Enhancement of S-Adenosylmethionine-Dependent Methylation by Integrating Methanol Metabolism with 5-Methyl-Tetrahydrofolate Formation in Escherichia coli
Abstract
1. Introduction
2. Results
2.1. Deregulation of SAM Biosynthesis
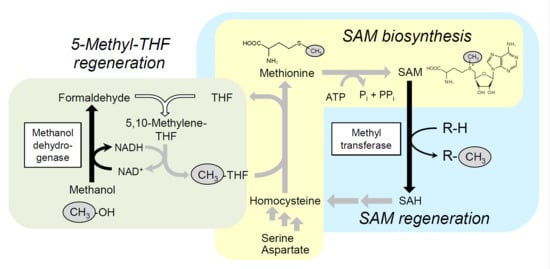
2.2. Integration of Methanol Metabolic Pathway for 5-Methyl-THF Formation into SAM-Dependent Methylation System
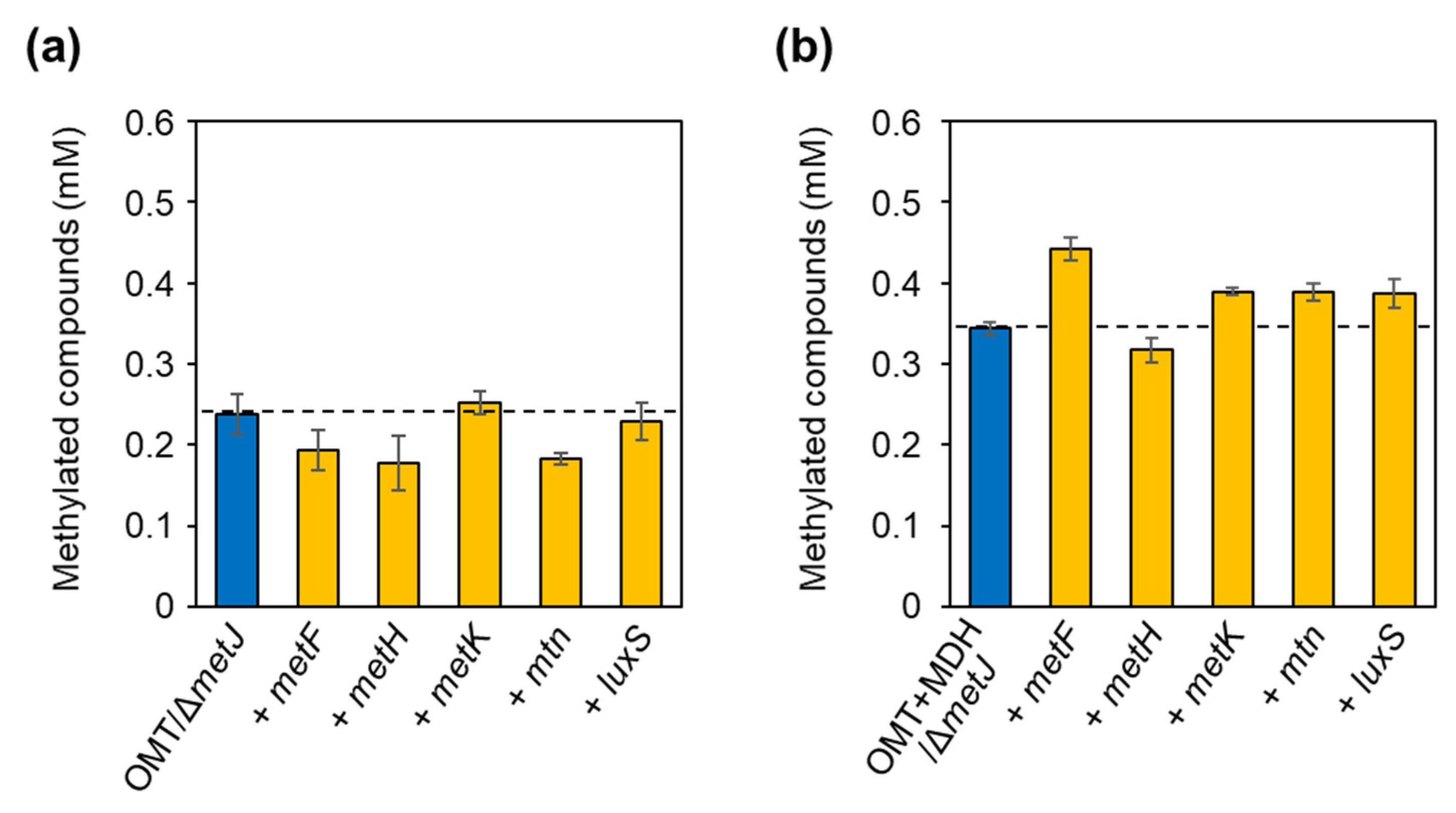
2.3. Alleviation of Bottlenecks in the SAM Regeneration Pathway
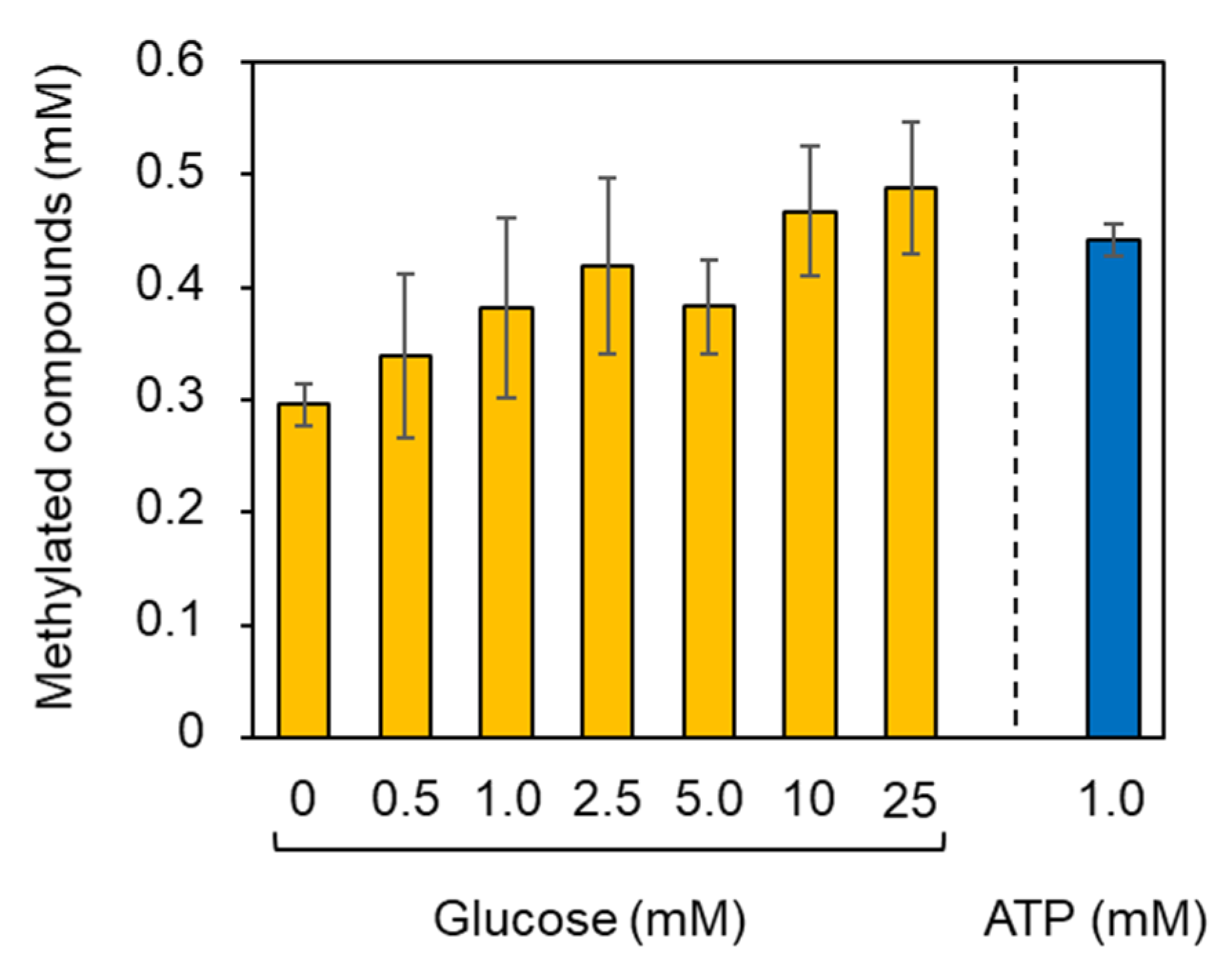
2.4. Methylation Reaction Using Glucose as a Carbon/Energy Source for ATP Regeneration
3. Discussion
4. Materials and Methods
4.1. Strains and Media
4.2. Plasmid Construction
4.3. Genome Editing
4.4. Methylation Reaction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, X.; Saba, T.; Yiu, H.H.P.; Howe, R.F.; Anderson, J.A.; Shi, J. Cofactor NAD(P)H regeneration inspired by heterogeneous pathways. Chem 2017, 2, 621–654. [Google Scholar] [CrossRef]
- Okano, K.; Honda, K.; Taniguchi, H.; Kondo, A. De novo design of biosynthetic pathways for bacterial production of bulk chemicals and biofuels. FEMS Microbiol. Lett. 2018, 365, fny215. [Google Scholar] [CrossRef] [PubMed]
- Uppada, V.; Bhaduri, S.; Noronha, S.B. Cofactor regeneration—An important aspect of biocatalysis. Curr. Sci. 2014, 106, 946–957. [Google Scholar]
- Lin, B.; Tao, Y. Whole-cell biocatalysts by design. Microb. Cell Fact. 2017, 16, 106. [Google Scholar] [CrossRef]
- Andexer, J.N.; Richter, M. Emerging enzymes for ATP regeneration in biocatalytic processes. ChemBioChem 2015, 16, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Struck, A.W.; Thompson, M.L.; Wong, L.S.; Micklefield, J. S-Adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem 2012, 13, 2642–2655. [Google Scholar] [CrossRef]
- Barreiro, E.J.; Kümmerle, A.E.; Fraga, C.A.M. The methylation effect in medicinal chemistry. Chem. Rev. 2011, 111, 5215–5246. [Google Scholar] [CrossRef]
- Schönherr, H.; Cernak, T. Profound methyl effects in drug discovery and a call for new C-H methylation reactions. Angew. Chem. Int. Ed. Engl. 2013, 52, 12256–12267. [Google Scholar] [CrossRef]
- Liao, C.; Seebeck, F.P. S-adenosylhomocysteine as a methyl transfer catalyst in biocatalytic methylation reactions. Nat. Catal. 2019, 2, 696–701. [Google Scholar] [CrossRef]
- Ogawa, H.; Gomi, T.; Fujioka, M. Serine hydroxymethyltransferase and threonine aldolase: Are they identical? Int. J. Biochem. Cell Biol. 2000, 32, 289–301. [Google Scholar] [CrossRef]
- Xu, Y.; Meng, H.; Ren, J.; Zeng, A.P. Formaldehyde formation in the glycine cleavage system and its use for an aldolase-based biosynthesis of 1,3-prodanediol. J. Biol. Eng. 2020, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Marx, C.J.; Van Dien, S.J.; Lidstrom, M.E. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 2005, 3, e16. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Park, Y.; Yi, Y.S.; Lee, Y.; Jo, G.; Park, J.C.; Ahn, J.H.; Lim, Y. Characterization of an O-methyltransferase from Streptomyces avermitilis MA-4680. J. Microbiol. Biotechnol. 2010, 20, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Y.; Chen, C.T.; Liu, J.T.J.; Bogorad, I.W.; Damoiseaux, R.; Liao, J.C. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1. Appl. Microbiol. Biotechnol. 2016, 100, 4969–4983. [Google Scholar] [CrossRef]
- Zhou, W.; Tsai, A.; Dattmore, D.A.; Stives, D.P.; Chitrakar, I.; D’Alessandro, A.M.; Patil, S.; Hicks, K.A.; French, J.B. Crystal structure of E. coli PRPP synthetase. BMC Struct. Biol. 2019, 19, 1. [Google Scholar] [CrossRef]
- Hershey, H.V.; Gutstein, R.; Taylor, M.W. Cloning and restriction map of the E. coli apt gene. Gene 1982, 19, 89–92. [Google Scholar] [CrossRef]
- Brune, M.; Schumann, R.; Wittinghofer, F. Cloning and sequencing of the adenylate kinase gene (adk) of Escherichia coli. Nucleic Acids Res. 1985, 13, 7139–7151. [Google Scholar] [CrossRef]
- Mordhorst, S.; Siegrist, J.; Müller, M.; Richter, M.; Andexer, J.N. Catalytic alkylation using a cyclic S-adenosylmethionine regeneration system. Angew. Chem. Int. Ed. 2017, 56, 4037–4041. [Google Scholar] [CrossRef]
- Kunjapur, A.M.; Hyun, J.C.; Prather, K.L. Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell Fact. 2016, 15, 61. [Google Scholar] [CrossRef]
- Luo, Z.W.; Cho, J.S.; Lee, S.Y. Microbial production of methyl anthranilate, a grape flavor compound. Proc. Natl. Acad. Sci. USA 2019, 116, 10749–10756. [Google Scholar] [CrossRef] [PubMed]
- Usuda, Y.; Kurahashi, O. Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 3228–3234. [Google Scholar] [CrossRef] [PubMed]
- Cress, B.F.; Leitz, Q.D.; Kim, D.C.; Amore, T.D.; Suzuki, J.Y.; Linhardt, R.J.; Koffas, M.A.G. CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Fact. 2017, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Krog, A.; Heggeset, T.M.B.; Müller, J.E.N.; Kupper, C.E.; Schneider, O.; Vorholt, J.A.; Ellingsen, T.E.; Brautaset, T. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS ONE 2013, 8, e59188. [Google Scholar] [CrossRef]
- Keltjens, J.T.; Pol, A.; Reimann, J.; Op den Camp, H.J.M. PQQ-dependent methanol dehydrogenases: Rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 2014, 98, 6163–6183. [Google Scholar] [CrossRef]
- Couderc, R.; Baratti, J. Oxidation of methanol by the yeast, Pichia pastoris. Purification and properties of the alcohol oxidase. Agric. Biol. Chem. 1980, 44, 2279–2289. [Google Scholar] [CrossRef]
- Müller, J.E.N.; Meyer, F.; Litsanov, B.; Kiefer, P.; Potthoff, E.; Heux, S.; Quax, W.J.; Wendisch, V.F.; Brautaset, T.; Portais, J.C.; et al. Engineering Escherichia coli for methanol conversion. Metab. Eng. 2015, 28, 190–201. [Google Scholar] [CrossRef]
- Meyer, F.; Keller, P.; Hartl, J.; Gröninger, O.G.; Kiefer, P.; Vorholt, J.A. Methanol-essential growth of Escherichia coli. Nat. Commun. 2018, 9, 1508. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Interrante, M.F.; Smolke, C.D. Complete biosynthesis of opioids in yeast. Science 2018, 349, 1095–1100. [Google Scholar] [CrossRef]
- Nakagawa, A.; Matsumura, E.; Koyanagi, T.; Katayama, T.; Kawano, N.; Yoshimatsu, K.; Yamamoto, K.; Kumagai, H.; Sato, F.; Minami, H. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 2016, 7, 10390. [Google Scholar] [CrossRef] [PubMed]


| Strain or Plasmid | Relevant Description | Source |
|---|---|---|
| Escherichia coli strain | ||
| TG1 | supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rK– mK–) [F’ traD36 proAB | Zymo Research |
| lacIqZΔM15] | ||
| BL21(DE3) | F– ompT hsdSB (rB– mB–) gal dcm (DE3) | Novagen |
| ΔmetJ | BL21 (DE3) ΔmetJ | This study |
| Plasmid | ||
| pETDuet-1 | Dual T7 promoters, pBR322 ori, bla, lacI | Novagen |
| pETD-OMT | pETDuet-1 derivative for expressing omt5 from S. avermitilis MA-4680 | This study |
| pETD-OMT-MDH | pETDuet-1 derivative for co-expressing omt5 and mdh2 (A26V, A31V, | This study |
| A169V) from C. necator N-1 | ||
| pACYCDuet-1 | Dual T7 promoter, p15A ori, cat, lacI | Novagen |
| pACD-MetF | pACYCDuet-1 derivative for expressing endogenous metF | This study |
| pACD-MetH | pACYCDuet-1 derivative for expressing endogenous metH | This study |
| pACD-MetK | pACYCDuet-1 derivative for expressing endogenous metK | This study |
| pACD-Mtn | pACYCDuet-1 derivative for expressing endogenous mtn | This study |
| pACD-LuxS | pACYCDuet-1 derivative for expressing endogenous luxS | This study |
| pCas | rep101(Ts), kan, Pcas-cas9, ParaB-Red, lacIq, Ptrc-sgRNA-pMB1 | [14] |
| pTargetF | pMB1 ori, aadA | [14] |
| pTarget-metJ sgRNA | pMB1 ori, aadA, metJ-sgRNA | This study |
| pTarget-ΔmetJ | pMB1 ori, aadA, metJ-sgRNA, metJ-donor DNA | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okano, K.; Sato, Y.; Inoue, S.; Kawakami, S.; Kitani, S.; Honda, K. Enhancement of S-Adenosylmethionine-Dependent Methylation by Integrating Methanol Metabolism with 5-Methyl-Tetrahydrofolate Formation in Escherichia coli. Catalysts 2020, 10, 1001. https://doi.org/10.3390/catal10091001
Okano K, Sato Y, Inoue S, Kawakami S, Kitani S, Honda K. Enhancement of S-Adenosylmethionine-Dependent Methylation by Integrating Methanol Metabolism with 5-Methyl-Tetrahydrofolate Formation in Escherichia coli. Catalysts. 2020; 10(9):1001. https://doi.org/10.3390/catal10091001
Chicago/Turabian StyleOkano, Kenji, Yu Sato, Shota Inoue, Shizuka Kawakami, Shigeru Kitani, and Kohsuke Honda. 2020. "Enhancement of S-Adenosylmethionine-Dependent Methylation by Integrating Methanol Metabolism with 5-Methyl-Tetrahydrofolate Formation in Escherichia coli" Catalysts 10, no. 9: 1001. https://doi.org/10.3390/catal10091001
APA StyleOkano, K., Sato, Y., Inoue, S., Kawakami, S., Kitani, S., & Honda, K. (2020). Enhancement of S-Adenosylmethionine-Dependent Methylation by Integrating Methanol Metabolism with 5-Methyl-Tetrahydrofolate Formation in Escherichia coli. Catalysts, 10(9), 1001. https://doi.org/10.3390/catal10091001