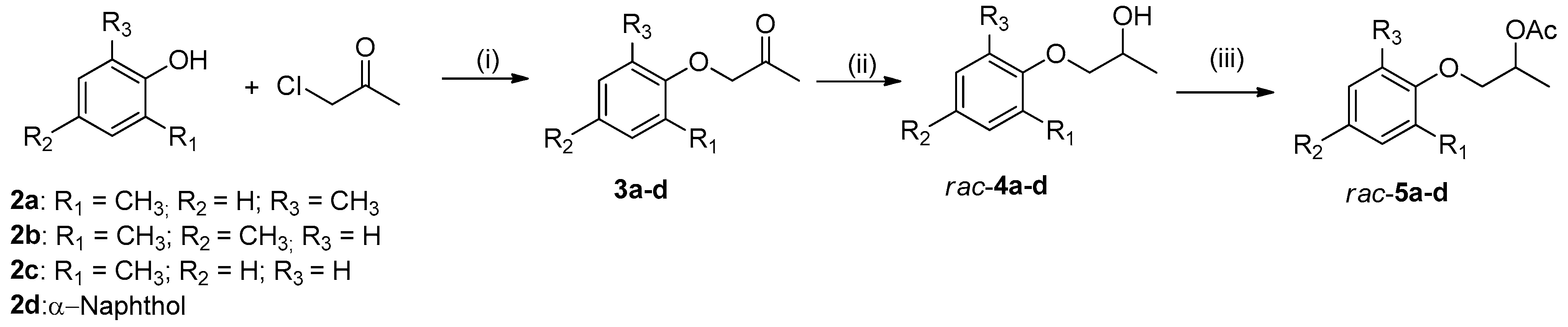
2.1. Preparation of Alcohols rac-4a-d and Their Corresponding Acetates rac-5a-d
2.2. Screening of Lipases for the Kinetic Resolution of Acetate rac-5a via Hydrolysis
2.3. Influence of the Co-Solvent on the KR of Acetate rac-5a, via Hydrolysis, Catalyzed by Lipases
2.4. Kinetic Resolution of rac-4a via Acetylation Reaction, Catalyzed by Lipases
2.5. Reuse Study of TLL Immobilized on Immobead 150 in KR of rac-4a, via Acetylation Reaction
2.6. KR Study of Acetates rac-5b-d via Hydrolysis Reaction in the Presence of Lipases
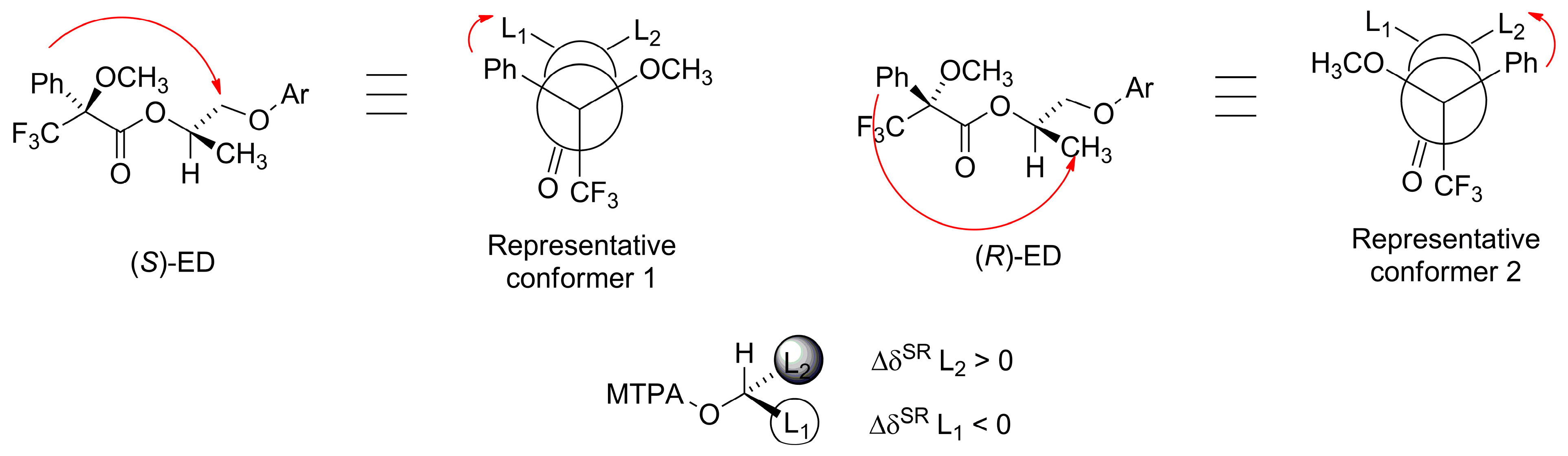
2.7. Determination of the Absolute Configurations of the New Mexiletine Analogs Intermediates by 1H NMR Spectroscopy, Using the Mosher Method
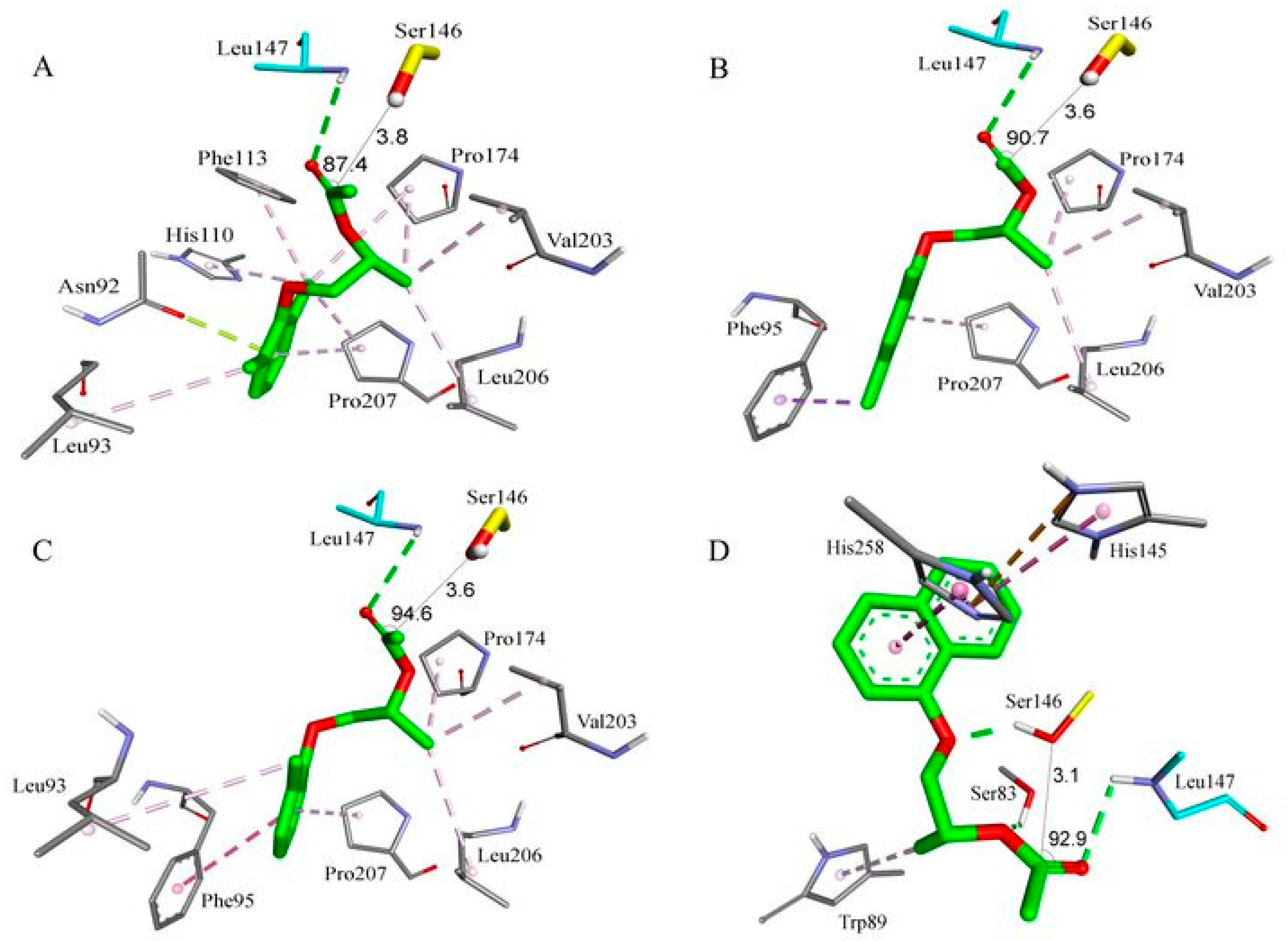
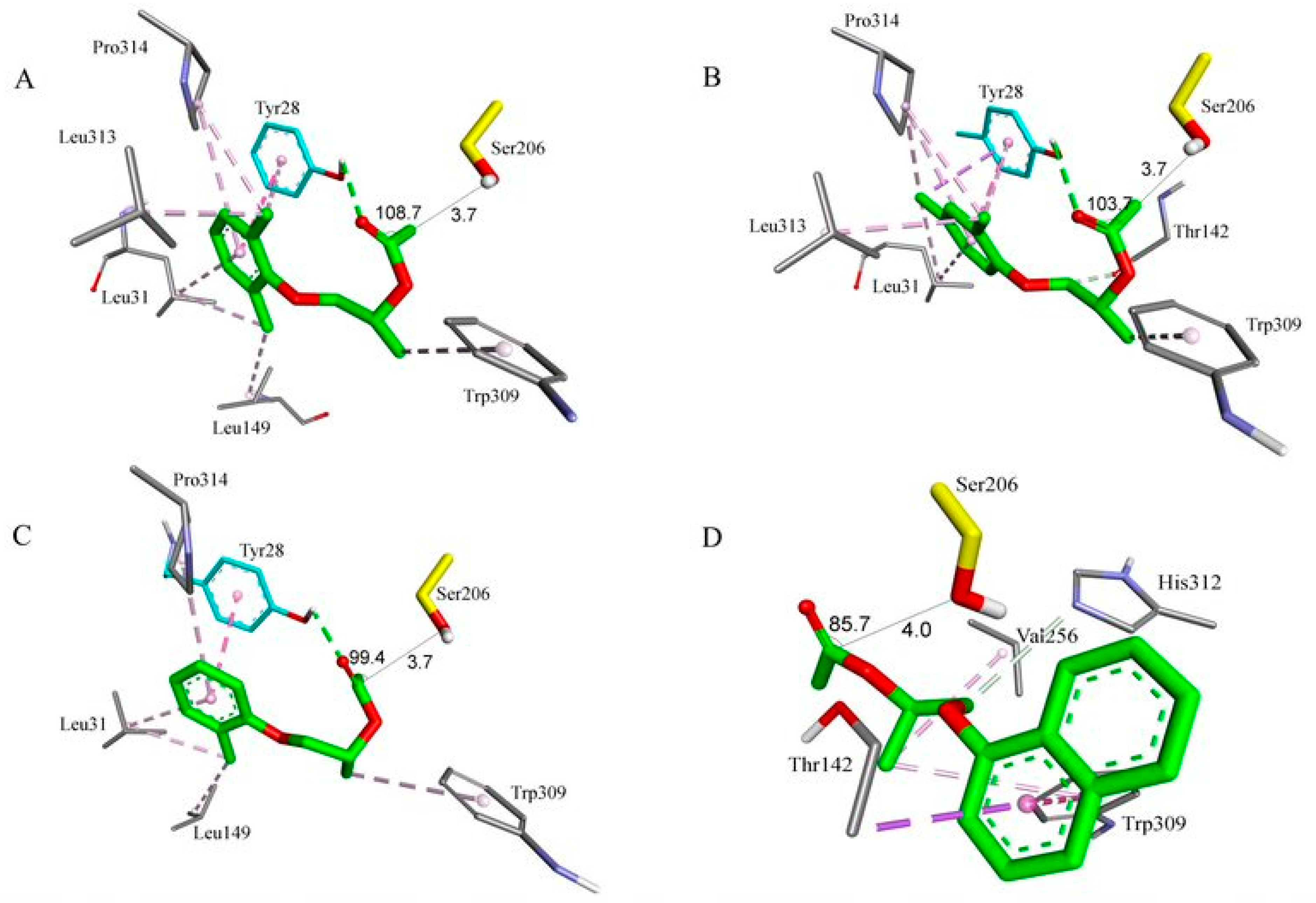
2.8. Molecular Docking Study
3. Materials and Methods
3.1. Enzymes
3.2. Chemical Materials
3.3. Analysis
3.4. Procedure for Immobilizing Lipase from Thermomyces lanuginosus
3.5. General Procedure for the Synthesis of Ketones (3a-d)
3.6. General Procedure for the Synthesis of Racemic Alcohols (rac-4a-d)
3.7. General Procedure for the Synthesis of Racemic Acetates (rac-5a-d)
3.8. General Procedure for the Synthesis of Mosher Esters
3.9. General Procedure for the Enzymatic Kinetic Resolution of Acetates rac-5a-d, via Hydrolysis, Catalyzed by Lipases
3.10. Procedure for the Enzymatic Kinetic Resolution of Alcohol rac-4a, via Acetylation Reaction, Catalyzed by Lipases
3.11. Procedure for the Reuse Study of TLL Immobilized on Immobead 150 in the Kinetic Resolution of Alcohol rac-4a via Acetylation Reaction
3.12. Molecular Docking Study
3.12.1. Computation
3.12.2. Methodology for Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shukla, E.; Bendre, A.D.; Gaikwad, S.M. Hydrolases: The Most Diverse Class of Enzymes, 1st ed.; Haider, S., Haider, A., Catalá, A., Eds.; Intechopen: London, UK, 2022; pp. 1–16. [Google Scholar]
- Delgado-Garcia, M.; Valdivia-Urdiales, B.; Aguilar-González, C.N.; Contreras-Esquivel, J.C.; Rodriguez-Herrera, R. Halophilic hydrolases as a new tool for the biotechnological industries. J. Sci. Food Agric. 2012, 92, 2575–2580. [Google Scholar] [CrossRef] [PubMed]
- Fasim, A.; More, V.S.; More, S.S. Large-scale production of enzymes for biotechnology uses. Curr. Opin. Biotechnol. 2021, 69, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Ramnath, L.; Sithole, B.; Govinden, R. Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can. J. Microbiol. 2017, 63, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Fact. 2020, 19, 169–211. [Google Scholar]
- Rodrigues, R.C.; Virgen-Ortiz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, A.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar] [PubMed] [Green Version]
- Rajendran, A.; Palanisamy, A.; Thangavelu, V. Lipase catalyzed ester synthesis for food processing industries. Braz. Arch. Biol. Technol. 2009, 52, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Gotor-Fernández, V.; Brieva, R.; Gotor, V. Lipases: Useful biocatalysts for the preparation of pharmaceuticals. J. Mol. Catal. B Enzym. 2006, 40, 111–120. [Google Scholar] [CrossRef]
- Carvalho, A.C.L.M.; Fonseca, T.S.; de Mattos, M.C.; de Oliveira, M.C.F.; de Lemos, T.L.G.; Molinari, F.; Romano, D.; Serra, I. Recent advances in lipase-mediated preparation of pharmaceuticals and their intermediates. Int. J. Mol. Sci. 2015, 16, 29682–29716. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.P.; Brown, M.J.B.; Diaz-Rodriguez, A.; Lloyd, R.C.; Roiban, G.-D. Biocatalysis: A pharma perspective. Adv. Synth. Catal. 2019, 361, 2421–2432. [Google Scholar] [CrossRef] [Green Version]
- Solano, D.M.; Hoyos, P.; Hernáiz, M.J.; Alcántara, A.R.; Sánchez-Montero, J.M. Industrial biotransformations in the synthesis of building blocks leading to enantiopure drugs. Bioresour. Technol. 2012, 114, 196–207. [Google Scholar] [CrossRef]
- Contesini, F.J.; Davanço, M.G.; Borin, G.P.; Vanegas, K.G.; Cirino, J.P.G.; de Melo, R.R.; Mortensen, U.H.; Hildén, K.; Campos, D.R.; Carvalho, P.O. Advances in recombinant lipases: Production, engineering, immobilization and application in pharmaceutical industry. Catalysts 2020, 10, 1032. [Google Scholar] [CrossRef]
- Faber, K.; Fessner, W.-D.; Turner, N.J. Biocatalysis: Ready to master increasing complexity. Adv. Synth. Catal. 2019, 361, 2373–2376. [Google Scholar] [CrossRef] [Green Version]
- Bezborodov, A.M.; Zagustina, N.A. Enzymatic biocatalysis in chemical synthesis of pharmaceuticals. Appl. Biochem. Microbiol. 2016, 52, 237–249. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, H.; Ang, E.L.; Zhao, H. Biocatalysis for the synthesis of pharmaceuticals and pharmaceutical intermediates. Bioorg. Med. Chem. 2018, 26, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- An, J.; No, Y.; Xu, Y. Structural insights into alcohols dehydrogenases catalyzing asymmetric reductions. Crit. Rev. Biotechnol. 2019, 39, 366–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-S.; de Souza, F.Z.R. Enzymatic synthesis of enantiopure alcohols: Current state and perspectives. RSC Adv. 2019, 9, 2102–2115. [Google Scholar] [CrossRef] [Green Version]
- Araujo, D.M.F.; Vieira, G.A.B.; de Mattos, M.C.; Lemos, T.L.G.; de Oliveira, M.C.F.; Melo, V.M.M.; de Gonzalo, G.; Gotor-Fernández, V.; Gotor, V. Chemoenzymatic preparation of a biologically active naphthoquinone from Tabebuia impetiginosa using lipases or alcohol dehydrogenases. J. Mol. Catal. B Enzym. 2009, 61, 279–283. [Google Scholar] [CrossRef]
- Fonseca, T.S.; da Silva, M.R.; de Oliveira, M.C.F.; de Lemos, T.L.G.; Marques, R.A.; de Mattos, M.C. Chemoenzymatic synthesis of rasagiline mesylate using lipases. Appl. Catal. A Gen. 2015, 492, 76–82. [Google Scholar] [CrossRef]
- Galvão, W.S.; Pinheiro, B.B.; Gonçalves, L.R.B.; de Mattos, M.C.; Fonseca, T.S.; Regis, T.; Zampieri, D.; dos Santos, J.C.S.; Costa, L.S.; Correa, M.A.; et al. Novel nanohybrid biocatalyst: Application in the kinetic resolution of secondary alcohols. J. Mater. Sci. 2018, 53, 14121–14137. [Google Scholar] [CrossRef]
- Vega, K.B.; Cruz, D.M.V.; Oliveira, A.R.T.; da Silva, M.R.; de Lemos, T.L.G.; Oliveira, M.C.F.; Bernardo, R.D.S.; de Souza, J.R.; Zanatta, G.; Nasário, F.D.; et al. Chemoenzymatic Synthesis of Apremilast: A Study Using Ketoreductases and Lipases. J. Braz. Chem. Soc. 2021, 32, 1100–1110. [Google Scholar] [CrossRef]
- Catalano, A.; Carocci, A. Antiarrhythmic Mexiletine: A Review on Synthetic Routes to Racemic and Homochiral Mexiletine and its Enantioseparation. Curr. Med. Chem. 2016, 23, 3227–3244. [Google Scholar] [CrossRef]
- Wu, W.-P.; Nordmark, J.; Wiesenfeld-Hallin, Z.; Xu, X.-J. Lack of stereoselectivity for the antiallodynic effect of mexiletine in spinally injured rats. Eur. J. Pain 2000, 4, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Loughhead, D.G.; Flippin, L.A.; Weikert, R.J. Synthesis of Mexiletine Stereoisomers and Related Compounds via SNAr Nucleophilic Substitution of a Cr(CO)3-Complexed Aromatic Fluoride. J. Org. Chem. 1999, 64, 3373–3375. [Google Scholar] [CrossRef]
- Carocci, A.; Catalano, A.; Corbo, F.; Duranti, A.; Amoroso, R.; Franchini, C.; Lentini, G.; Tortorella, V. Stereospecific synthesis of mexiletine and related compounds: Mitsunobu versus Williamson reaction. Tetrahedron Asymmetry 2000, 11, 3619–3634. [Google Scholar] [CrossRef]
- Franchini, C.; Carocci, A.; Catalano, A.; Cavalluzzi, M.M.; Corbo, F.; Lentini, G.; Scilimati, A.; Tortorella, P.; Camerino, D.C.; De Luca, A. Optically Active Mexiletine Analogues as Stereoselective Blockers of Voltage-Gated Na+ Channels. J. Med. Chem. 2003, 46, 5238–5248. [Google Scholar] [CrossRef] [PubMed]
- Catalano, A.; Budriesi, R.; Bruno, C.; Di Mola, A.; Defrenza, I.; Cavalluzzi, M.M.; Micucci, M.; Carocci, A.; Franchini, C.; Lentini, G. Searching for new antiarrhythmic agents: Evaluation of meta-hydroxymexiletine enantiomers. Eur. J. Med. Chem. 2013, 65, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Fesko, K.; Steiner, K.; Breinbauer, R.; Schwab, H.; Schürmann, M.; Strohmeier, G.A. Investigation of one-enzyme systems in the ω-transaminase-catalyzed synthesis of chiral amines. J. Mol. Catal. B Enzym. 2013, 96, 103–110. [Google Scholar] [CrossRef]
- Andrade, L.H.; Kroutil, W.; Jamison, T.F. Continuous Flow Synthesis of Chiral Amines in Organic Solvents: Immobilization of E. coli Cells Containing Both ω-Transaminase and PLP. Org. Lett. 2014, 16, 6092–6095. [Google Scholar] [CrossRef]
- Carocci, A.; Franchini, C.; Lentini, G.; Loiodice, F.; Tortorella, V. Facile entry to (-)-(R)- and (+)-(S)-mexiletine. Chirality 2000, 12, 103–106. [Google Scholar] [CrossRef]
- Sasikumar, M.; Nikalje, M.D.; Muthukrishnan, M. A convenient synthesis of enantiomerically pure (R)-mexiletine using hydrolytic kinetic resolution method. Tetrahedron Asymmetry 2009, 20, 2814–2817. [Google Scholar] [CrossRef]
- da Silva, M.R.; de Mattos, M.C.; de Oliveira, M.C.F.; de Lemos, T.L.G.; Ricardo, N.M.P.; de Gonzalo, G.; Lavandera, I.; Gotor-Fernández, V.; Gotor, V. Asymmetric chemoenzymatic synthesis of N-acetyl-α-amino esters based on lipase-catalyzed kinetic resolution through interesterification reactions. Tetrahedron 2014, 70, 2264–2271. [Google Scholar] [CrossRef]
- Bezerra, F.A.; Lima, G.C.; Carvalho, A.C.L.M.; Vega, K.B.; Oliveira, M.C.F.; de Lemos, T.L.G.; dos Santos, J.C.S.; Gonçalves, L.R.B.; Rios, N.S.; Fernandez-Lafuente, R.; et al. Chemoenzymatic synthesis of both enantiomers of propafenone hydrochloride through lipase-catalyzed process. Mol. Catal. 2022, 529, 112540. [Google Scholar] [CrossRef]
- Faber, K. Biotransformations in Organic Chemistry, 7th ed.; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Kazlauskas, R.J.; Weissfloch, A.N.E.; Rappaport, A.T.; Cuccia, L.A. A rule to predict which enantiomer of a secondary alcohol reacts faster in reactions catalyzed by cholesterol esterase, lipase from Pseudomonas cepacia, and lipase from Candida rugosa. J. Org. Chem. 1991, 56, 2656–2665. [Google Scholar] [CrossRef]
- Byrne, F.P.; Jin, S.; Paggiola, G.; Petchey, T.H.M.; Clark, J.H.; Farmer, T.J.; Hunt, A.J.; McElroy, C.R.; Sherwood, J. Tools and techniques for solvent selection: Green solvent selection guides. Sustain. Chem. Process. 2016, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Kanerva, L.T.; Vihanto, J.; Halme, M.H.; Loponen, J.M.; Euranto, E.K. Solvents effects in lipase-catalysed transesterification reactions. Acta Chem. Scand. 1990, 44, 1032–1035. [Google Scholar] [CrossRef] [Green Version]
- Gogoi, S.; Pathak, M.G.; Dutta, A.; Dutta, N.N. Porcine pancreas lipase catalyzed synthesis of lauryl laurate in organic solvent media: A kinetic study. Ind. J. Biochem. Biophys. 2008, 45, 192–197. [Google Scholar]
- Lima, G.V.; da Silva, M.R.; Fonseca, T.S.; de Lima, L.B.; de Oliveira, M.C.F.; de Lemos, T.L.G.; Zampieri, D.; dos Santos, J.C.S.; Rios, N.S.; Gonçalves, L.B.R.; et al. Chemoenzymatic synthesis of (S)-Pindolol using lipases. Appl. Catal. A General 2017, 546, 7–14. [Google Scholar] [CrossRef]
- Seco, J.M.; Quiñoá, E.; Riguera, R. The Assignment of Absolute Configuration by NMR. Chem. Rev. 2004, 104, 17–117. [Google Scholar] [CrossRef]
- Mathpati, A.C.; Bhanage, B.M. Combined docking and molecular dynamics study of lipase catalyzed kinetic resolution of 1-phenylethanol in organic solvents. J. Mol. Catal. B Enzym. 2016, 133, 5119–5127. [Google Scholar] [CrossRef]
- Escorcia, A.M.; Molina, D.; Daza, M.; Doerr, M. Acetylation of (R,S)-propranolol catalyzed by Candida antarctica lipase B: An experimental and computational study. J. Mol. Catal. B Enzym. 2013, 98, 21–29. [Google Scholar] [CrossRef]
- Fonseca, T.S.; Vega, K.B.; da Silva, M.R.; de Oliveira, M.C.F.; de Lemos, T.L.G.; Contente, M.L.; Molinari, F.; Cespuli, M.; Fortuna, S.; Gardossi, L.; et al. Lipase Mediated enzymatic kinetic resolution of phenylethyl halohydrins acetates: A case of study and rationalization. Mol. Catal. 2020, 485, 110819. [Google Scholar] [CrossRef]
- Bruice, T.C.; Lightstone, F.C. Ground state and transition state contributions to the rates of intramolecular and enzymatic reactions. Acc. Chem. Res. 1999, 32, 127–136. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Savage, H.; Verma, C.S.; Turkenburg, J.P.; Lawson, D.M.; Svendsen, A.; Patkar, S. Structural origins of the interfacial activation in Thermomyces (Humicola) lanuginose lipase. Biochemistry 2000, 39, 15071–15082. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Angkawidjaja, C.; Matsumura, H.; Koga, Y.; Takano, K.; Kanaya, S. X-ray Crystallographic and MD simulation studies on the mechanism of interfacial activation of a family I.3 lipase with two lids. J. Mol. Biol. 2010, 400, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schimidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Borowiecki, P.; Paprocki, D.; Dudzik, A.; Plenkiewicz, J. Chemoenzymatic synthesis of proxyphylline enantiomer. J. Org. Chem. 2016, 81, 380–395. [Google Scholar] [CrossRef]
- Svendsen, A. Understanding Enzymes Function, Design, Engineering, and Analysis, 1st ed.; Pan Stanford Publishing: Singapore, 2016. [Google Scholar]
- Hur, S.; Bruice, T.C. The near attack conformation approach to the study of the chorismite to prephenate reaction. Proc. Natl. Acad. Sci. USA 2003, 100, 12015–12020. [Google Scholar] [CrossRef] [Green Version]
- Bezerra, R.M.; Neto, D.M.A.; Galvão, W.S.; Rios, N.S.; Carvalho, A.C.L.M.; Correa, M.A.; Bohn, F.; Fernandez-Lafuente, R.; Fechine, P.B.A.; de Mattos, M.C.; et al. Design a lipase-nano particle biocatalysts and it use in the kinetic resolution of medicament precursors. Biochem. Eng. J. 2017, 125, 104–115. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cousins, K.R. Computer review of ChemDraw Ultra 12.0. J. Am. Chem. Soc. 2011, 133, 8388. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]


| Entry a | Lipase | ees (%) b | eep (%) b | c (%) c | E d |
|---|---|---|---|---|---|
| 1 | P. fluorescens | >99 | >99 | 50 | >200 |
| 2 e | TLL | >99 | >99 | 50 | >200 |
| 3 | P. camemberti | 1 | 29 | 03 | 2 |
| 4 f | C. antarctica B | >99 | 98 | 50 | >200 |
| 5 | C. rugosa | 23 | 57 | 28 | 5 |
| 6 f | R. miehei | 95 | 86 | 52 | 49 |
| 7 | B. cepacia | 90 | 86 | 51 | 41 |
| 8 g | B. cepacia | >99 | 93 | 51 | 135 |
| 9 | SPMN@APTES-TLL | 18 | >99 | 15 | >200 |
| 10 | SPMN@PEI-TLL | 50 | >99 | 34 | >200 |
| 11 | SPMN@APTES-GA | 5 | >99 | 17 | 13 |
| 12 | SPMN@PEI-GA | 20 | >99 | 17 | >200 |
| Lipase | Entry a | Co-solvent | ees (%) b | eep (%) b | c (%) c | E d |
|---|---|---|---|---|---|---|
| P. fluorescens | 1 | None | 96 | >99 | 49 | >200 |
| 2 | Acetonitrile | >99 | >99 | 50 | >200 | |
| 3 | IPA | >99 | 98 | 50 | >200 | |
| 4 | Diethyl ether | 05 | >99 | 05 | >200 | |
| 5 | THF | 84 | >99 | 46 | >200 | |
| TLL immobilized on Immobead 150 | 6 | None | >99 | >99 | 50 | >200 |
| 7 | Acetonitrile | >99 | >99 | 50 | >200 | |
| 8 | IPA | >99 | >99 | 50 | >200 | |
| 9 | Diethyl ether | >99 | >99 | 50 | >200 | |
| 10 | THF | 87 | >99 | 47 | >200 | |
| TLL immobilized onto SPMN@PEI | 11 | None | 06 | >99 | 06 | >200 |
| 12 | Acetonitrile | 50 | >99 | 34 | >200 | |
| 13 | IPA | 12 | >99 | 11 | >200 | |
| 14 | Diethyl ether | >99 | >99 | 50 | >200 | |
| 15 | THF | >99 | >99 | 50 | >200 | |
| CAL-B immobilized on anionic resin | 16 | None | 55 | >99 | 36 | >200 |
| 17 | Acetonitrile | >99 | 98 | 50 | >200 | |
| 18 | IPA | >99 | >99 | 50 | >200 | |
| 19 | Diethyl ether | 71 | >99 | 42 | >200 | |
| 20 | THF | 88 | >99 | 47 | >200 |
| Entry a | Solvent | Time (min) | ees (%) b | eep (%) b | c (%) c | E d |
|---|---|---|---|---|---|---|
| 1 | 1,4-dioxane | 30 | 39 | 48 | 45 | 4 |
| 2 e | toluene | 15 | 94 | 95 | 50 | 139 |
| 3 | THF | 180 | 95 | 92 | 51 | 89 |
| 4 f | n-hexane | 20 | 57 | 45 | 56 | 4.5 |
| Substrate | Lipase | Entry a | Co-Solvent | ees (%) b | eep (%) b | c (%) c | E d |
|---|---|---|---|---|---|---|---|
| rac-5b | TLL | 1 | None | 38 | 98 | 72 | 9 |
| 2 | Acetonitrile | >99 | >99 | 47 | >200 | ||
| P. fluorescens | 3 | None | 90 | >99 | 52 | 99 | |
| 4 | Acetonitrile | 98 | 98 | 50 | >200 | ||
| rac-5c | TLL | 5 | None | 94 | 94 | 50 | 115 |
| 6 | Acetonitrile | 92 | 98 | 48 | >200 | ||
| P. fluorescens | 7 | None | >99 | 75 | 57 | 36 | |
| 8 | Acetonitrile | 92 | 87 | 49 | 68 | ||
| rac-5d | TLL | 9 | None | 45 | 2 | 4 | 3 |
| 10 | Acetonitrile | 43 | >99 | 30 | >200 | ||
| 11 e | Acetonitrile | 84 | >99 | 54 | 59 | ||
| P. fluorescens | 12 | None | 69 | 12 | 15 | 6 | |
| 13 | Acetonitrile | 43 | >99 | 30 | >200 | ||
| 14 e | Acetonitrile | 95 | >99 | 51 | >200 |
| Starting Alcohol | Entry | (S)-ED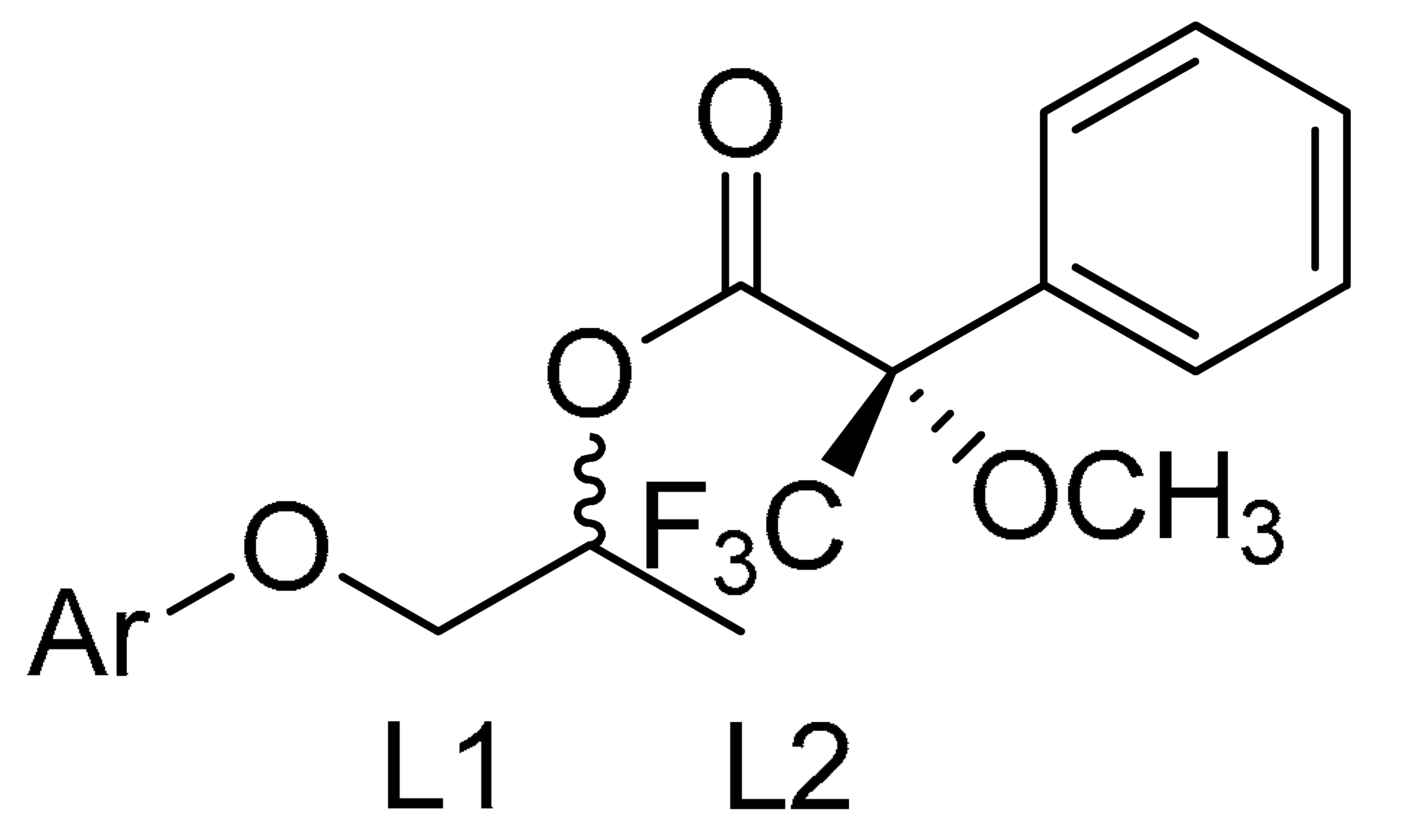 | (R)-ED | ∆δSR |
|---|---|---|---|---|
| 4b | 1 | L1 = 3.98 | L1 = 4.02 | −0.04 |
| 2 | L2 = 1.49 | L2 = 1.43 | 0.06 | |
| 4c | 3 | L1 = 4.04 | L1 = 4.08 | −0.04 |
| 4 | L2 = 1.44 | L2 = 1.40 | 0.04 | |
| 4d | 5 | L1 = 4.20 | L1 = 4.32 | −0.12 |
| 6 | L2 = 1.56 | L2 = 1.48 | 0.08 |
| Substrate | Entry | Lipase | NAC a | Binding Energy (kcal.mol−1) b | Distance (Å) c | Angle (°) d | Hydrogen Bond with the Carbonyl Oxygen |
|---|---|---|---|---|---|---|---|
| (R)-5a | 1 | TLL | 8 | −5.04 | 3.8 | 87.4 | O:NHLeu147 |
| 2 | P. fluorescens | 11 | −4.14 | 3.7 | 108.7 | O:OHTyr28 | |
| (S)-5a | 3 | TLL | 1 | −4.72 | 3.4 | 109.8 | - |
| 4 | P. fluorescens | 5 | −3.95 | 3.5 | 91.7 | O:OHTyr28 | |
| (R)-5b | 5 | TLL | 6 | −5.15 | 3.6 | 90.7 | O:NHLeu147 |
| 6 | P. fluorescens | 1 | −4.18 | 3.7 | 103.7 | O:OHTyr28 | |
| (S)-5b | 7 | TLL | 0 | - | - | - | - |
| 8 | P. fluorescens | 0 | - | - | - | - | |
| (R)-5c | 9 | TLL | 2 | −4.61 | 3.6 | 94.6 | O:NHLeu147 |
| 10 | P. fluorescens | 6 | −3.49 | 3.7 | 99.4 | O:OHTyr28 | |
| (S)-5c | 11 | TLL | 1 | −4.83 | 3.9 | 89.4 | - |
| 12 | P. fluorescens | 5 | −4.13 | 3.1 | 86.5 | - | |
| (R)-5d | 13 | TLL | 1 | −5.02 | 3.1 | 92.9 | O:NHLeu147 |
| 14 | P. fluorescens | 1 | −3.41 | 4.0 | 85.7 | - | |
| (S)-5d | 15 | TLL | 0 | - | - | - | - |
| 16 | P. fluorescens | 0 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, A.C.L.d.M.; de Oliveira, B.R.; Lima, G.V.; Negreiro, J.M.; Oliveira, M.C.F.; de Lemos, T.L.G.; da Silva, M.R.; Fonseca, T.d.S.; Bezerra, R.M.; dos Santos, J.C.S.; et al. Resolution of Racemic Aryloxy-Propan-2-yl Acetates via Lipase-Catalyzed Hydrolysis: Preparation of Enantiomerically Pure/Enantioenriched Mexiletine Intermediates and Analogs. Catalysts 2022, 12, 1566. https://doi.org/10.3390/catal12121566
Carvalho ACLdM, de Oliveira BR, Lima GV, Negreiro JM, Oliveira MCF, de Lemos TLG, da Silva MR, Fonseca TdS, Bezerra RM, dos Santos JCS, et al. Resolution of Racemic Aryloxy-Propan-2-yl Acetates via Lipase-Catalyzed Hydrolysis: Preparation of Enantiomerically Pure/Enantioenriched Mexiletine Intermediates and Analogs. Catalysts. 2022; 12(12):1566. https://doi.org/10.3390/catal12121566
Chicago/Turabian StyleCarvalho, Ana Caroline Lustosa de Melo, Bruna Rocha de Oliveira, Gledson Vieira Lima, Jonatas Martins Negreiro, Maria Conceição Ferreira Oliveira, Telma Leda Gomes de Lemos, Marcos Reinaldo da Silva, Thiago de Sousa Fonseca, Rayanne Mendes Bezerra, Jose Cleiton Sousa dos Santos, and et al. 2022. "Resolution of Racemic Aryloxy-Propan-2-yl Acetates via Lipase-Catalyzed Hydrolysis: Preparation of Enantiomerically Pure/Enantioenriched Mexiletine Intermediates and Analogs" Catalysts 12, no. 12: 1566. https://doi.org/10.3390/catal12121566
APA StyleCarvalho, A. C. L. d. M., de Oliveira, B. R., Lima, G. V., Negreiro, J. M., Oliveira, M. C. F., de Lemos, T. L. G., da Silva, M. R., Fonseca, T. d. S., Bezerra, R. M., dos Santos, J. C. S., Gonçalves, L. R. B., Rios, N. S., Zanatta, G., & de Mattos, M. C. (2022). Resolution of Racemic Aryloxy-Propan-2-yl Acetates via Lipase-Catalyzed Hydrolysis: Preparation of Enantiomerically Pure/Enantioenriched Mexiletine Intermediates and Analogs. Catalysts, 12(12), 1566. https://doi.org/10.3390/catal12121566











