Sesquiterpene Cyclases from the Basidiomycete Cerrena unicolor
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Genes
2.2. Heterologous Production and Functional Characterization
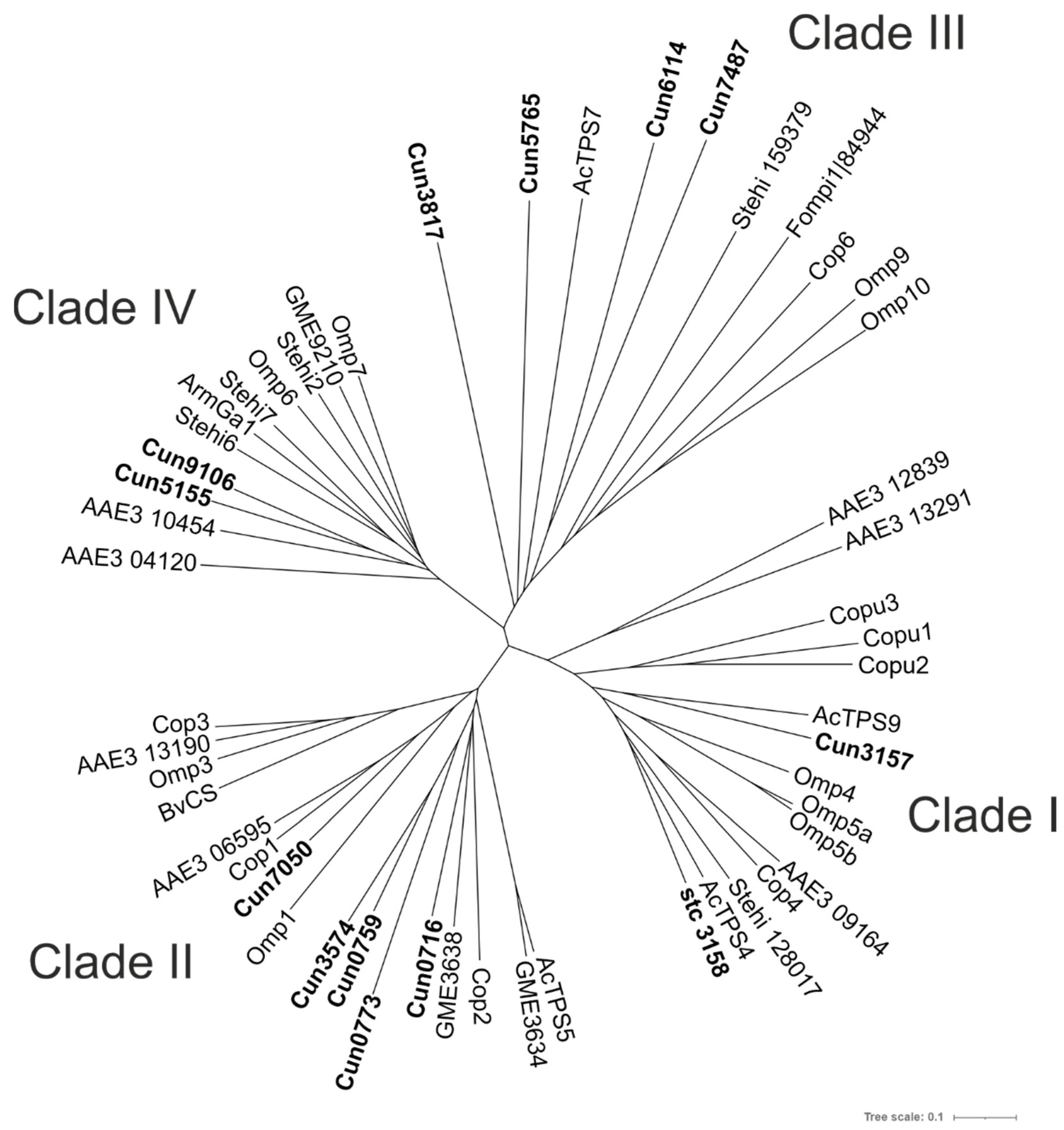
2.3. Phylogenetic Determination
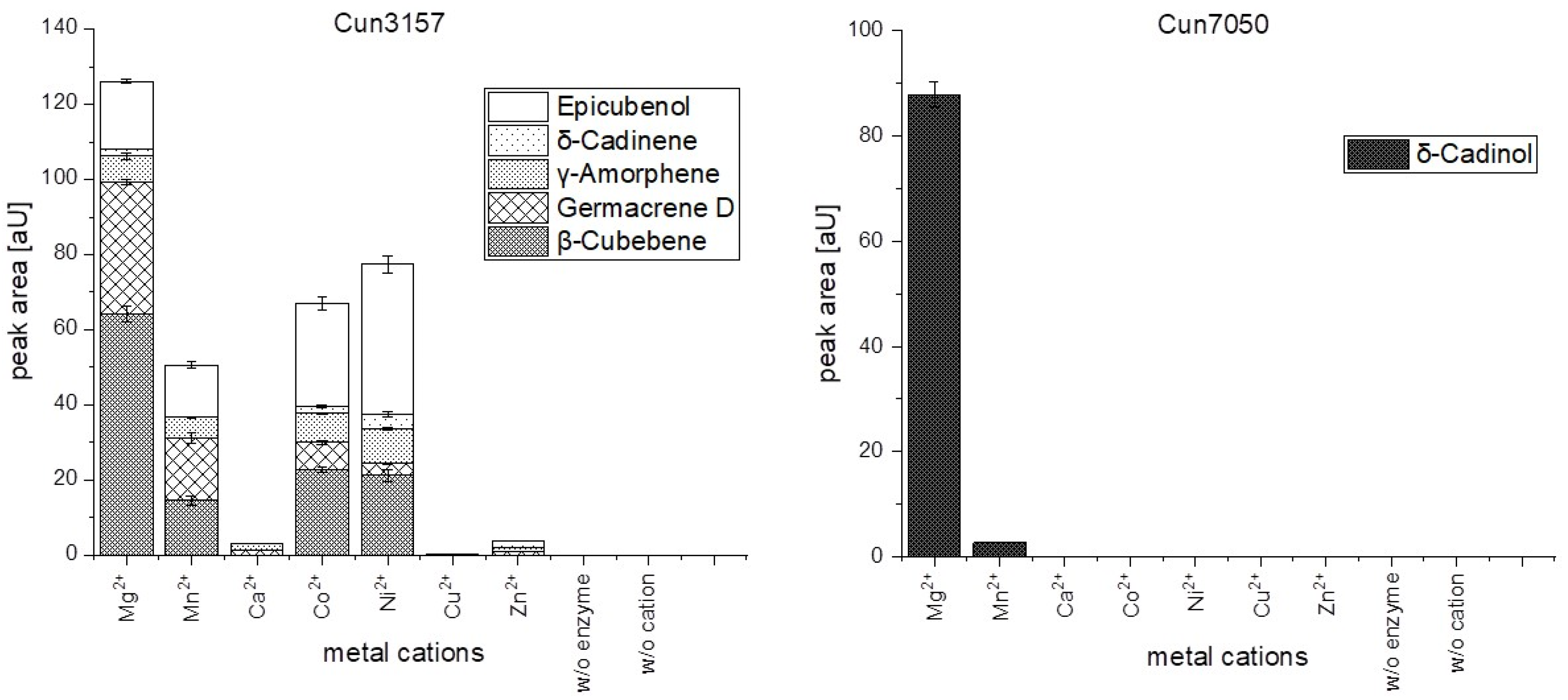
2.4. Temperature Optimum and Metal Ion Dependence of Cun3157 and Cun7050
3. Materials and Methods
3.1. Chemicals, Reagents, and Strains
3.2. Gene Identification
3.3. Cloning of pCOLD I_STS Constructs
3.4. Cultivation and Expression
3.5. Enzyme Purification
3.6. Biocatalysis
3.7. Product Analysis
3.8. Phylogenetic Analysis
3.9. Semi-Native PAGE
3.10. Peptide Mass Fingerprinting
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhong, J.-J.; Xiao, J.-H. Secondary Metabolites from Higher Fungi: Discovery, Bioactivity, and Bioproduction. Adv. Biochem. Eng. Biotechnol. 2009, 113, 79–150. [Google Scholar] [CrossRef]
- Pateraki, I.; Heskes, A.M.; Hamberger, B. Cytochromes P450 for Terpene Functionalisation and Metabolic Engineering. Adv. Biochem. Eng. Biotechnol. 2015, 148, 107–139. [Google Scholar] [CrossRef]
- Faylo, J.L.; Ronnebaum, T.A.; Christianson, D.W. Assembly-Line Catalysis in Bifunctional Terpene Synthases. Acc. Chem. Res. 2021, 54, 3780–3791. [Google Scholar] [CrossRef]
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef] [Green Version]
- Alves, M.J.; Ferreira, I.; Dias, J.; Teixeira, V.; Martins, A.; Pintado, M.M. A Review on Antimicrobial Activity of Mushroom (Basidiomycetes) Extracts and Isolated Compounds. Planta Medica 2012, 78, 1707–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehbub, M.; Lei, J.; Franco, C.; Zhang, W. Marine Sponge Derived Natural Products between 2001 and 2010: Trends and Opportunities for Discovery of Bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [Green Version]
- Fidyt, K.; Fiedorowicz, A.; Strzadala, L.; Szumny, A. β-caryophyllene and β-caryophyllene oxide-natural compounds of anticancer and analgesic properties. Cancer Med. 2016, 5, 3007–3017. [Google Scholar] [CrossRef]
- Leonhardt, R.-H.; Berger, R.G. Nootkatone. Signal. Pathw. Transl. 2014, 148, 391–404. [Google Scholar]
- Tan, L.T.H.; Lee, L.H.; Yin, W.F.; Chan, C.K.; Kadir, H.A.; Chan, K.G.; Goh, B.H. Traditional Uses, Phytochemistry, and Bioactivities ofCananga odorata(Ylang-Ylang). Evid.-Based Complement. Altern. Med. 2015, 2015, 1–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moniodis, J.; Jones, C.G.; Renton, M.; Plummer, J.A.; Barbour, E.L.; Ghisalberti, E.L.; Bohlmann, J. Sesquiterpene Variation in West Australian Sandalwood (Santalum spicatum). Molecules 2017, 22, 940. [Google Scholar] [CrossRef] [Green Version]
- Chemat, F.; Vian, M.A.; Cravotto, G. Green Extraction of Natural Products: Concept and Principles. Int. J. Mol. Sci. 2012, 13, 8615–8627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-L.; Tian, T.; Alonso-Gutierrez, J.; Garabedian, B.; Wang, S.; Baidoo, E.E.K.; Benites, V.; Chen, Y.; Petzold, C.J.; Adams, P.D.; et al. Renewable production of high density jet fuel precursor sesquiterpenes from Escherichia coli. Biotechnol. Biofuels 2018, 11, 285. [Google Scholar] [CrossRef]
- Bian, G.; Deng, Z.; Liu, T. Strategies for terpenoid overproduction and new terpenoid discovery. Curr. Opin. Biotechnol. 2017, 48, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Quin, M.; Flynn, C.M.; Schmidt-Dannert, C. Traversing the fungal terpenome. Nat. Prod. Rep. 2014, 31, 1449–1473. [Google Scholar] [CrossRef]
- Schmidt-Dannert, C. Biosynthesis of Terpenoid Natural Products in Fungi. Signal. Pathw. Transl. 2014, 148, 19–61. [Google Scholar] [CrossRef]
- Kachlishvili, E.; Metreveli, E.; Elisashvili, V. Modulation of Cerrena unicolor laccase and manganese peroxidase production. SpringerPlus 2014, 3, 463. [Google Scholar] [CrossRef] [Green Version]
- Michniewicz, A.; Ullrich, R.; Ledakowicz, S.; Hofrichter, M. The white-rot fungus Cerrena unicolor strain 137 produces two laccase isoforms with different physico-chemical and catalytic properties. Appl. Microbiol. Biotechnol. 2006, 69, 682–688. [Google Scholar] [CrossRef]
- Strauß, E. Effektoren der Terpenbiosynthese bei Basidiomycota in Submerskultur. Bachelor Thesis, Leibniz-University Hannover, Hanover, Germany, 2018. [Google Scholar]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, X.; Orban, A.; Shukal, S.; Birk, F.; Too, H.-P.; Rühl, M. Agrocybe aegerita Serves As a Gateway for Identifying Sesquiterpene Biosynthetic Enzymes in Higher Fungi. ACS Chem. Biol. 2020, 15, 1268–1277. [Google Scholar] [CrossRef]
- Christianson, D.W. Structural Biology and Chemistry of the Terpenoid Cyclases. Chem. Rev. 2006, 106, 3412–3442. [Google Scholar] [CrossRef]
- Rabe, P.; Schmitz, T.; Dickschat, J.S. Mechanistic investigations on six bacterial terpene cyclases. Beilstein J. Org. Chem. 2016, 12, 1839–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickschat, J.S. Bacterial terpene cyclases. Nat. Prod. Rep. 2016, 33, 87–110. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cai, Y.-S.; Yuan, Y.; Bian, G.; Ye, Z.; Deng, Z.; Liu, T. Genome mining in Trichoderma viride J1-030: Discovery and identification of novel sesquiterpene synthase and its products. Beilstein J. Org. Chem. 2019, 15, 2052–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawrzyn, G.T.; Quin, M.; Choudhary, S.; Gallego, F.L.; Schmidt-Dannert, C. Draft Genome of Omphalotus olearius Provides a Predictive Framework for Sesquiterpenoid Natural Product Biosynthesis in Basidiomycota. Chem. Biol. 2012, 19, 772–783. [Google Scholar] [CrossRef] [Green Version]
- Mischko, W.; Hirte, M.; Fuchs, M.; Mehlmer, N.; Brück, T.B. Identification of sesquiterpene synthases from the Basidiomycota Coniophora puteana for the efficient and highly selective β-copaene and cubebol production in E. coli. Microb. Cell Factories 2018, 17, 164. [Google Scholar] [CrossRef]
- Quin, M.B.; Flynn, C.M.; Wawrzyn, G.T.; Choudhary, S.; Schmidt-Dannert, C. Mushroom Hunting by Using Bioinformatics: Application of a Predictive Framework Facilitates the Selective Identification of Sesquiterpene Synthases in Basidiomycota. ChemBioChem 2013, 14, 2480–2491. [Google Scholar] [CrossRef] [Green Version]
- Misiek, M.; Hoffmeister, D. Processing sites involved in intron splicing of Armillaria natural product genes. Mycol. Res. 2008, 112, 216–224. [Google Scholar] [CrossRef]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [Green Version]
- Agger, S.; Lopez-Gallego, F.; Schmidt-Dannert, C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 2009, 72, 1181–1195. [Google Scholar] [CrossRef] [Green Version]
- Bayram, O.; Braus, G.H. Coordination of secondarymetabolism and development in fungi: The velvet familyof regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merhej, J.; Richard-Forget, F.; Barreau, C. The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet. Biol. 2011, 48, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-L.; Ma, L.-T.; Lee, Y.-R.; Shaw, J.-F.; Wang, S.-Y.; Chu, F.-H. Differential Gene Expression Network in Terpenoid Synthesis of Antrodia cinnamomea in Mycelia and Fruiting Bodies. J. Agric. Food Chem. 2017, 65, 1874–1886. [Google Scholar] [CrossRef] [PubMed]
- Engels, B.; Heinig, U.; Grothe, T.; Stadler, M.; Jennewein, S. Cloning and Characterization of an Armillaria gallica cDNA Encoding Protoilludene Synthase, Which Catalyzes the First Committed Step in the Synthesis of Antimicrobial Melleolides. J. Biol. Chem. 2011, 286, 6871–6878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Yang, Y.-L.; Zeng, J.; Zhang, L.; Ding, Z.-H.; Zeng, Y. Identification and Characterization of a δ-Cadinol Synthase Potentially Involved in the Formation of Boreovibrins in Boreostereum vibrans of Basidiomycota. Nat. Prod. Bioprospect. 2016, 6, 167–171. [Google Scholar] [CrossRef] [Green Version]
- Yap, H.-Y.Y.; Muria-Gonzalez, M.J.; Kong, B.-H.; Stubbs, K.A.; Tan, C.-S.; Ng, S.-T.; Tan, N.-H.; Solomon, P.S.; Fung, S.-Y.; Chooi, Y.-H. Heterologous expression of cytotoxic sesquiterpenoids from the medicinal mushroom Lignosus rhinocerotis in yeast. Microb. Cell Factories 2017, 16, 103. [Google Scholar] [CrossRef] [Green Version]
- Ronnebaum, T.A.; Gardner, S.M.; Christianson, D.W. An Aromatic Cluster in the Active Site of epi-Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways. Biochemistry 2020, 59, 4744–4754. [Google Scholar] [CrossRef] [PubMed]
- Vattekkatte, A.; Garms, S.; Brandt, W.; Boland, W. Enhanced structural diversity in terpenoid biosynthesis: Enzymes, substrates and cofactors. Org. Biomol. Chem. 2018, 16, 348–362. [Google Scholar] [CrossRef] [Green Version]
- Croteau, R. Biosynthesis and catabolism of monoterpenoids. Chem. Rev. 1987, 87, 929–954. [Google Scholar] [CrossRef]
- Frick, S.; Nagel, R.; Schmidt, A.; Bodemann, R.R.; Rahfeld, P.; Pauls, G.; Brandt, W.; Gershenzon, J.; Boland, W.; Burse, A. Metal ions control product specificity of isoprenyl diphosphate synthases in the insect terpenoid pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 4194–4199. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| STS | Molecular Mass (kDa) 1 | Sequence Coverage (%) | Score | Matches |
|---|---|---|---|---|
| Cun3157 | 40.4 | 70 | 40,853 | 603 |
| Cun7050 | 38.8 | 67 | 15,743 | 256 |
| Cun3574 | 38.8 | 86 | 30,151 | 568 |
| Cun3158 | 38.2 | 58 | 12,608 | 229 |
| Cun0759 | 43.0 | 69 | 18,445 | 339 |
| Cun9106 | 45.7 | 62 | 8345 | 188 |
| Cun5155 | 38.8 | 72 | 10,039 | 213 |
| Cun3817 | 46.4 | 45 | 5341 | 102 |
| Cun0773 | 38.2 | 46 | 4622 | 100 |
| Cun0716 | 38.3 | 65 | 12,289 | 288 |
| STS | Cyclic Terpene Products | RI Polar (Lit.) | RI Non-Polar (Lit.) | Product Ratio (%) |
|---|---|---|---|---|
| Cun3157 | β-Cubebene A | 1547 (1540) | 1385 (1388) | 47.4 |
| Germacrene D A | 1720 (1719) | 1478 (1476) | 23.8 | |
| γ-Amorphene A | 1488 (1491) | 10.4 | ||
| δ-Cadinene A | 1764 (1762) | 1515 (1521) | 4.3 | |
| Epicubenol A | 2084 (2072) | 1625 (1625) | 14.1 | |
| Cun7050 | δ-Cadinol A | 2212 (2205) | 1644 (1647) | 100 |
| Cun3574 | α-Copaene A | 1495 (1502) | 1372 (1375) | 100 |
| Cun3158 | β-Cubebene A | 1546 (1540) | 1390 (1388) | 13.0 |
| Germacrene D A | 1720 (1719) | 1477 (1476) | 14.4 | |
| γ-Amorphene A | 1488 (1491) | 11.0 | ||
| δ-Cadinene A | 1765 (1762) | 1515 (1521) | 61.6 | |
| Cun5765 | none | |||
| Cun0759 | α-Muurolene A | 1729 (1711) | 1495 (1500) | 100 |
| Cun9106 | unknown | 1512 | 1372 | 100 |
| Cun5155 | Aromadendrene B | 1632 (1608) | 1432 (1442) | 100 |
| Cun6114 | none | |||
| Cun3817 | γ-Cadinene A | 1767 (1745) | 1511 (1513) | 100 |
| Cun0773 | Germacrene D A | 1718 (1707) | 1478 (1476) | 100 |
| Cun0716 | α-Muurolene A | 1730 (1727) | 1497 (1500) | 7.9 |
| δ-Cadinol A | 2209 (2205) | 1646 (1647) | 92.1 | |
| Cun7487 | none | |||
| Cun0802 | none |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Püth, N.; Ersoy, F.; Krings, U.; Berger, R.G. Sesquiterpene Cyclases from the Basidiomycete Cerrena unicolor. Catalysts 2021, 11, 1361. https://doi.org/10.3390/catal11111361
Püth N, Ersoy F, Krings U, Berger RG. Sesquiterpene Cyclases from the Basidiomycete Cerrena unicolor. Catalysts. 2021; 11(11):1361. https://doi.org/10.3390/catal11111361
Chicago/Turabian StylePüth, Nils, Franziska Ersoy, Ulrich Krings, and Ralf G. Berger. 2021. "Sesquiterpene Cyclases from the Basidiomycete Cerrena unicolor" Catalysts 11, no. 11: 1361. https://doi.org/10.3390/catal11111361
APA StylePüth, N., Ersoy, F., Krings, U., & Berger, R. G. (2021). Sesquiterpene Cyclases from the Basidiomycete Cerrena unicolor. Catalysts, 11(11), 1361. https://doi.org/10.3390/catal11111361







