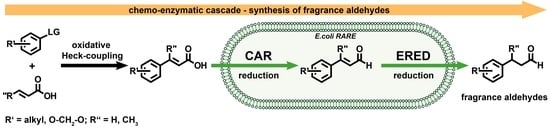
Chemo-Enzymatic Cascade for the Generation of Fragrance Aldehydes
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gröger, H.; Hummel, W. Combining the “two worlds” of chemocatalysis and biocatalysis towards multi-step one-pot processes in aqueous media. Curr. Opin. Chem. Biol. 2014, 19, 171–179. [Google Scholar] [CrossRef]
- Rudroff, F.; Mihovilovic, M.D.; Gröger, H.; Snajdrova, R.; Iding, H.; Bornscheuer, U.T. Opportunities and challenges for combining chemo- and biocatalysis. Nat. Catal. 2018, 1, 12–22. [Google Scholar] [CrossRef]
- Larsson, A.L.E.; Persson, B.A.; Bäckvall, J.-E. Enzymatic Resolution of Alcohols Coupled with Ruthenium-Catalyzed Racemization of the Substrate Alcohol. Angew. Chemie Int. Ed. Engl. 1997, 36, 1211–1212. [Google Scholar] [CrossRef]
- Modern Carbonyl Olefination; Takeda, T. (Ed.) Wiley: Hoboken, NJ, USA, 2003; ISBN 9783527306343. [Google Scholar]
- Rabbani, G. A Concise Introduction of Perkin Reaction. Org. Chem. Curr. Res. 2018, 7, 2. [Google Scholar] [CrossRef]
- Heck, R.F.; Nolley, J.P. Palladium-catalyzed vinylic hydrogen substitution reactions with aryl, benzyl, and styryl halides. J. Org. Chem. 1972, 37, 2320–2322. [Google Scholar] [CrossRef]
- Christoffel, F.; Ward, T.R. Palladium-Catalyzed Heck Cross-Coupling Reactions in Water: A Comprehensive Review. Catal. Lett. 2018, 148, 489–511. [Google Scholar] [CrossRef]
- Ourailidou, M.E.; Dockerty, P.; Witte, M.; Poelarends, G.J.; Dekker, F.J. Metabolic alkene labeling and in vitro detection of histone acylation via the aqueous oxidative Heck reaction. Org. Biomol. Chem. 2015, 13, 3648–3653. [Google Scholar] [CrossRef] [Green Version]
- Ourailidou, M.E.; Van Der Meer, J.Y.; Baas, B.J.; Jeronimus-Stratingh, M.; Gottumukkala, A.L.; Poelarends, G.J.; Minnaard, A.J.; Dekker, F.J. Aqueous oxidative heck reaction as a Protein-labeling strategy. ChemBioChem 2014, 15, 209–212. [Google Scholar] [CrossRef] [Green Version]
- Ohtaka, A.; Okagaki, T.; Hamasaka, G.; Uozumi, Y.; Shinagawa, T.; Shimomura, O.; Nomura, R. Application of “Boomerang” Linear Polystyrene-Stabilized Pd Nanoparticles to a Series of C-C Coupling Reactions in Water. Catalysts 2015, 5, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Askevold, B.; Mikula, H.; Kohler, R.H.; Pirovich, D.; Weissleder, R. Nano-palladium is a cellular catalyst for in vivo chemistry. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Gaikwad, D.S.; Undale, K.A.; Patil, D.B.; Pore, D.M.; Kamble, A.A. Triton X-100 stabilized Pd nanoparticles and their catalytic application in one-pot sequential Heck and Hiyama coupling in water. Res. Chem. Intermed. 2018, 44, 265–275. [Google Scholar] [CrossRef]
- He, A.; Li, T.; Daniels, L.; Fotheringham, I.; Rosazza, J.P.N. Nocardia sp. carboxylic acid reductase: Cloning, expression, and characterization of a new aldehyde oxidoreductase family. Appl. Environ. Microbiol. 2004, 70, 1874–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, A.; Stürmer, R.; Hauer, B.; Rosche, B. Stereospecific Alkyne Reduction: Novel Activity of Old Yellow Enzymes. Angew. Chemie Int. Ed. 2007, 46, 3316–3318. [Google Scholar] [CrossRef] [PubMed]
- Padhi, S.K.; Bougioukou, D.J.; Stewart, J.D. Site-saturation mutagenesis of tryptophan 116 of Saccharomyces pastorianus old yellow enzyme uncovers stereocomplementary variants. J. Am. Chem. Soc. 2009, 131, 3271–3280. [Google Scholar] [CrossRef] [PubMed]
- Kunjapur, A.M.; Tarasova, Y.; Prather, K.L.J. Synthesis and Accumulation of Aromatic Aldehydes in an Engineered Strain of Escherichia coli. J. Am. Chem. Soc. 2014, 136, 11644–11654. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.M.; Atsumi, S. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab. Eng. 2014, 25, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Bühler, B.; Bollhalder, I.; Hauer, B.; Witholt, B.; Schmid, A. Use of the two-liquid phase concept to exploit kinetically controlled multistep biocatalysis. Biotechnol. Bioeng. 2003, 81, 683–694. [Google Scholar] [CrossRef]
- Jakoblinnert, A.; Rother, D. A two-step biocatalytic cascade in micro-aqueous medium: Using whole cells to obtain high concentrations of a vicinal diol. Green Chem. 2014, 16, 3472–3482. [Google Scholar] [CrossRef] [Green Version]
- Horvat, M.; Winkler, M. In Vivo Reduction of Medium- to Long-Chain Fatty Acids by Carboxylic Acid Reductase (CAR) Enzymes: Limitations and Solutions. ChemCatChem 2020, 12, 5076–5090. [Google Scholar] [CrossRef]
- Ressmann, A.K.; Schwendenwein, D.; Leonhartsberger, S.; Mihovilovic, M.D.; Bornscheuer, U.T.; Winkler, M.; Rudroff, F. Substrate-Independent High-Throughput Assay for the Quantification of Aldehydes. Adv. Synth. Catal. 2019, 361, 2538–2543. [Google Scholar] [CrossRef] [Green Version]
- Schwendenwein, D.; Ressmann, A.K.; Doerr, M.; Höhne, M.; Bornscheuer, U.T.; Mihovilovic, M.D.; Rudroff, F.; Winkler, M. Random Mutagenesis-Driven Improvement of Carboxylate Reductase Activity using an Amino Benzamidoxime-Mediated High-Throughput Assay. Adv. Synth. Catal. 2019, 361, 2544–2549. [Google Scholar] [CrossRef] [Green Version]
- Schwendenwein, D.; Fiume, G.; Weber, H.; Rudroff, F.; Winkler, M. Selective Enzymatic Transformation to Aldehydes in vivo by Fungal Carboxylate Reductase from Neurospora crassa. Adv. Synth. Catal. 2016, 358, 3414–3421. [Google Scholar] [CrossRef]
- Sheldon, R.A. The greening of solvents: Towards sustainable organic synthesis. Curr. Opin. Green Sustain. Chem. 2019, 18, 13–19. [Google Scholar] [CrossRef]
- Henderson, R.K.; Jiménez-González, C.; Constable, D.J.C.; Alston, S.R.; Inglis, G.G.A.; Fisher, G.; Sherwood, J.; Binks, S.P.; Curzons, A.D. Expanding GSK’s solvent selection guide—Embedding sustainability into solvent selection starting at medicinal chemistry. Green Chem. 2011, 13, 854–862. [Google Scholar] [CrossRef]
- Byrne, F.P.; Jin, S.; Paggiola, G.; Petchey, T.H.M.; Clark, J.H.; Farmer, T.J.; Hunt, A.J.; Robert McElroy, C.; Sherwood, J. Tools and techniques for solvent selection: Green solvent selection guides. Sustain. Chem. Process. 2016, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Maugeri, Z.; Rother, D. Application of Imine Reductases (IREDs) in Micro-Aqueous Reaction Systems. Adv. Synth. Catal. 2016, 358, 2745–2750. [Google Scholar] [CrossRef]
- Luquin, E.; Pérez-Lorenzo, E.; Aymerich, M.S.; Mengual, E. Two-color Fluorescence Labeling in Acrolein-fixed Brain Tissue. J. Histochem. Cytochem. 2010, 58, 359–368. [Google Scholar] [CrossRef] [Green Version]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Studier, F.W. Stable Expression Clones and Auto-Induction for Protein Production in E. coli. In Structural Genomics; Humana Press: Totowa, NJ, USA, 2014; Volume 1091, pp. 14–32. ISBN 978-1-62703-690-0. [Google Scholar]
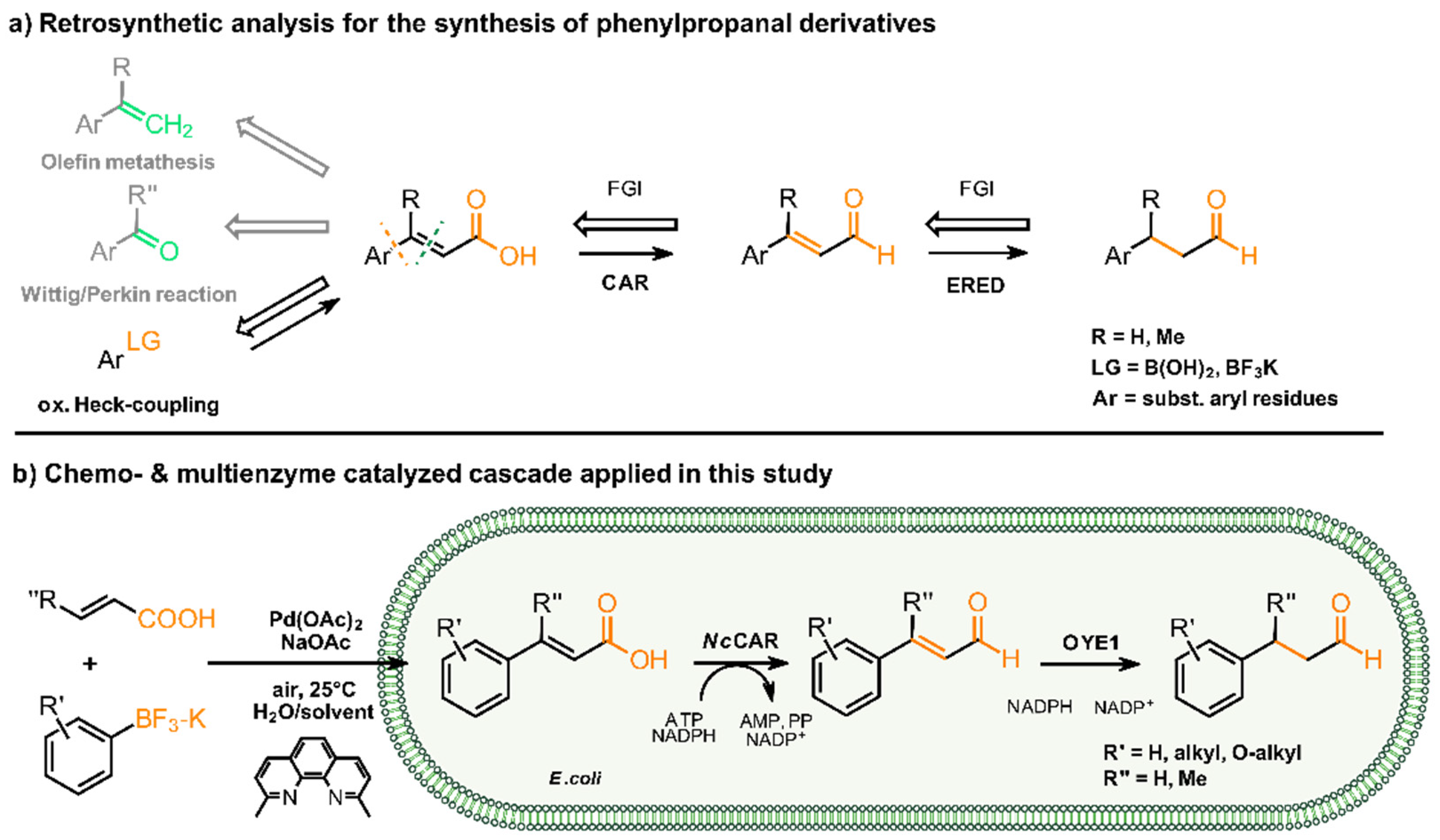
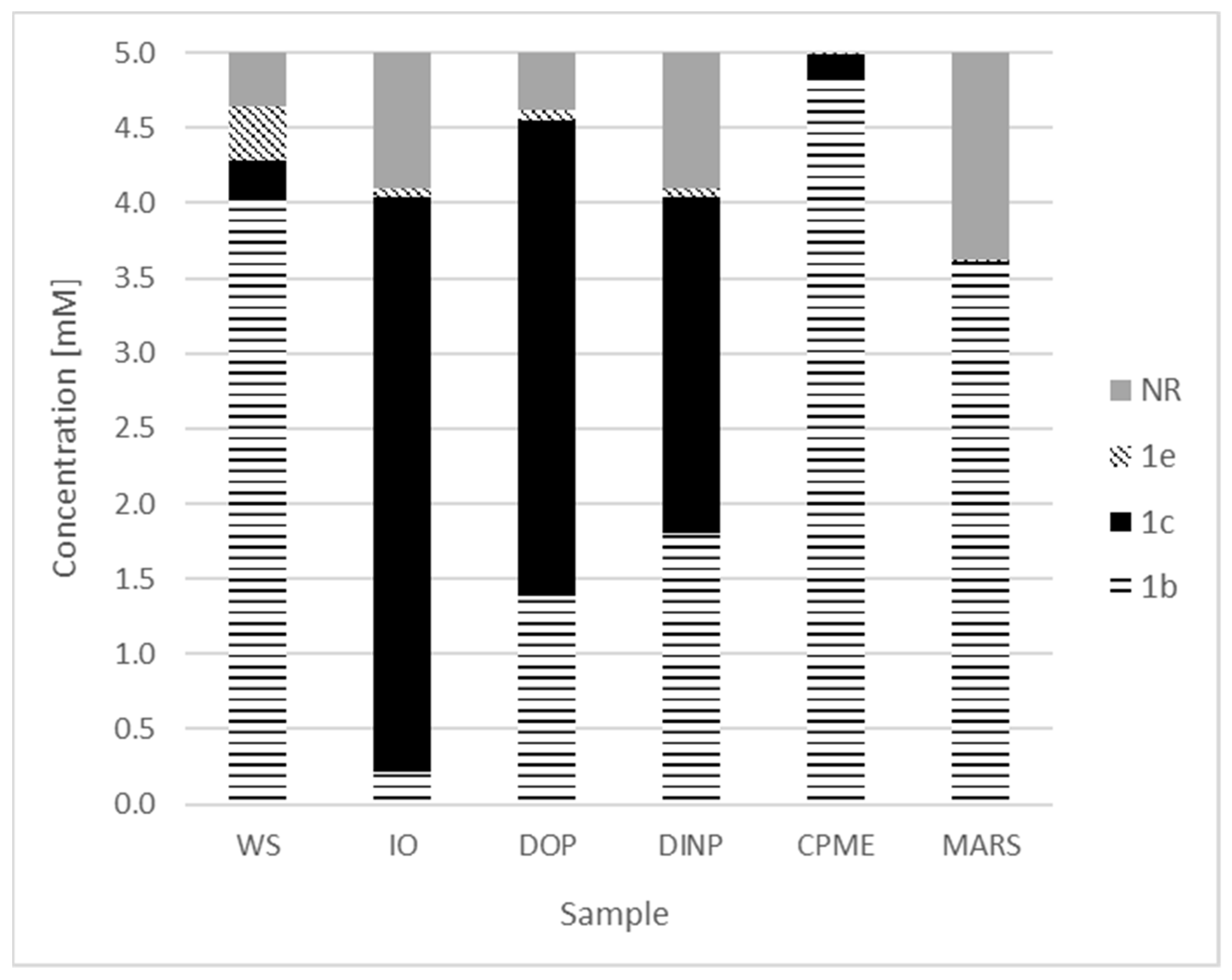
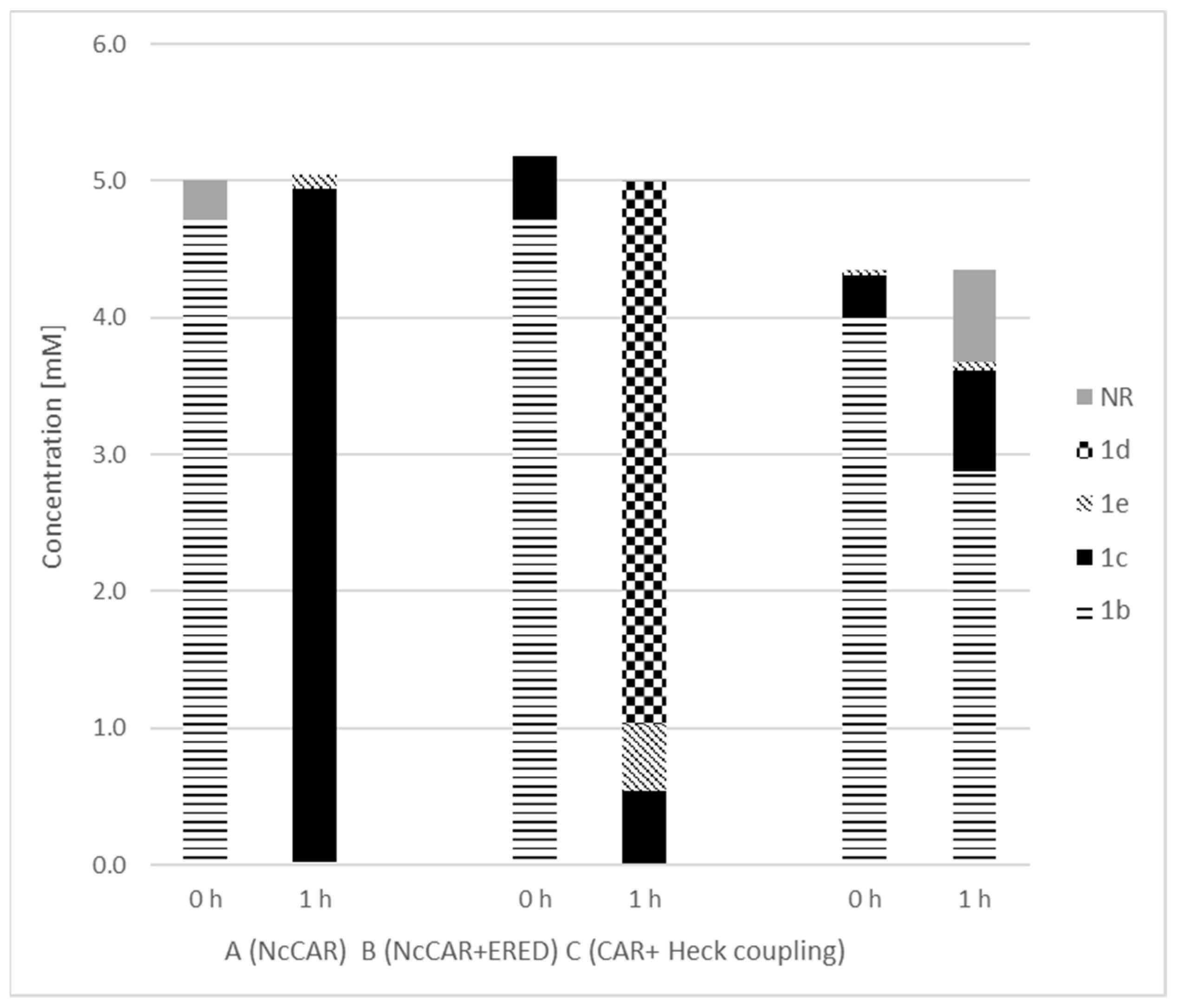
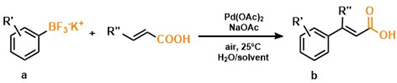
| Entry | Product | Yield [%] |
|---|---|---|
| 1 | 1b R″ = H, R′ = H | 76.0 ± 1.0 |
| 2 | 2b R″ = CH3, R′ = H | 61.0 ± 4.0 |
| 3 | 3b R″ = H, R′ = 3,4-O-CH2-O | 73.0 ± 1.0 |
| 4 | 4b R″ = CH3, R′ = 3,4-O-CH2-O | 49.0 ± 2.0 |
| 5 | 5b R″ = H, R′= 4-tBu | 37.0 ± 3.0 |
| 6 | 6b R″ = CH3, R′ = 4-tBu | 8.0 ± 0.4 |
| 7 | 7b R″ = H, R′ = 4-isoBu | 30.0 ± 1.0 |
| 8 | 8b R″ = CH3, R′ = 4-isoBu | 11.0 ± 1.0 |
| Entry | Substrate | Activity [U/mg] |
|---|---|---|
| 1 | 1b Cinnamic acid | 0.92 ± 0.07 |
| 2 | 2b 3-Methyl cinnamic acid | 0.31 ± 0.03 |
| 3 | Acrylic acid | 0.35 ± 0.03 |
| 4 | Crotonic acid | 0.47 ± 0.01 |
| Entry | Cinnamic Acid [mM] | Acrylic Acid [mM] | Pd(II)Acetate/Necocuproine | Conversion [%] |
|---|---|---|---|---|
| 1 | 5 | 0 | 0 | 99 |
| 2 | 5 | 5 | 0 | 46 |
| 3 | 5 | 10 | 0 | 4 |
| 4 | 5 | 0 | 6 mol% | 93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwendenwein, D.; Ressmann, A.K.; Entner, M.; Savic, V.; Winkler, M.; Rudroff, F. Chemo-Enzymatic Cascade for the Generation of Fragrance Aldehydes. Catalysts 2021, 11, 932. https://doi.org/10.3390/catal11080932
Schwendenwein D, Ressmann AK, Entner M, Savic V, Winkler M, Rudroff F. Chemo-Enzymatic Cascade for the Generation of Fragrance Aldehydes. Catalysts. 2021; 11(8):932. https://doi.org/10.3390/catal11080932
Chicago/Turabian StyleSchwendenwein, Daniel, Anna K. Ressmann, Marcello Entner, Viktor Savic, Margit Winkler, and Florian Rudroff. 2021. "Chemo-Enzymatic Cascade for the Generation of Fragrance Aldehydes" Catalysts 11, no. 8: 932. https://doi.org/10.3390/catal11080932
APA StyleSchwendenwein, D., Ressmann, A. K., Entner, M., Savic, V., Winkler, M., & Rudroff, F. (2021). Chemo-Enzymatic Cascade for the Generation of Fragrance Aldehydes. Catalysts, 11(8), 932. https://doi.org/10.3390/catal11080932