An Evolutionary Marker of the Ribokinase Superfamily Is Responsible for Zinc-Mediated Regulation of Human Pyridoxal Kinase
Abstract
1. Introduction
2. Results and Discussion
2.1. Real-Value Evolutionary Trace (rvET) Identifies a Three-Residue Motif that Varies between the Major Ribokinase Superfamily Groups
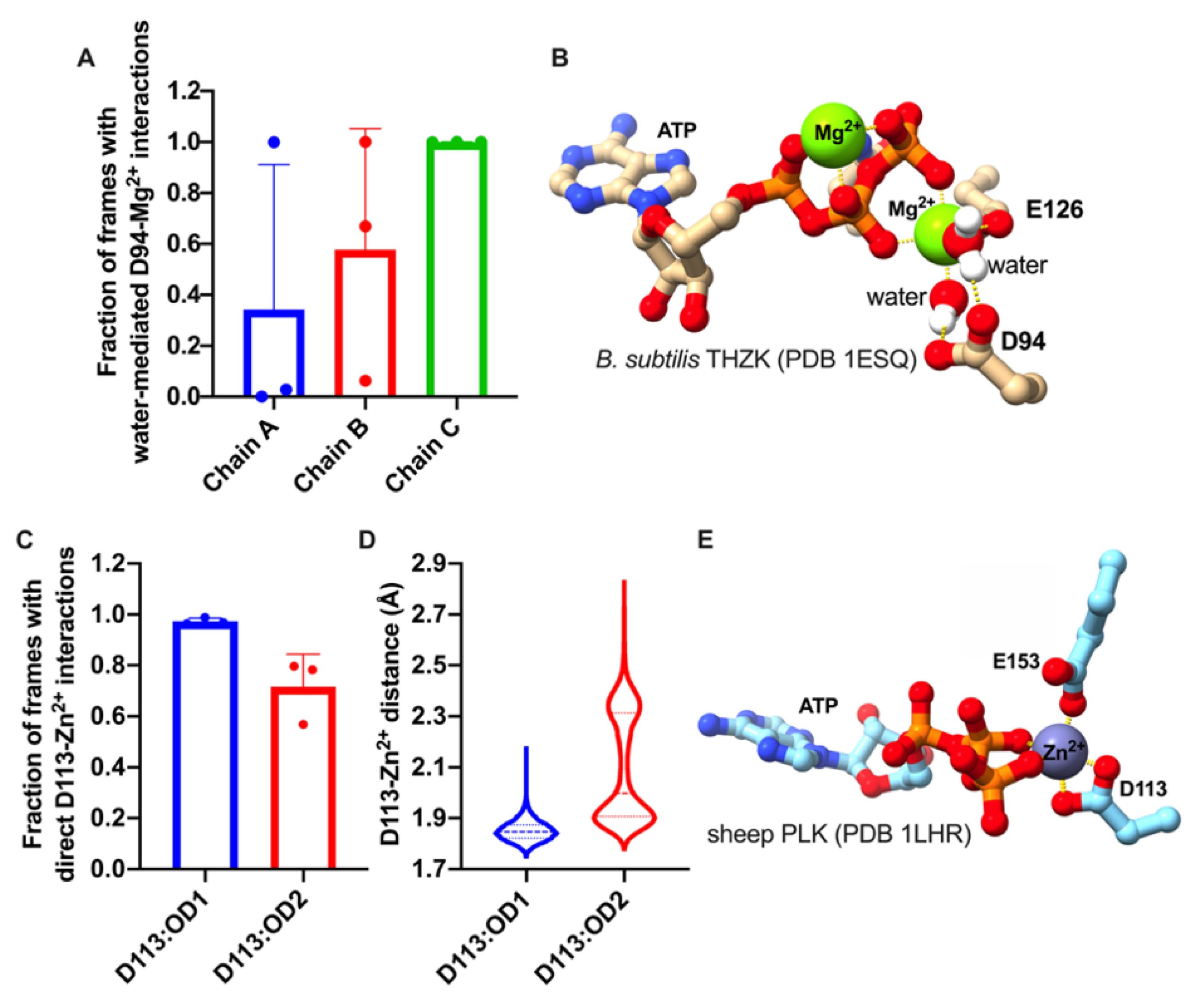
2.2. The DPV Motif Establishes Direct or Water-Mediated Interactions with Divalent Metals in the Active Site
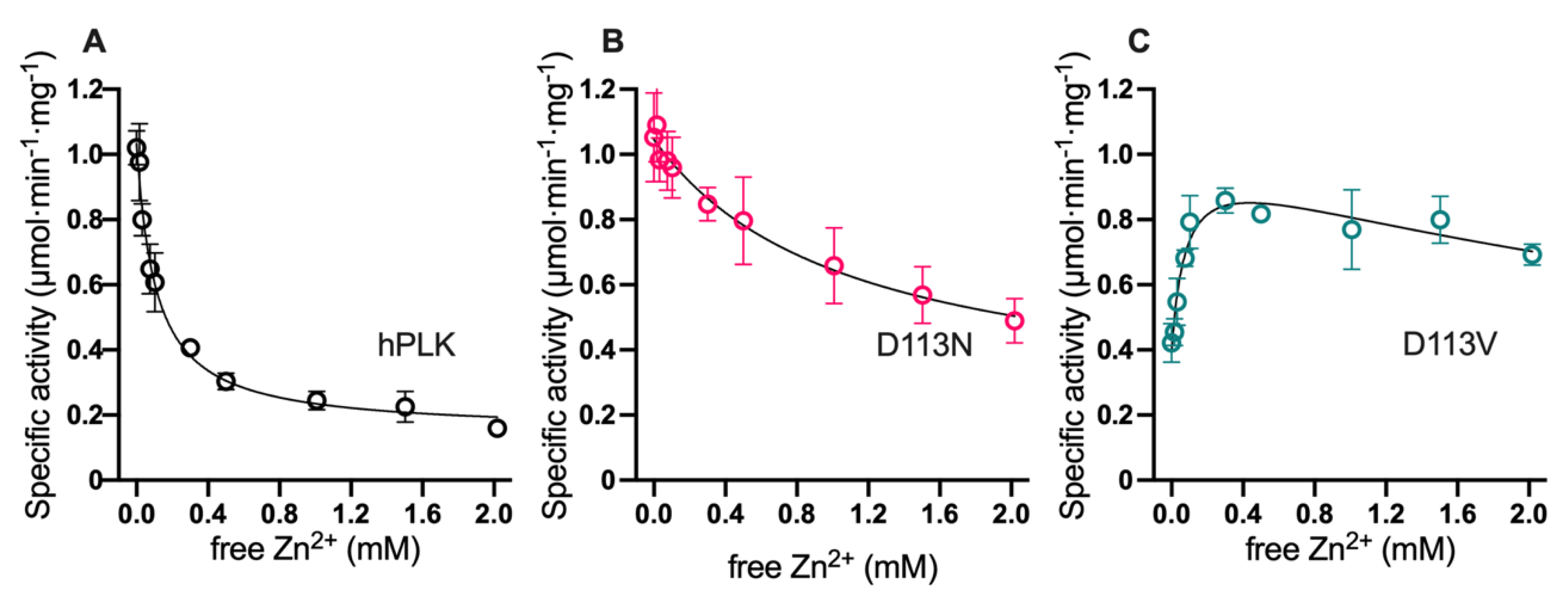
2.3. DPV Motif is Responsible for the Free Divalent Cation Inhibition of Human Pyridoxal Kinase (hPLK)
3. Materials and Methods
3.1. Evolutionary Trace Analysis
3.2. Molecular Dynamics
3.3. Overexpression and Purification of hPLK
3.4. Enzyme Activity Assays
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guixé, V.; Merino, F. The ADP-dependent sugar kinase family: Kinetic and evolutionary aspects. IUBMB Life 2009, 61, 753–761. [Google Scholar] [CrossRef]
- Sigrell, J.A.; Cameron, A.D.; Jones, T.A.; Mowbray, S.L. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 å resolution: Insights into a new family of kinase structures. Structure 1998, 6, 183–193. [Google Scholar] [CrossRef]
- Cabrera, R.; Ambrosio, A.L.B.; Garratt, R.C.; Guixé, V.; Babul, J. Crystallographic Structure of Phosphofructokinase-2 from Escherichia coli in Complex with Two ATP Molecules. Implications for Substrate Inhibition. J. Mol. Biol. 2008, 383, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Pardo, J.A.; Alegre-Cebollada, J.; Ramírez-Sarmiento, C.A.; Fernandez, J.M.; Guixé, V. Identifying Sequential Substrate Binding at the Single-Molecule Level by Enzyme Mechanical Stabilization. ACS Nano 2015, 9, 3996–4005. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Sarmiento, C.A.; Baez, M.; Zamora, R.A.; Balasubramaniam, D.; Babul, J.; Komives, E.A.; Guixé, V. The Folding Unit of Phosphofructokinase-2 as Defined by the Biophysical Properties of a Monomeric Mutant. Biophys. J. 2015, 108, 2350–2361. [Google Scholar] [CrossRef][Green Version]
- Herrera-Morande, A.; Castro-Fernández, V.; Merino, F.; Ramírez-Sarmiento, C.A.; Fernández, F.J.; Vega, M.C.; Guixé, V. Protein topology determines substrate-binding mechanism in homologous enzymes. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2869–2878. [Google Scholar] [CrossRef]
- Maj, M.C.; Singh, B.; Gupta, R.S. Pentavalent ions dependency is a conserved property of adenosine kinase from diverse sources: Identification of a novel motif implicated in phosphate and magnesium ion binding and substrate inhibition. Biochemistry 2002, 41, 4059–4069. [Google Scholar] [CrossRef]
- Parducci, R.E.; Cabrera, R.; Baez, M.; Guixé, V. Evidence for a catalytic Mg2+ ion and effect of phosphate on the activity of Escherichia coli phosphofructokinase-2: Regulatory properties of a ribokinase family member. Biochemistry 2006, 45, 9291–9299. [Google Scholar] [CrossRef]
- Quiroga-Roger, D.; Babul, J.; Guixé, V. Role of monovalent and divalent metal cations in human ribokinase catalysis and regulation. BioMetals 2015, 28, 401–413. [Google Scholar] [CrossRef]
- Gandhi, A.K.; Ghatge, M.S.; Musayev, F.N.; Sease, A.; Aboagye, S.O.; di Salvo, M.L.; Schirch, V.; Safo, M.K. Kinetic and structural studies of the role of the active site residue Asp235 of human pyridoxal kinase. Biochem. Biophys. Res. Commun. 2009, 381, 12–15. [Google Scholar] [CrossRef]
- Merino, F.; Rivas-Pardo, J.A.; Caniuguir, A.; García, I.; Guixé, V. Catalytic and regulatory roles of divalent metal cations on the phosphoryl-transfer mechanism of ADP-dependent sugar kinases from hyperthermophilic archaea. Biochimie 2012, 94, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Abarca-Lagunas, M.J.; Rivas-Pardo, J.A.; Ramírez-Sarmiento, C.A.; Guixé, V. Dissecting the functional roles of the conserved NXXE and HXE motifs of the ADP-dependent glucokinase from Thermococcus litoralis. FEBS Lett. 2015, 589, 3271–3276. [Google Scholar] [CrossRef] [PubMed]
- Safo, M.K.; Musayev, F.N.; Hunt, S.; di Salvo, M.L.; Scarsdale, N.; Schirch, V. Crystal structure of the PdxY Protein from Escherichia coli. J. Bacteriol. 2004, 186, 8074–8082. [Google Scholar] [CrossRef] [PubMed]
- Campobasso, N.; Mathews, I.I.; Begley, T.P.; Ealick, S.E. Crystal Structure of 4-Methyl-5-β-hydroxyethylthiazole Kinase from Bacillus subtilis at 1.5 Å Resolution. Biochemistry 2000, 39, 7868–7877. [Google Scholar] [CrossRef]
- Newman, J.A.; Das, S.K.; Sedelnikova, S.E.; Rice, D.W. The Crystal Structure of an ADP Complex of Bacillus subtilis Pyridoxal Kinase Provides Evidence for the Parallel Emergence of Enzyme Activity During Evolution. J. Mol. Biol. 2006, 363, 520–530. [Google Scholar] [CrossRef]
- Navarro, F.; Ramírez-Sarmiento, C.A.; Guixé, V. Catalytic and regulatory roles of species involved in metal–nucleotide equilibriums in human pyridoxal kinase. BioMetals 2013, 26, 805–812. [Google Scholar] [CrossRef]
- Mihalek, I.; Reš, I.; Lichtarge, O. A Family of Evolution–Entropy Hybrid Methods for Ranking Protein Residues by Importance. J. Mol. Biol. 2004, 336, 1265–1282. [Google Scholar] [CrossRef]
- Cao, P.; Gong, Y.; Tang, L.; Leung, Y.-C.; Jiang, T. Crystal structure of human pyridoxal kinase. J. Struct. Biol. 2006, 154, 327–332. [Google Scholar] [CrossRef]
- Li, M.H.; Kwok, F.; Chang, W.R.; Lau, C.K.; Zhang, J.P.; Lo, S.C.L.; Jiang, T.; Liang, D.C. Crystal structure of brain pyridoxal kinase, a novel member of the ribokinase superfamily. J. Biol. Chem. 2002, 277, 46385–46390. [Google Scholar] [CrossRef]
- Safo, M.K.; Musayev, F.N.; di Salvo, M.L.; Hunt, S.; Claude, J.-B.; Schirch, V. Crystal Structure of Pyridoxal Kinase from the Escherichia coli pdxK Gene: Implications for the Classification of Pyridoxal Kinases. J. Bacteriol. 2006, 188, 4542–4552. [Google Scholar] [CrossRef]
- Cheng, G.; Bennett, E.M.; Begley, T.P.; Ealick, S.E. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure 2002, 10, 225–235. [Google Scholar] [CrossRef]
- Jeyakanthan, J.; Thamotharan, S.; Velmurugan, D.; Rao, V.S.N.; Nagarajan, S.; Shinkai, A.; Kuramitsu, S.; Yokoyama, S. New structural insights and molecular-modelling studies of 4-methyl-5-β-hydroxyethylthiazole kinase from Pyrococcus horikoshii OT3 ( Ph ThiK). Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Currie, M.A.; Merino, F.; Skarina, T.; Wong, A.H.Y.; Singer, A.; Brown, G.; Savchenko, A.; Caniuguir, A.; Guixé, V.; Yakunin, A.F.; et al. ADP-dependent 6-Phosphofructokinase from Pyrococcus horikoshii OT3. J. Biol. Chem. 2009, 284, 22664–22671. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, H.; Sakuraba, H.; Kobe, T.; Kujime, A.; Katunuma, N.; Ohshima, T. Crystal structure of the ADP-dependent glucokinase from Pyrococcus horikoshii at 2.0-Å resolution: A large conformational change in ADP-dependent glucokinase. Protein Sci. 2009, 11, 2456–2463. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Fushinobu, S.; Jeong, J.J.; Yoshioka, I.; Koga, S.; Shoun, H.; Wakagi, T. Crystal structure of an ADP-dependent glucokinase from Pyrococcus furiosus: Implications for a sugar-induced conformational change in ADP-dependent kinase. J. Mol. Biol. 2003, 331, 871–883. [Google Scholar] [CrossRef]
- Tokarz, P.; Wiśniewska, M.; Kamiński, M.M.; Dubin, G.; Grudnik, P. Crystal structure of ADP-dependent glucokinase from Methanocaldococcus jannaschii in complex with 5-iodotubercidin reveals phosphoryl transfer mechanism. Protein Sci. 2018, 27, 790–797. [Google Scholar] [CrossRef]
- Mathews, I.I.; Erion, M.D.; Ealick, S.E. Structure of human adenosine kinase at 1.5 Å resolution. Biochemistry 1998, 37, 15607–15620. [Google Scholar] [CrossRef]
- Arnfors, L.; Hansen, T.; Schönheit, P.; Ladenstein, R.; Meining, W. Structure of Methanocaldococcus jannaschii nucleoside kinase: An archaeal member of the ribokinase family. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 1085–1097. [Google Scholar] [CrossRef]
- Mathews, I.I.; McMullan, D.; Miller, M.D.; Canaves, J.M.; Elsliger, M.-A.; Floyd, R.; Grzechnik, S.K.; Jaroszewski, L.; Klock, H.E.; Koesema, E.; et al. Crystal structure of 2-keto-3-deoxygluconate kinase (TM0067) from Thermotoga maritima at 2.05 Å resolution. Proteins Struct. Funct. Bioinforma. 2007, 70, 603–608. [Google Scholar] [CrossRef]
- Musayev, F.N.; di Salvo, M.L.; Ko, T.-P.; Gandhi, A.K.; Goswami, A.; Schirch, V.; Safo, M.K. Crystal Structure of human pyridoxal kinase: Structural basis of M + and M 2+ activation. Protein Sci. 2007, 16, 2184–2194. [Google Scholar] [CrossRef]
- Di Salvo, M.L.; Hunt, S.; Schirch, V. Expression, purification, and kinetic constants for human and Escherichia coli pyridoxal kinases. Protein Expr. Purif. 2004, 36, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, R.; Babul, J.; Guixé, V. Ribokinase family evolution and the role of conserved residues at the active site of the PfkB subfamily representative, Pfk-2 from Escherichia coli. Arch. Biochem. Biophys. 2010, 502, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Grishin, N.V. PROMALS3D: Multiple Protein Sequence Alignment Enhanced with Evolutionary and Three-Dimensional Structural Information. In Multiple Sequence Alignment Methods; Humana Press: Totowa, NJ, USA, 2014; Volume 1079, pp. 263–271. ISBN 978-1-62703-646-7. [Google Scholar]
- Lua, R.C.; Wilson, S.J.; Konecki, D.M.; Wilkins, A.D.; Venner, E.; Morgan, D.H.; Lichtarge, O. UET: A database of evolutionarily-predicted functional determinants of protein sequences that cluster as functional sites in protein structures. Nucleic Acids Res. 2016, 44, D308–D312. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2016; pp. 23–54. [Google Scholar]
- Zhang, Z.; Schäffer, A.A.; Miller, W.; Madden, T.L.; Lipman, D.J.; Koonin, E.V.; Altschul, S.F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998, 26, 3986–3990. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinforma. 2014, 47, 5.6.1–5.6.32. [Google Scholar] [CrossRef]
- Pineda, T.; Churchich, J.E. Reversible unfolding of pyridoxal kinase. J. Biol. Chem. 1993, 268, 20218–20222. [Google Scholar]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016. University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Meagher, K.L.; Redman, L.T.; Carlson, H.A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 2003, 24, 1016–1025. [Google Scholar] [CrossRef]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Abercrombie, D.M.; Martin, D.L. Inhibition of pyridoxal kinase by the pyridoxal-gamma-aminobutyrate imine. J. Biol. Chem. 1980, 255, 79–84. [Google Scholar] [PubMed]
- Storer, A.C.; Cornish-Bowden, A. Concentration of MgATP2- and other ions in solution. Calculation of the true concentrations of species present in mixtures of associating ions. Biochem. J. 1976, 159, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Castro-Fernandez, V.; Herrera-Morande, A.; Zamora, R.; Merino, F.; Gonzalez-Ordenes, F.; Padilla-Salinas, F.; Pereira, H.M.; Brandão-Neto, J.; Garratt, R.C.; Guixe, V. Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases. J. Biol. Chem. 2017, 292, 15598–15610. [Google Scholar] [CrossRef] [PubMed]
- Castro-Fernandez, V.; Bravo-Moraga, F.; Ramirez-Sarmiento, C.A.; Guixe, V. Emergence of pyridoxal phosphorylation through a promiscuous ancestor during the evolution of hydroxymethyl pyrimidine kinases. FEBS Lett. 2014, 588, 3068–3073. [Google Scholar] [CrossRef]
- Reddick, J.J.; Kinsland, C.; Nicewonger, R.; Christian, T.; Downs, D.M.; Winkler, M.E.; Begley, T.P. Overexpression, purification and characterization of two pyrimidine kinases involved in the biosynthesis of thiamin: 4-amino-5-hydroxymethyl-2-methylpyrimidine kinase and 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase. Tetrahedron 1998, 54, 15983–15991. [Google Scholar] [CrossRef]
- McCormick, D.B.; Snell, E.E. Pyridoxal phosphokinases. II. Effects of inhibitors. J. Biol. Chem. 1961, 236, 2085–2088. [Google Scholar]

| Enzyme | Species | PDB ID | Family | Refs. |
|---|---|---|---|---|
| PLK | human | 2F7K | ATP-dependent vitamin kinase | [18] |
| PLK | Ovis aries | 1LHP | [19] | |
| PLK | Escherichia coli (pdxK) | 2DDM | [20] | |
| PLK | Escherichia coli (pdxY) | 1TD2 | [13] | |
| PLK | Lactobacillus plantarum | 3H74 | ||
| HMPK | Salmonella typhimurium | 1JXH | [21] | |
| HMPK | Thermus thermophilus | 1UB0 | ||
| HMPK/PLK | Bacillus subtilis | 2I5B | [15] | |
| HMPK | Bacteroides thetaiotaomicron | 3MBH | ||
| THZK | Bacillus subtilis | 1EKQ | [14] | |
| THZK | Pyrococcus horikoshii | 3HPD | [22] | |
| THZK | Enterococcus faecalis | 3DZV | ||
| Phosphofructokinase (PFK) | Pyrococcus horikoshii | 1U2X | ADP-dependent sugar kinase (GK/PFK) | [23] |
| Glucokinase (GK) | Pyrococcus horikoshii | 1L2L | [24] | |
| Glucokinase (GK) | Pyrococcus furiosus | 1UA4 | [25] | |
| GK/PFK | Methanocaldococcus jannaschii | 5OD2 | [26] | |
| Adenosine kinase | human | 1BX4 | ATP-dependent sugar kinase (Ribokinase family) | [27] |
| Putative ribokinase | Agrobacterium tumefaciens | 2RBC | ||
| Nucleoside kinase | Methanocaldococcus jannaschii | 2C49 | [28] | |
| Carbohydrate kinase | Thermotoga maritima | 1VK4 | ||
| Nucleoside kinase | Chlorobium tepidum | 3KD6 | ||
| Ribokinase | human | 2FV7 | ||
| 2-keto-3-deoxygluconate kinase | Thermotoga maritima | 2AFB | [29] | |
| Fructokinase | Bacteroides thetaiotaomicron | 2QHP | ||
| Phosphofructokinase | Listeria innocua | 3IE7 | ||
| Phosphofructokinase | Escherichia coli (Pfk-2) | 3CQD | [3] | |
| Putative ribokinase | Escherichia coli | 3IN1 |
| Enzyme | Species | Acc. Code | Enzyme | Species | Acc. Code |
|---|---|---|---|---|---|
| THZK | B. subtilis | P39593 | THZK | S. epidermidis | Q21VL4 |
| THZK | B. velezensis | A7ZA59 | THZK | S. haemolyticus | Q466J4 |
| THZK | B. licheniformis | Q65DJ9 | THZK | R. ferrireducens | Q2LWX9 |
| THZK | Geobacillus sp. | C5D2N0 | THZK | M. barkeri | B1I1S3 |
| THZK | B. cereus | Q81IG9 | THZK | S. aciditrophicus | A3CS46 |
| THZK | B. brevis | C0ZGJ5 | THZK | C. Desulforudis | Q6LPQ4 |
| THZK | B. pumilus | A8FIR7 | THZK | M. marisnigri | C0QWJ2 |
| THZK | G.thermodenitrificans | A4IN26 | THZK | P. profundum | A4J233 |
| THZK | A. flavithermus | B7GKQ7 | THZK | B.hyodysenteriae | A7IA08 |
| THZK | B. clausii | Q5WDW4 | THZK | D. reducens | Q87JW7 |
| THZK | Exiguobacterium sp. | C4KZE2 | HMPK | M. boonei | P55882 |
| THZK | H. somni | Q0I129 | HMPK | V.parahaemolyticus | P56904 |
| THZK | L. monocytogenes | B8DET2 | HMPK | S. enterica | Q6GEY2 |
| THZK | C. subterraneus | Q8R807 | HMPK | S. meliloti | P44697 |
| THZK | A. succinogenes | A6VPG7 | HMPK | S. aureus | O25515 |
| THZK | M. thermoacetica | Q2RGX6 | HMPK | H. influenzae | O31620 |
| THZK | H. influenzae | Q57233 | HMPK | H. pylori | Q9ZBR6 |
| THZK | D. psychrophila | Q6AQZ6 | PLK | B. subtilis | P77150 |
| THZK | T. pseudethanolicus | B0KBA7 | PLK | S. coelicolor | Q7N3W7 |
| THZK | P. multocida | P57931 | PLK | E. coli | Q6D5V1 |
| THZK | B. halodurans | Q9K7L2 | PLK | P. laumondii | Q66A50 |
| THZK | G. parasuis | B8F7P4 | PLK | P.atrosepticum | Q51892 |
| THZK | D. aliphaticivorans | B8FJW2 | PLK | Y. pseudotuberculosis | B7V753 |
| THZK | C. tetani | Q893Q9 | PLK | P. mirabilis | A5WB73 |
| THZK | M. arvoryzae | Q0W1L9 | PLK | P. aeruginosa | B0UUD2 |
| THZK | C. aggregans | B8G7I8 | PLK | P. putida | Q5E345 |
| THZK | D. oleovorans | A8ZV98 | PLK | H. somni | Q141E8 |
| THZK | S. fumaroxidans | A0LP26 | PLK | A. fischeri | Q65UE8 |
| THZK | C. beijerinckii | A6M1W7 | PLK | P. xenovorans | A5UA83 |
| THZK | B. subtilis | B8I3J5 | PLK | M. succiniciproducens | Q63SC2 |
| THZK | B. velezensis | Q2FNE1 | PLK | H. influenzae | A6VNE5 |
| THZK | R. cellulolyticum | B2UY66 | PLK | B. pseudomallei | Q6LP62 |
| THZK | M. hungatei | A4FX33 | PLK | A. succinogenes | Q7MGA4 |
| THZK | C. botulinum | A8MK93 | PLK | P. profundum | Q1AYE5 |
| THZK | M. maripaludis | Q9UZQ4 | PLK | V. vulnificus | Q1J237 |
| THZK | A. oremLandii | B1HX32 | PLK | R. xylanophilus | Q9RYX0 |
| THZK | P. abyssi | Q8ESJ1 | PLK | D. geothermalis | Q0BSF0 |
| THZK | L. sphaericus | C1CD69 | PLK | D. radiodurans | A8A2R4 |
| THZK | O. iheyensis | Q890C2 | PLK | G. bethesdensis | P40192 |
| THZK | S. pneumoniae | Q49Z40 | PLK | E. coli | Q7W6K7 |
| THZK | L. plantarum | Q8ESZ2 | PLK | S. enterica | Q1LFU5 |
| THZK | S. saprophyticus | A5UUL2 | PLK | B. parapertussis | Q2L1P5 |
| THZK | O. iheyensis | A2SU11 | PLK | C. metallidurans | O46560 |
| THZK | Roseiflexus sp. | Q8U191 | PLK | B. avium | Q8W1X2 |
| THZK | M. labreanum | B3QT42 | PLK | S. scrofa | O01824 |
| THZK | P. furiosus | Q5HMC9 | PLK | A. thaliana | Q55EK9 |
| THZK | C. thalassium | Q4L7X4 |
| Free Zn2+ | hPLK | D113N | D113V | |||
|---|---|---|---|---|---|---|
| 0.1 mM | 0.5 mM | 0.1 mM | 0.5 mM | 0.1 mM | 0.5 mM | |
| KM,app, pyridoxal (mM) | 1.44 ± 0.35 | 4.09 ± 0.76 | 0.55 ± 0.10 | 0.11 ± 0.03 | 2.34 ± 0.55 | 1.08 ± 0.12 |
| KM,app, ZnATP (µM) | 20 ± 2 | 57 ± 12 | 36 ± 5 | 8 ± 1 | 13 ± 1 | 5 ± 1 |
| Vmax (µmol·min−1·mg−1) | 0.62 ± 0.04 | 0.70 ± 0.06 | 1.28 ± 0.06 | 0.66 ± 0.03 | 0.85 ± 0.08 | 0.77 ± 0.03 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Sarmiento, C.A.; Engelberger, F.; Guixé, V. An Evolutionary Marker of the Ribokinase Superfamily Is Responsible for Zinc-Mediated Regulation of Human Pyridoxal Kinase. Catalysts 2020, 10, 555. https://doi.org/10.3390/catal10050555
Ramírez-Sarmiento CA, Engelberger F, Guixé V. An Evolutionary Marker of the Ribokinase Superfamily Is Responsible for Zinc-Mediated Regulation of Human Pyridoxal Kinase. Catalysts. 2020; 10(5):555. https://doi.org/10.3390/catal10050555
Chicago/Turabian StyleRamírez-Sarmiento, César A., Felipe Engelberger, and Victoria Guixé. 2020. "An Evolutionary Marker of the Ribokinase Superfamily Is Responsible for Zinc-Mediated Regulation of Human Pyridoxal Kinase" Catalysts 10, no. 5: 555. https://doi.org/10.3390/catal10050555
APA StyleRamírez-Sarmiento, C. A., Engelberger, F., & Guixé, V. (2020). An Evolutionary Marker of the Ribokinase Superfamily Is Responsible for Zinc-Mediated Regulation of Human Pyridoxal Kinase. Catalysts, 10(5), 555. https://doi.org/10.3390/catal10050555






